Abstract
Indirect pathogenicity (IP), the commensal protection of antibiotic-sensitive pathogens by resistant microorganisms of low intrinsic virulence, can prevent the eradication of polymicrobial infections. The contributions of antibiotic resistance mechanisms and biofilm structure to IP within polymicrobial biofilms were investigated using a model two-member consortium. Escherichia coli ATCC 33456 was transformed with vectors conferring either ampicillin or spectinomycin resistance, creating two distinct populations with different resistance mechanisms. Each strain alone or the consortium was grown as biofilms in flow cells and planktonically in chemostats. Comparisons in survival and activity were made on the basis of MICs and minimum biofilm preventative concentrations, a newly introduced descriptor. In ampicillin-containing medium, commensal interactions were evident during both modes of cultivation, but the sensitive strain experienced a greater benefit in the chemostat, indicating that the biofilm environment limited the commensal interaction between the Ampr and Sptr strains. In spectinomycin-containing medium, growth of the sensitive strain in chemostats and biofilms was not aided by the resistant strain. However, green fluorescent protein expression by the sensitive strain was greater in mixed-population biofilms (9% ± 1%) than when the strain was grown alone (2% ± 0%). No comparable benefit was evident during growth in the chemostat, indicating that the biofilm structure contributed to enhanced activity of the sensitive strain.
The concept of indirect pathogenicity (IP) describes the circumstance where, in a mixed infection, an antibiotic-resistant microorganism of low intrinsic virulence protects an antibiotic-sensitive pathogen from eradication. IP was recognized during treatment failure of polymicrobial infections, and numerous examples involving beta-lactamase-producing bacteria have been reported (3, 15, 16). IP is a form of commensalism, which is an interactive association where one organism benefits while the other is not affected (2). In the case of beta-lactamase-producing bacteria, susceptible antibiotics including penicillins and cephalosporins are inactivated via hydrolysis of the cyclic amide bond (25), leading to commensal protection of the sensitive strain. The occurrence of IP in microbial biofilms, or surface-attached communities of microorganisms, could have broad significance, because diverse kinds of polymicrobial infections are biofilm based, including device-related infections (8), ventilator-associated pneumonia (1), wounds (9), and abscesses (4). Presently, little information is available on the role of biofilm structure in the severity of IP or the development of IP in the presence of non-beta-lactam antibiotics.
In a polymicrobial infection exhibiting IP, the efficiency of antibiotic inactivation by a resistant microorganism should determine the extent of the commensal protection provided to the sensitive microorganism. To investigate the role of antibiotic detoxification mechanisms in IP development in biofilms, a two-member model consortium based on Escherichia coli ATCC 33456 was established. Two strains of E. coli ATCC 33456 were constructed, each with different mechanisms of antibiotic resistance. One strain harbored a plasmid encoding beta-lactamase, conferring resistance to the beta-lactam antibiotic ampicillin. The other strain had a plasmid carrying spectinomycin adenyltransferase, encoding resistance to the aminocyclitol spectinomycin (13). Ampicillin and spectinomycin were selected for use in the model because of (i) the sensitivity of E. coli ATCC 33456 to both in the absence of a heterologous resistance gene, (ii) the ability of these antibiotics to be permanently inactivated by the protein product of their respective resistance genes, and (iii) differences in the energetics of the resistance mechanisms. The two plasmids belonged to the same incompatibility group, preventing their exchange between the two populations. Additionally, the ampicillin-resistant strain expressed green fluorescent protein, allowing it to be distinguished from the other strain following counterstaining with a red dye. The use of two subpopulations derived from a single parent strain was intended to reduce the number of variables in the system that could influence biofilm structure and organization.
