Abstract
The Streptococcus agalactiae bacteriophage B30 endolysin contains three domains: cysteine, histidine-dependent amidohydrolase/peptidase (CHAP), Acm glycosidase, and the SH3b cell wall binding domain. Truncations and point mutations indicated that the Acm domain requires the SH3b domain for activity, while the CHAP domain is responsible for nearly all the cell lysis activity.
Streptococcus agalactiae (group B Streptococcus [GBS]) is a pathogen that infects human neonates, primarily through exposure in the birth canal of the mother (23), and causes mastitis in dairy cattle (20). In two recent studies, 44% of 202 bacterial isolates and 57% of 811 isolates from bovine mastitis exhibited resistance to at least one antibiotic (5, 14). Alternative antimicrobial agents for use against pathogens, including streptococci, are attracting much interest in part due to the increased incidence of antibiotic resistance and to the fact that mastitis is the most common reason for antimicrobial use in dairy herds (3, 4, 6, 9, 18, 21).
The cell lysis activity of bacteriophage endolysins makes them good candidates for protein antimicrobial agents. The endolysin of GBS phage B30 and a homolog that was 99% identical were recently characterized (3, 12). This endolysin contains two peptidoglycan hydrolase domains and an SH3b cell wall binding domain (11, 22) (Fig. 1), and the purified endolysin is active against many different species of streptococci. The enzymatic activities of the cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) (1, 15) and Acm (acetylmuramidase) (8) domains have been characterized previously. Each hydrolase domain degrades peptidoglycan preparations independent of the other hydrolase domain. Moreover, the CHAP endopeptidase cleaves between the d-alanyl-l-alanyl moieties between the peptidoglycan stem peptide and the cross bridge (12).
FIG. 1.
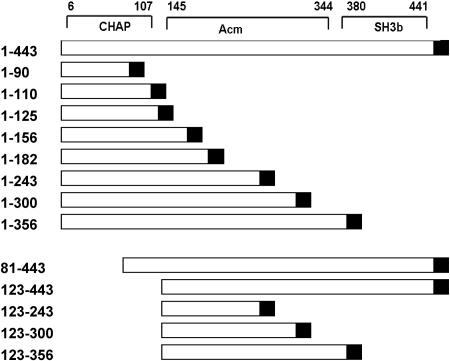
Phage B30 endolysin (443-amino-acid) conserved domains and deletion constructs. The schematic diagram at the top indicates the locations of the NH2-terminal CHAP endopeptidase, Acm glycosidase, C-terminal SH3b cell wall binding domain, and the six-His tag (solid box) in the pET21a-derived construct. The numbers indicate the initial and final amino acids in the deletion constructs and the C-terminal six-His tag.
The aim of this study was to define the functional domains of the two B30 peptidoglycan hydrolase activities. A series of deletion mutants were created, and their lytic activities against mastitis-causing pathogens and lactic acid bacteria were determined.
C-terminal truncations of the B30 endolysin.
All the constructs described in this paper were derived from the full-length (443-codon) phage B30 endolysin gene previously subcloned (pSD101) into pET21a (Novagen), with addition of a C-terminal six-His tag (12) (Fig. 1). A series of C-terminal deletion mutants were created by PCR subcloning using pSD101 as the template. Two forward primers, NdeF and BglF (Table 1), that contained either a unique NdeI site or a unique BglII site immediately 5′ of the endolysin coding sequences were synthesized. Reverse primers were designed to introduce an XhoI site at specific amino acids, including amino acids 90 (90R), 110 (110R), 125 (125R), 156 (156R), 182 (182R), 243 (243R), 300 (300R), and 356 (356R). The PCR product was either (i) TA cloned into pGEM-T, in which the appropriate fragment was isolated and ligated into similarly digested pET21a, or (ii) purified on an agarose gel, doubly digested with either NdeI and XhoI or BglII and XhoI, gel purified again, and ligated into similarly digested pET21a. The constructs were then transformed into either Escherichia coli INVαF′ or E. coli DH5α, isolated, characterized, and retransformed into E. coli BL21(DE3) for protein expression (Fig. 1).
TABLE 1.