To shed light on the role of biofilm structure in IP, the two-member consortium was cultivated in the presence of steady-state antibiotic concentrations in flow cells (biofilm growth) and in chemostats (planktonic growth). The extent of commensal protection conferred by each antibiotic resistance mechanism for each cultivation mode was determined by comparing the growth of the sensitive organism alone with its growth in the presence of the resistant strain. By considering both types of cultivation, it was possible to determine whether the biofilm environment contributed to commensal interactions affecting the survival and activity of antibiotic-sensitive bacteria.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Two antibiotic-resistant populations of E. coli ATCC 33456 (26) were prepared. One population was transformed with the plasmid pEGFP (Clontech, Palo Alto, CA), a pUC19-based vector conferring ampicillin resistance by the bla gene and green fluorescent protein (GFP) production via the egfp gene under control of the constitutive Plac promoter. The second population was transformed with the vector pUCSpec, a pUC18-derived plasmid that confers spectinomycin resistance via the AAD(9) determinant (13). In this research, E. coli ATCC 33456 pEGFP is referred to as the “Ampr” strain and is resistant to ampicillin and sensitive to spectinomycin. E. coli ATCC 33456 pUCSpec is referred to as the “Sptr” strain and is resistant to spectinomycin and sensitive to ampicillin. Inocula were prepared from clonal populations that were stored at −82°C. Prior to use in flow cell experiments, the Ampr strain was grown overnight at 37°C on Luria-Bertani (LB) agar plates containing 400 ppm ampicillin, and the Sptr strain was grown on LB agar plates containing 100 ppm spectinomycin. For chemostat experiments, inocula of the Ampr or Sptr strain were grown overnight in a shaking incubator at 37°C and 200 rpm in LB broth containing either 400 ppm ampicillin or 100 ppm spectinomycin, respectively.
Chemicals.
Ampicillin and spectinomycin were obtained from Sigma (St. Louis, MO). Antibiotics were dissolved in ultrapure water, sterilized with 0.20-μm-pore filters to create stock solutions, and stored at −20°C. Nitrocefin was obtained from Calbiochem (San Diego, CA). SYTO 59 was purchased from Molecular Probes, Inc. (Eugene, OR).
Biofilm cultivation.
Biofilms were cultivated using parallel-plate flow cells according to a previously described technique (10). Briefly, cells were inoculated into medium reservoirs containing 200 ml LB broth to an optical density at 600 nm (OD600) of 0.03 (0.015 of each strain in mixed-population biofilms) and were recirculated through the flow cell at 0.84 ml min−1 for 2 h to allow surface colonization. Antibiotics were present during recirculation at the same concentrations that were used for continuous (steady-state) flow, described below. After recirculation, the system was switched to continuous flow for 46 h (medium flow rate of 0.35 ml min−1), introducing LB broth with antibiotics as required to the flow cell. The selected medium flow rate replaced the volume of the flow cell approximately once per minute and was chosen such that the antibiotic concentration in the bulk fluid approximated steady-state conditions. Biofilms were grown for 48 h in order to allow GFP fluorescence to develop and the biofilm structure to mature and also to permit the cultivation of a large number of biofilms. Each growth condition was repeated at least in triplicate. Biofilms were imaged by confocal laser scanning microscopy (CLSM) as described below. Following CLSM imaging, biofilm cells were displaced from flow cells by introducing air into the channels and were resuspended in 1 ml sterile 50 mM phosphate buffer (pH = 7.2) using a pipette. Visual inspection of flow cells by microscopy following biofilm displacement indicated that greater than 99 percent of the cells not directly adhered to the glass substratum were recovered.
Confocal laser scanning microscopy.
Prior to imaging, biofilms were rinsed with sterile 50 mM potassium phosphate buffer (pH = 7.2; no autofluorescence detected) for 10 min and then stained with 20 μM SYTO 59 for 15 min and subsequently rinsed with sterile 50 mM potassium phosphate buffer for another 10 min. Intact biofilms were imaged nondestructively using a Zeiss LSM 510 confocal laser scanning microscope (Zeiss, Thornwood, NY) equipped with a Fluor 40× oil immersion lens. Samples were excited simultaneously at wavelengths of 488 nm and 523 nm. Four image stacks of each biofilm were taken at different locations throughout the flow cell, using a 1-μm z-step increment.
COMSTAT analysis.
Quantitative analysis of CLSM images of biofilms was conducted using the digital image analysis program COMSTAT (12). For COMSTAT analysis, the following settings were used: pixel intensity threshold of 30; minimum colony size of 100 pixels, representing a cluster of five cells.
Chemostat experiments.