Primers used in this study
| Primer | Sequence | Site |
|---|---|---|
| NdeF | 5′-GCACTACATATGGCAACTTATCAAGAATATAAAAG-3′ | NdeI |
| BglF | 5′-TCCGGCGTAGAGGATCGAGAT-3′ | BgllI |
| 90R | 5′-TGCTACATGCTCGAGAGGCGT-3′ | XhoI |
| 110R | 5′-TAGCACCCTCGAGATTTTG-3′ | XhoI |
| 156R | 5′-GCCGATACCTCGAGAAAGTAATC-3′ | XhoI |
| 182R | 5′-TCGGATACCTCGAGAATCGT-3′ | XhoI |
| 243R | 5′-TGCGGAATCTCGAGAGTCAAT-3′ | XhoI |
| 300R | 5′-TGGATAACCCTCGAGCCAAA-3′ | XhoI |
| 356R | 5′-CGCTGCAGCTCGAGATCAACCTTAGGTATATCCATTTTGCT-3′ | XhoI |
| 81F | 5′-TTGCTATTCATATGGTTGTTG-3′ | NdeI |
| 123F | 5′-CTTGTACATATGCCATATTC-3′ | NdeI |
| ddfusionlysinR | 5′-TTCCTTTCGGGCTTTACTAGTAGCCGGATCTCAGTG-3′ | MboI |
Plate lysis assays were performed to test the activity of these constructs. E. coli cells grown in 100 ml of Superbroth (Becton Dickinson) were first induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) and then pelleted, washed with lysin buffer A (LBA) (50 mM ammonium acetate, 10 mM CaCl2, 1 mM dithiothreitol; pH 6.2), and frozen at −80°C. Thawed cell pellets were resuspended in 2 ml lysin buffer A and disrupted with six 5-s sonication pulses on ice with 5-s rest periods between pulses. Lysates were clarified by centrifugation for 30 min at 16,000 × g in a microcentrifuge at 4°C, filtered (Millex 0.22-μm filter), and stored at −80°C. Ten microliters of filtered lysate was spotted directly onto tryptic soy agar plates (0.7% agar) containing 5% 50×-concentrated heat-killed (60°C for 30 s) or viable mid-log-phase target bacteria (S. agalactiae, Streptococcus dysgalactiae, Streptococcus uberis, or Staphylococcus aureus) and incubated overnight at 4°C.
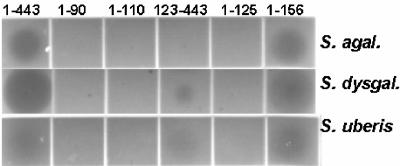
C-terminal truncations 1-156, 1-182, 1-243, 1-300, and 1-356 (Fig. 1) lysed the three major streptococcal mastitis pathogens (S. agalactiae, S. dysgalactiae, and S. uberis U.S. Department of Agriculture mastitis isolates) in the plate lysis assay (Table 2 and Fig. 2). No construct lysed S. aureus (data not shown). The 1-90 construct that bisected the predicted endopeptidase domain (amino acids 6 to 107) was inactive. Constructs 1-110 and 1-125, which included the entire predicted CHAP domain, were also inactive in the plate lysis assay. Therefore, the “functional” C terminus of the CHAP domain is between amino acids 125 and 156, up to 50 amino acids beyond the conserved domain sequences. These additional sequences might be necessary for correct folding.
TABLE 2.
Plate lysis assay results for B30 endolysin truncations
| Construct | Plate lysis assay resultsa
|
||
|---|---|---|---|
| S. agalactiae | S. dysgalactiae | S. uberis | |
| 1-443 | +++ | +++ | ++ |
| 1-90 | − | − | − |
| 1-110 | − | − | − |
| 1-125 | − | − | − |
| 1-156 | +++ | +++ | ++ |
| 1-182 | +++ | +++ | ++ |
| 1-243 | +++ | +++ | ++ |
| 1-300 | +++ | +++ | ++ |
| 1-356 | +++ | +++ | ++ |
| 81-443 | ++ | ++ | ++ |
| 123-443 | + | ++ | + |
| 123-243 | − | − | − |
| 123-300 | − | − | − |
| 123-356 | − | + | − |
+++, clearing in <2 h; ++, clearing overnight; +, weak clearing overnight; −, no clearing overnight.
FIG. 2.
Plate lysis assay of the B30 endolysin and selected truncations. Ten-microliter portions of E. coli extracts harboring B30-derived proteins were spotted onto tryptic soy agar plates containing mid-log-phase cultures of the pathogens S. agalactiae (S. agal.), S. dysgalactiae (S. dysgal.), and S. uberis.
It is interesting that the degree of lytic activity observed on the streptococcal mastitis pathogens S. agalactiae and S. uberis was less than that observed on group C Streptococcus (S. dysgalactiae). Factors affecting substrate susceptibility and affinity, such as the substrate binding site of the B30 endolysin, as well as the cell wall structures of these species, might help explain the interspecies differences in peptidoglycan hydrolase activity. However, the CHAP domain does not require the SH3b domain for cell lysis activity with any of the streptococci tested or for the greater hydrolase activity on S. dysgalactiae, and both the 1-443 and 1-156 constructs exhibited higher activity (Fig. 2).
N-terminal truncations of the B30 endolysin.
To define the functional N terminus of the Acm glycosidase domain, two N-terminal deletion mutants of the B30 endolysin were created by PCR subcloning (see above) using NdeI and XhoI sites. An engineered NdeI site (CATATG) with an in-frame ATG translation initiation codon was introduced at either codon 80 or codon 122 by PCR amplification with forward primers 81F and 123F and reverse primer ddfusionlysinR (sequence located in the vector pSD101 3′ of the six-His tag of the B30 endolysin coding sequences) (Table 1). Proteins derived from both truncations (81-443 and 123-443) exhibited weak lysis activity in the plate assay (Table 2 and Fig. 2) with all three streptococcal pathogens tested.
The shortest N-terminal truncation (the 123-443 truncation) was used to construct C-terminal deletion mutants to define the C terminus of the functional glycosidase domain. Deletions at the C terminus were constructed in the 123-443 vector utilizing primers employed previously (243R, 300R, and 356R) to make the B30 C-terminal deletion mutants (Fig. 1). Deletions 123-243 and 123-300 removed all of the SH3b domain (amino acids 380 to 441; http://smart.embl-heidelberg.de/smart/show_many_proteins.pl) and most of the predicted glycosidase domain. These constructs were inactive in plate lysis assays (Table 2). Construct 123-356 contained the entire predicted Acm domain (amino acids 145 to 344) (12) but lacked the SH3b domain. The 123-356 construct exhibited weak lysis of S. dysgalactiae (Fig. 2 and Table 2). However, this construct was not active against the B30 host strain S. agalactiae and was only weakly lytic with S. uberis, suggesting that the SH3b domain is critical for Acm domain activity.
Activities of the purified B30-derived proteins with mastitis-causing pathogens.
Representative Ni-purified truncated proteins 1-156, 1-182, and 1-356 and full-length recombinant protein 1-443 (Fig. 3A) were used in a turbidity assay with three bacterial strains (Fig. 3B). Proteins were purified from E. coli extracts prepared essentially as described above from 500-ml induced cultures of BL21(DE3) harboring the pSD101-derived constructs, washed, and resuspended in 10 ml lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole; pH 8) with 15 10-s sonication pulses on ice with 10-s rest periods between pulses and with 30 min of centrifugation at 6,800 rpm in an HS4 Sorvall rotor (8,500 × g) at 4°C. Clarified extracts were batch incubated with 5 ml of Ni-nitrilotriacetic acid slurry (QIAGEN) for 1 h at 4°C with gentle rolling. The slurry was washed and the protein was eluted as described by the manufacturer. Protein eluates were desalted in Micro Bio-Spin 30 columns (Bio-Rad) prior to protein determination with the bicinchoninic acid protein assay (Pierce).
FIG. 3.