Chemostats were made from 250-ml sidearm flasks. Flasks were sealed with a rubber stopper containing a 3.2-mm-inside-diameter Pharmed tube that extended to the bottom of the flask for influent media and another tube for the intake of air, which passed through a 0.20-μm-pore-size filter. Sterile LB broth with antibiotics as required was contained in 2-liter Pyrex bottles incubated in a 37°C water bath and pumped into the chemostat using a peristaltic pump (Cole Parmer) through autoclaved 3.2-mm-inside-diameter Pharmed tubing. The sidearm flask was located on a heated magnetic stir plate at a setting that maintained a temperature of 37°C ± 0.5°C. A magnetic stir bar kept the flask contents well mixed, and effluent flowed out of the sidearm into a sterile, hooded funnel leading to a waste vessel. Samples were collected by extending a sterile microcentrifuge tube held by flame-sterilized tweezers into the sterile hood to collect effluent. A Bunsen burner was stationed next to the chemostat to maintain aseptic conditions. At each time point, a sample was collected to measure turbidity and a second sample was collected, centrifuged, resuspended in 10% glycerol, and stored at −82°C for analysis by plate count. The cell densities of cultures that were stored frozen at −82°C prior to dilution plate counting were 83% ± 6% of those of cultures that were not frozen prior to plating. Freezer storage did not alter the specific fluorescence of GFP-containing cells. Dilution rates were set to maintain the sensitive strain at 55 percent (Sptr strain) or 60 percent (Ampr strain) of its maximum growth rate in LB medium. With ampicillin in the medium, the pump flow was maintained at 4.5 ml min−1, corresponding to a complete reactor displacement every 64 min. With spectinomycin in the medium, the pump flow was maintained at 3.8 ml min−1, corresponding to a complete reactor displacement every 76 min. The inoculum concentration of the resistant strain was always an OD600 of 0.40, corresponding to 2 × 108 CFU ml−1. The inoculum concentration of the sensitive strain was always 0.04. Preliminary work with ampicillin-containing medium indicated that when the sensitive strain was inoculated at higher initial optical densities, the population size declined and stabilized at an OD600 of approximately 0.04. Chemostats were run in at least duplicate for each condition.
Determination of MIC and MBPC.
Antibiotic MIC determinations were performed as described by Jorgensen and Turnidge (14). The concentration of antibiotic required to prevent the formation of biofilm by viable cells adhering to the flow cell substratum during the recirculation phase was designated the minimum biofilm preventative concentration (MBPC). MBPCs were determined by measuring the concentration at which biofilm biomass as calculated by COMSTAT equaled 0 μm3 μm−2, indicating that only cells which adhered to the flow cell substratum during its inoculation were present. At the MBPC, the areal cell density was 3.5 × 103 CFU mm−2.
Nitrocefin assay.
The beta-lactamase potential of the Ampr strain during growth in biofilms and chemostats was measured using the chromogenic substrate nitrocefin (22). Briefly, Ampr cells were resuspended in sterile 50 mM phosphate buffer, adjusted to an OD600 of 0.40, and incubated with 0.1 mM nitrocefin for 5 min at 37°C. Activity was measured by using a spectrophotometer (486 nm, as per the manufacturer's recommendation) at 0 and 5 min. All assays were carried out in at least triplicate, and statistical significance was determined using Student's t test.
Flow cytometry.
Flow cytometry was performed with a Becton-Dickinson FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). Cells were washed and resuspended in sterile 50 mM phosphate buffer to an OD600 of 0.03 prior to analysis. Samples were excited at a wavelength of 488 nm with an argon laser, and 10,000 events were collected. Fluorescence was measured using logarithmic gain, and side scatter (SSC) was measured using linear gain. Six-micrometer-diameter beads (BD Biosciences, San Jose, CA) were used as size controls.
Plate counts.
Cells recovered from biofilms or chemostats were resuspended in sterile 50 mM phosphate buffer, serially diluted, and plated on LB agar plates supplemented with antibiotics as necessary. Plates were incubated overnight at 37°C. Biofilms resuspended to an OD600 of 1.0 corresponded to an average cell density of 7.7 × 108 CFU ml−1 (n = 8), whereas the average cell density of suspensions of chemostat cells with an OD600 of 1.0 corresponded to 6.4 × 108 CFU ml−1 (n = 10).
Growth kinetics.
Specific growth rates were determined from hourly measurements of optical densities (600 nm) during growth of each strain in LB broth in batch culture. Triplicate test tubes were inoculated to an initial OD600 of 0.03 from liquid cultures in mid-log-phase growth.