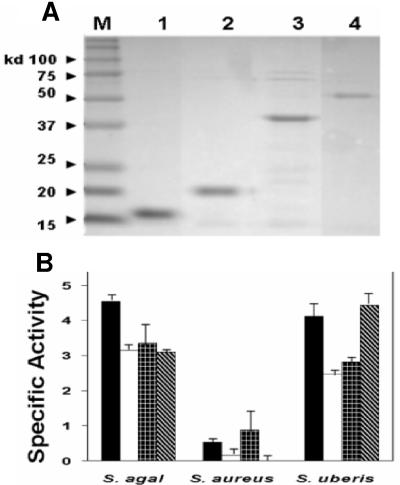
Protein purification analysis and turbidity assay with selected B30 endolysin truncated proteins. (A) Representative SDS-PAGE gel containing B30 endolysin and derived proteins purified with nickel affinity columns. Purified proteins were analyzed by standard 15% SDS-PAGE performed with Tris-glycine buffer at 131 V for 1.5 h in a Bio-Rad Mini-PROTEAN 3 gel apparatus according to the manufacturer's instructions. Gels were stained with BioSafe Coomassie blue stain (Bio-Rad) for 1 h and then rinsed in distilled water overnight. Lane M, molecular mass protein standards (Kaleidoscope protein standards; Bio-Rad); lane 1, 1-156; lane 2, 1-182; lane 3, 1-356; lane 4, full-length 1-443 protein. (B) Turbidity assay results with three bacterial strains obtained by using 100 μg of each B30 endolysin-derived protein. Black bars, 1-156; white bars, 1-182; plaid bars, 1-356; striped bars, full-length 1-443. The turbidity data are the results of three independent experiments performed with three unique preparations of purified protein. Specific activity is expressed in OD600/hour/milligram. S. agal., S. agalactiae.
For the turbidity assay, the target cells were grown in brain heart infusion to the mid-log phase (optical density at 600 nm [OD600], 0.4 to 0.6), pelleted, and resuspended in LBA to an OD600 of ∼2.0. Assays were initiated by addition of target cells to a final OD600 of approximately 1.0 to 1.2 to LBA with 100 μg of B30 endolysin-derived proteins. Changes in OD600 were recorded for 1 h. Changes in OD600 for untreated cells were subtracted from endolysin sample values prior to calculation of the activity. The results were expressed as the change in OD600/hour/milligram of total protein. All values represent three experiments (performed on three different days), and all samples were assayed on each day. The protein was purified and quantified, and the turbidity assays were performed on the same day due to the instability of some of the truncated proteins (7).
The turbidity assays indicated that the 1-156, 1-182, and 1-356 truncated proteins and full-length endolysin 1-443 exhibited similar activities against both mastitis pathogens (S. agalactiae and S. uberis) but little or no activity against S. aureus (Fig. 3B). This is consistent with findings obtained previously with E. coli extracts harboring the 1-443 and 1-182 constructs (7).
The activities of construct 1-443, 1-356, and 1-182 purified proteins (100 μg) were shown to be within the linear range of the assay (data not shown). Despite high lytic activity with 100 μg of the CHAP domain protein fragments, the same amount of 123-443 harboring the entire Acm and SH3b domains was not active in the turbidity assay with S. agalactiae (data not shown).
Activities of the mutated B30-derived proteins against S. agalactiae.
To confirm the activity results obtained with protein truncations, single-amino-acid mutations (12) were also tested (Fig. 4). Point mutants are expected to have a minimal effect on protein conformation and stability (10). It was shown previously that C26S and H91A mutations resulted in a complete loss of endopeptidase activity and that a D158A mutation led to a loss of almost 90% of the glycosidase activity (12). These mutant proteins were purified with an Ni column, analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Fig. 4A), and used in turbidity assays with S. agalactiae (Fig. 4B). Both CHAP domain mutations (C26S and H91A) completely abolished all lytic activity. However, inactivation of the Acm domain (D158A) did not reduce the level of lysis activity from the wild-type activity level. Identical results were obtained when S. uberis was tested (data not shown). Consequently, the CHAP endopeptidase was responsible for almost 100% of the lysis activity, and based on the deletion analysis described above, the C-terminal SH3b domain is not required. At this time, it is not known if the unnecessary SH3b domain, when “lysing from without,” is typical of GBS phage endolysins. Analyses of the binding properties of the C-terminal SH3b domain (by using fusions to the green fluorescent protein) are under way.
FIG. 4.