RESULTS
Growth kinetics.
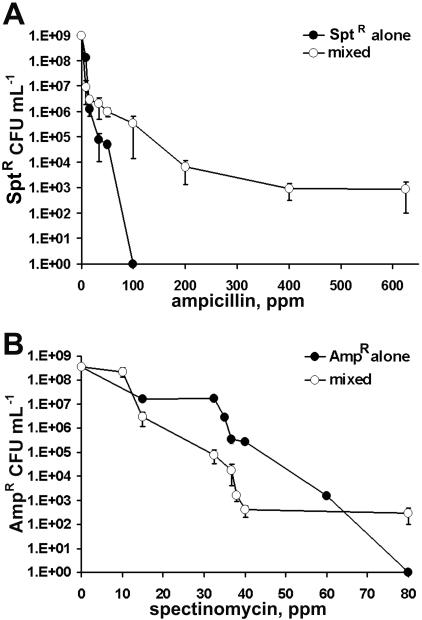
The specific growth rates of the Sptr and Ampr strains growing exponentially in LB broth without antibiotics were 1.7 h−1 and 1.3 h−1, respectively. The measured MIC for the Sptr strain exposed to ampicillin was 16 ppm (Fig. 1A), and the measured MIC for the Ampr strain exposed to spectinomycin was 30 ppm (Fig. 1B). These values were similar to MICs of 8 to 16 ppm reported elsewhere for ampicillin (6, 21) and 12.5 to 50 ppm for spectinomycin (18).
FIG. 1.
Antibiotic concentrations required to prevent planktonic or biofilm growth. MICs for each strain were determined in batch culture. The antibiotic concentration at which attached cells were unable to form a biofilm within 48 h was termed the MBPC. A: Sptr strain in ampicillin-containing medium. B: Ampr strain in spectinomycin-containing medium. Filled symbols, MIC; open symbols, MBPC.
Phenotype stability.
In order to assess the stability of the relevant phenotypes, single-strain biofilms were grown for 48 h in the absence of antibiotic selective pressure. Biofilm-grown cells of the Ampr strain were estimated by flow cytometry to be 97% ± 1% fluorescent. Biofilm-grown cells of the Sptr strain were plated on LB agar with and without spectinomycin and were determined to be 103% ± 16% resistant to spectinomycin (n = 7).
Exchange of antibiotic resistance determinants in biofilms.
To check if antibiotic resistance determinants were exchanged between the Ampr and Sptr strains during growth in biofilms, cells from resuspended mixed-population biofilms were cultured on plates containing either ampicillin or spectinomycin. Eight biofilms cultivated in ampicillin-containing medium and eight biofilms cultivated in spectinomycin-containing medium were tested. Cells that grew were inoculated into LB broth containing twice the MIC of the reciprocal antibiotic. Out of 16 biofilms that were screened, only 1 spontaneous mutant, originally grown in ampicillin, was found to be resistant to both antibiotics.
Nitrocefin assay of beta-lactamase potential.
The beta-lactamase potential of Ampr cells grown in chemostats or 48-h biofilms were compared. Cells from chemostats showed a greater potential (P < 0.042) to cleave nitrocefin than biofilms cells (per 1 × 108 cells): 0.75 ± 0.17 ppm min−1 in the chemostat and 0.49 ± 0.08 ppm min−1 in biofilms.
Commensal protection in biofilms.
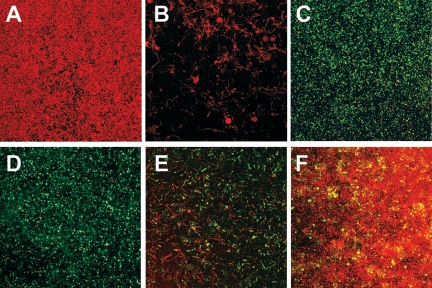
The MBPC for the Sptr strain was 50 ppm ampicillin (Fig. 1A), and the MBPC for the Ampr strain was 40 ppm spectinomycin (Fig. 1B). In the absence of ampicillin, the Sptr strain developed robust biofilms with an overall spongiform morphology, with individual cells exhibiting the typical coccobacillus shape (Fig. 2A). The average thickness of these biofilms was 50 μm. Exposure to sublethal concentrations of ampicillin caused the formation of filamentous structures (Fig. 2B), and these biofilms had a maximum height of approximately 6 μm at ampicillin concentrations greater than 13 ppm. The Ampr strain formed an extensive biofilm of fluorescent cells in the absence of antibiotics (Fig. 2D). Concentrations of spectinomycin greater than 20 ppm caused deformation of cells into filaments and a reduction in GFP expression (Fig. 2E and 3). Ampr biofilms were approximately 35 μm thick through 35 ppm spectinomycin but then decreased in size until the MBPC of 40 ppm was reached.
FIG. 2.
Prevention of morphological damage to antibiotic-sensitive bacteria during growth with a resistant strain. Biofilms of each strain alone or the two strains grown together were imaged by CLSM. Representative images of the substratum are shown. A: Sptr strain in LB broth. B: Sptr strain in LB broth plus 8 ppm ampicillin. C: Ampr and Sptr strains in LB broth plus 8 ppm ampicillin. D: Ampr strain in LB broth. E: Ampr strain in LB broth plus 20 ppm spectinomycin. F: Ampr and Sptr strains in LB broth plus 20 ppm spectinomycin. Magnification, ×400.
FIG. 3.
Flow cytometry scatter plots of Ampr biofilms grown in spectinomycin-containing medium. Representative flow cytometry scatter plots showing the distribution of fluorescent events in Ampr biofilms grown alone in medium containing subinhibitory concentrations of spectinomycin. x axis: SSC. y axis: GFP fluorescence (FL1-H).
The ability of the resistant strain to prevent morphological damage to the sensitive strain in mixed-population biofilms was considered. In the presence of 8 ppm ampicillin, no abnormally shaped Sptr cells were evident in mixed biofilms (Fig. 2C), in contrast to results with their growth alone. In the presence of 20 ppm spectinomycin, morphological damage was minimal in mixed biofilms (Fig. 2F). The sensitive-cell population size decreased substantially with increasing antibiotic concentration (Fig. 4); thus, comparisons were made at low antibiotic concentrations where the sensitive population was adequately represented.
FIG. 4.
Commensal protection in biofilms. Antibiotic-sensitive organisms grown in biofilms either alone (filled symbols) or with the resistant strain (open symbols) were enumerated by plate counts. A: Sptr strain in ampicillin-containing medium. B: Ampr strain in spectinomycin-containing medium.
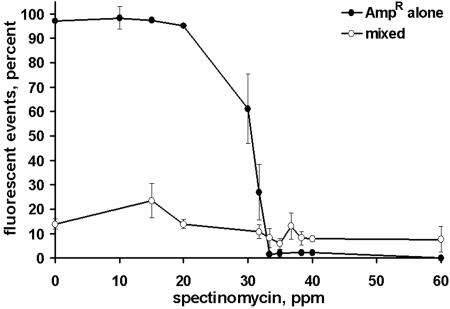
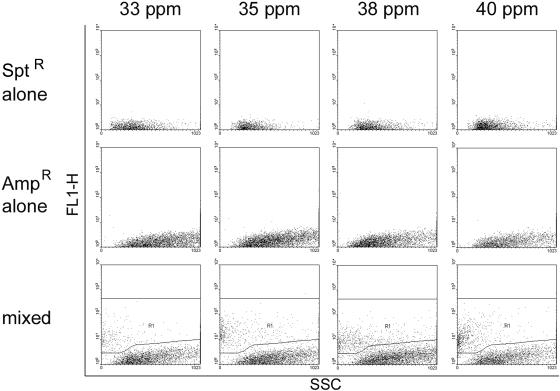
In biofilms exposed to ampicillin, the growth of the Sptr population was aided by growth with the Ampr strain. By 33 ppm ampicillin, the number of viable Sptr cells in single-strain biofilms was an order of magnitude lower than in mixed population biofilms, and no viable Sptr cells were recovered following continuous exposure to ampicillin concentrations greater than 100 ppm (Fig. 4A). In contrast, populations of the Sptr strain with cell densities greater than 103 CFU ml−1 were present in mixed-population biofilms exposed to 625 ppm ampicillin, 12.5 times greater than the Sptr strain MBPC. In biofilms exposed to spectinomycin, there was no evidence of commensal protection based on Ampr cell numbers (Fig. 4B). However, a higher percentage of Ampr cells producing GFP were evident in mixed-population biofilms than in single-strain Ampr biofilms (Fig. 5). Analysis by flow cytometry indicated a decrease in the size and brightness of the fluorescent subpopulation within single-strain Ampr biofilms as a function of increasing spectinomycin concentration, with a substantial loss of fluorescence at spectinomycin concentrations greater than 33.3 ppm. (Fig. 5; Fig. 6, middle row). In contrast, the fluorescent Ampr strain subpopulation in mixed-population biofilms stabilized at approximately 9% ± 1% of the total biofilm population by 33.3 ppm spectinomycin and was significantly larger than the fluorescent population in single-strain Ampr biofilms (2% ± 0% fluorescent) for spectinomycin concentrations of 33.3 ppm and higher (Fig. 5; Fig. 6, bottom row).