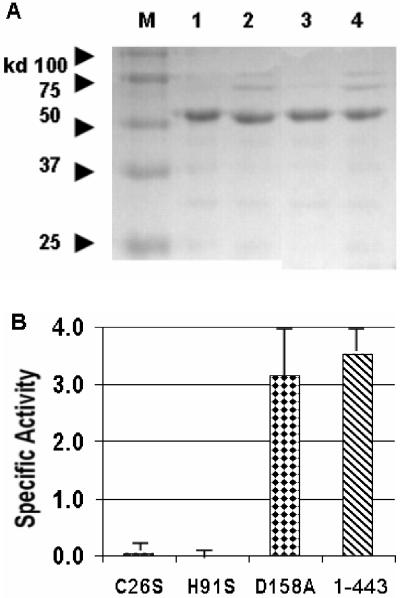
Protein purification analysis and turbidity assay with B30 endolysin site-directed mutants. (A) SDS-PAGE of the B30 endolysin and selected nickel column-purified mutant proteins. Lane M, protein standards; lane 1, C26S; lane 2, H91A; lane 3, D158A; lane 4, full-length 1-443 protein. (B) Turbidity assay results with S. agalactiae obtained by using 100 μg of each B30 endolysin and selected mutated proteins. The turbidity data are the results of two independent experiments. Specific activity is expressed in OD600/hour/milligram.
An aberrant protein conformation or instability resulting from single-amino-acid mutations of the CHAP domain is not likely to be responsible for the findings described above. Indeed, two different CHAP constructs (C26S and H91A) had the same effect, and both still exhibited glycosidase enzymatic activity with a prepared peptidoglycan substrate (12). Moreover, similar yields were obtained for both purified proteins and the wild-type endolysin (protein 1-443), as determined by SDS-PAGE analysis (Fig. 4A). Also, the specific lytic activity of the Acm domain mutant (D158A) was nearly identical to the specific activity of the wild-type 1-443 protein, suggesting that nearly all of the lytic activity is due to the nonmutant CHAP domain, which is consistent with the weak activity observed with the Acm domain-isolating deletion constructs.
Activity of the purified B30 endolysin and derived proteins on lactic acid bacteria and other streptococci.
The purified full-length B30 endolysin (protein 1-443) and the 1-156 CHAP protein harboring a C-terminal truncation were used in turbidity assays with lactic acid bacteria used in the manufacture of fermented dairy products, as well as other closely related streptococci. Clarified E. coli extracts containing the full-length 1-443 lysin and a large truncated endolysin (1-182) are known to be lytic for some lactic acid bacteria (7). The purified 1-443 endolysin exhibited a lytic profile similar to that of E. coli extracts harboring the same construct, and the specific activities were 1.3 OD600/mg/h for Streptococcus salivarius ATCC 25975 (7), 11.7 OD600/mg/h for Streptococcus thermophilus SMQ-301 (19), and 5.6 OD600/mg/h for Leuconostoc cremoris HER1286 (16). The shorter truncated protein, 1-156, was also purified and was shown to have similar lytic activities against related bacteria; the specific activities were 2.0 OD600/mg/h for S. thermophilus, 2.8 OD600/mg/h for S. salivarius, 7.9 OD600/mg/h for L. cremoris, and 1.6 OD600/mg/h for Streptococcus suis 89-999 (13). As shown previously for the full-length 1-443 protein and for the 1-182 truncation in E. coli extracts (7), the purified 1-443 and 1-156 proteins were not active against Leuconostoc mesenteroides HER1273 (17) or Lactococcus lactis IL-1403 (2).
In conclusion, we identified a 156-amino-acid protein fragment from a phage endolysin that contains a functional CHAP endolysin domain with strong antimicrobial activity against three major mastitis-causing streptococci. Although the B30 endolysin constructs lyse the milk-processing bacteria S. thermophilus and L. cremoris, they are also inactivated during pasteurization (7). Moreover, this antimicrobial peptide is likely to be degraded in the gut (9), further reducing putative food safety concerns should the constructs be produced in transgenic cow’s milk. We also describe a highly conserved Acm phage endolysin domain that does not exhibit significant lytic activity against GBS or other streptococci when they are exposed “from without.” The role of this seemingly silent domain in bacteriophage B30 requires further investigation.
Acknowledgments
This work was funded in part by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to S.M.) and by funds from Public Health Service grant AI054897 (to D.G.P.).