FIG. 5.
Flow cytometry analyses of Ampr populations grown alone or together with the Sptr strain in spectinomycin-containing medium. Ampr strain grown alone, filled circles. Ampr and Sptr strains grown together, open circles.
FIG. 6.
Representative scatter plots of biofilms grown in media containing 33.3 to 40 ppm spectinomycin. Top row: Sptr strain alone. Middle row: Ampr strain alone. Bottom row: mixed biofilms, with fluorescent populations indicated by the R1 region. x axis: SSC. y axis: GFP fluorescence (FL1-H).
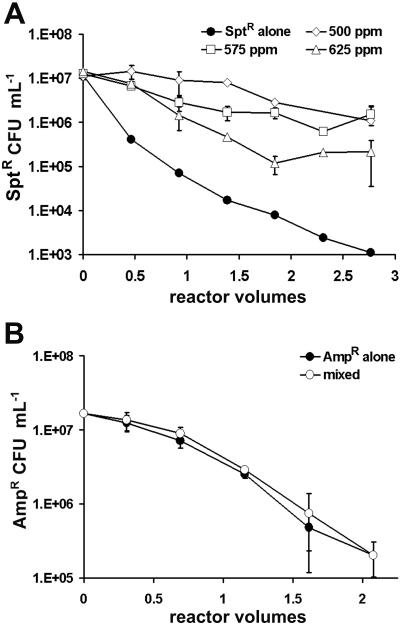
Commensal protection in chemostats.
The survival of the sensitive Sptr strain was enhanced by the presence of the resistant Ampr strain in the chemostat (Fig. 7A). In the absence of the Ampr strain, the Sptr strain was almost completely displaced from the chemostat in 64 min, corresponding to a single reactor volume replacement. In contrast, with the Ampr strain present, the Sptr strain population was approximately 3 orders of magnitude larger than the Sptr strain population grown alone after 3 h, a period of time corresponding to more than two reactor volume replacements (Fig. 7A). The Sptr strain survived in chemostats containing 625 ppm ampicillin, corresponding to 39 times the MIC. In mixed culture, the decline in the Sptr population as a function of increasing ampicillin concentration was nearly 2 orders of magnitude greater in biofilms than in the chemostat (Fig. 8). There was no evidence of enhanced survival of the Ampr strain in coculture with the Sptr strain in spectinomycin-containing medium (Fig. 7B), nor was there any beneficial effect on GFP fluorescence (data not shown). Populations of the Ampr strain declined steadily over a 3-h period to approximately 2 to 3% of their initial concentration. This behavior was observed in both 100 and 150 ppm spectinomycin, corresponding to 3.3 and 5 times the MIC, respectively.
FIG. 7.
Commensal protection in chemostats. Antibiotic-sensitive organisms grown in chemostats either alone (filled symbols) or with the resistant strain (open symbols) were enumerated by plate counts. A: Sptr strain in ampicillin-containing medium. B: Ampr strain in spectinomycin-containing medium.
FIG. 8.
Comparison of chemostat and biofilm commensalism in ampicillin-containing media. The log scale reduction of ampicillin-sensitive Sptr cells grown in coculture with Ampr cells was compared for chemostats (filled symbols) and biofilms (open symbols). Samples were taken from chemostats after three reactor volumes and from biofilms after 48 h. Cells were enumerated by dilution spread plating.