We thank Julie Samson for technical assistance with the enzymatic activity and Max Paape, USDA, for bacterial strains.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, Q., D. Nelson, S. Zhu, and V. A. Fischetti. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 49:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, M. L. 2000. Changing patterns of infectious disease. Nature 406:762-767. [DOI] [PubMed] [Google Scholar]
- 5.De Oliveira, A. P., J. L. Watts, S. A. Salmon, and F. M. Aarestrup. 2000. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J. Dairy Sci. 83:855-862. [DOI] [PubMed] [Google Scholar]
- 6.Donovan, D. M., D. E. Kerr, and R. J. Wall. 2005. Engineering disease resistant cattle. Transgenic Res. 14:563-567. [DOI] [PubMed] [Google Scholar]
- 7.Donovan, D. M., S. Dong, G. Garrett, G. M. Rousseau, S. Moineau, and D. G. Pritchard. 2006. Peptidoglycan hydrolase fusions maintain their parental specificities. Appl. Environ. Microbiol. 72:2988-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huard, C., G. Miranda, F. Wessner, A. Bolotin, J. Hansen, S. J. Foster, and M. P. Chapot-Chartier. 2003. Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149:695-705. [DOI] [PubMed] [Google Scholar]
- 9.Kerr, D. E., and O. Wellnitz. 2003. Mammary expression of new genes to combat mastitis. J. Anim. Sci. 81(Suppl. 3):38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pace, C. N., R. W. Alston, and K. L. Shaw. 2000. Charge-charge interactions influence the denatured state ensemble and contribute to protein stability. Protein Sci. 9:1395-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting, C. P., L. Aravind, J. Schultz, P. Bork, and E. V. Koonin. 1999. Eukaryotic signaling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289:729-745. [DOI] [PubMed] [Google Scholar]
- 12.Pritchard, D. G., S. Dong, J. R. Baker, and J. A. Engler. 2004. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 150:2079-2087. [DOI] [PubMed] [Google Scholar]
- 13.Quessy, S., J. D. Dubreuil, M. Caya, and R. Higgins. 1995. Discrimination of virulent and avirulent Streptococcus suis capsular type 2 isolates from different geographical origins. Infect. Immun. 63:1975-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajala-Schultz, P. J., K. L. Smith, J. S. Hogan, and B. C. Love. 2004. Antimicrobial susceptibility of mastitis pathogens from first lactation and older cows. Vet. Microbiol. 102:33-42. [DOI] [PubMed] [Google Scholar]
- 15.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of gamma-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 16.Saxelin, M.-L., E.-L. Nurmiaho-Lassila, V. T. Meriläinen, and R. I. Forsén. 1986. Ultrastructure and host specificity of bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and Leuconostoc cremoris from Finnish fermented milk “viili.” Appl. Environ. Microbiol. 52:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sozzi, T., J. M. Poulin, R. Maret, and R. Pousaz. 1978. Isolation of a bacteriophage of Leuconostoc mesenteroides from dairy products. J. Appl. Bacteriol. 44:159-161. [Google Scholar]
- 18.Thiel, K. 2004. Old dogma, new tricks—21st century phage therapy. Nat. Biotechnol. 22:31-36. [DOI] [PubMed] [Google Scholar]
- 19.Tremblay, D. M., and S. Moineau. 1999. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology 255:63-76. [DOI] [PubMed] [Google Scholar]
- 20.Van den Heever, L. W., and W. H. Giesecke. 1980. Experimental induction of bovine mastitis with human strains of group B streptococci (Streptococcus agalactiae). J. S. Afr. Vet. Assoc. 51:107-109. [PubMed] [Google Scholar]
- 21.Wall, R. J., A. M. Powell, M. J. Paape, D. E. Kerr, D. D. Bannerman, V. G. Pursel, K. D. Wells, N. Talbot, and H. W. Hawk. 2005. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nat. Biotechnol. 23:445-451. [DOI] [PubMed] [Google Scholar]
- 22.Whisstock, J. C., and A. M. Lesk. 1999. SH3 domains in prokaryotes. Trends Biochem. Sci. 24:132-133. [DOI] [PubMed] [Google Scholar]
- 23.Yow, M. D., L. J. Leeds, P. K. Thompson, E. O. Mason, Jr., D. J. Clark, and C. W. Beachler. 1980. The natural history of group B streptococcal colonization in the pregnant woman and her offspring. I. Colonization studies. Am. J. Obstet. Gynecol. 137:34-38. [DOI] [PubMed] [Google Scholar]