DISCUSSION
In well-mixed planktonic cultures, cells that cannot coaggregate move past one another and do not establish interactions that have a spatial component. In contrast, in biofilms, cells are primarily fixed in space and must contend with intercellular relationships in three dimensions. During growth in either ampicillin- or spectinomycin-containing medium, biofilm structure influenced the extent of commensal interactions between the consortium members. In ampicillin-containing medium, the biofilm environment reduced the commensal benefit to the Sptr strain in two ways. First, the cells competed for space. In the biofilm setting, the Ampr strain outcompeted the Sptr strain by accumulating in the limited available space, whereas in the chemostat the size of the Ampr population was restricted by washout, allowing a substantial population of the Sptr strain to persist (due to its faster specific growth rate). The faster initial growth of the Ampr strain in the biofilm also could have established a gradient of nutrients favoring its own growth (17). Second, the beta-lactamase activity was greater in the chemostat than in the biofilm. The difference in average beta-lactamase activity most likely occurred because the population in the chemostat was in the exponential growth phase, whereas the biofilm contained a subpopulation of older, less-active cells. Thus, while these data support the observations of others that IP occurs in biofilms (5), they demonstrate that the biofilm environment limited the commensal interaction between the Ampr and Sptr strains in the presence of ampicillin.
In spectinomycin-containing medium, no protection was afforded to the sensitive Ampr strain in the chemostat with respect to population size or GFP expression. Similarly, in terms of population size, the Ampr strain received no benefit from the Sptr strain in biofilms. However, a commensal interaction was evident in biofilms in terms of enhanced GFP expression in the Ampr strain. The commensal interaction likely resulted from the development of microenvironments with reduced spectinomycin concentrations resulting from the enzyme-catalyzed detoxification of spectinomycin by the Sptr strain. The reduction in Ampr cells with abnormal morphology in the commensal environment that was observed by CLSM also supports the concept of spectinomycin-reduced microenvironments within the mixed biofilm. Alternatively, the greater biomass that formed in the mixed biofilms may have also contributed to enhanced Ampr strain resistance to spectinomycin.
Two methods-related aspects of this work warrant discussion. First, the concept of MBPC, the concentration of antibiotic required to prevent biofilm formation, was introduced. Existing parameters emphasize the concentration of chemical required to eliminate biofilms already attached to a surface. For example, the minimum biofilm eradication concentration measures the concentration of antibiotic required to kill an already-established biofilm (7), and the minimum biofilm inhibitory concentration measures the antibiotic concentration required to inhibit growth of individual cells shed from an established biofilm (24). In contrast, the MBPC describes the concentration of an agent required to keep a surface free of biofilm. Second, GFP fluorescence was used to characterize the quality of the biofilm environment for E. coli activity. GFP fluorescence requires low levels of oxygen (11, 27), is pH dependent (23), and may be inhibited when cells are challenged with antibiotics. Thus, a decrease in GFP fluorescence may indicate suboptimal conditions for an aerobic neutrophile like E. coli. Conversely, the persistence of localized GFP fluorescence under inhibitory conditions, as was detected in mixed biofilms exposed to spectinomycin, indicated the presence of microenvironments with suitable conditions for Ampr strain fluorescence.
The mechanism of antibiotic resistance substantially influenced the extent of the commensal interaction between the Ampr and Sptr strains. In the case of ampicillin, detoxification by the TEM-1 beta-lactamase produced by the Ampr strain facilitated the growth of a significant population of sensitive bacteria. TEM-1 beta-lactamase has a high affinity for ampicillin (Km = 14 μM [∼5 ppm]) (20) and hydrolyzes ampicillin without an energy input, leading to efficient antibiotic inactivation. In contrast, the inactivation of spectinomycin by the Sptr strain provided a small benefit to spectinomycin-sensitive cells. The spectinomycin adenyltransferase AAD(9) determinant inactivates spectinomycin by adenylation at the 9-OH position in an ATP-consuming reaction (19), and the associated energy cost most likely inhibited extensive antibiotic detoxification. The potential for commensal interactions to cause IP in response to other antibiotics is largely uninvestigated to date. Antibiotics that are enzymatically degraded or modified could potentially be susceptible, including aminoglycosides, macrolides, chloramphenicol, and rifamycin (28). In the case of IP caused by beta-lactamase-producing bacteria, administration of amoxicillin and the beta-lactamase inhibitor clavulanic acid reduced the extent of treatment failure (4) and helped eliminate penicillin-sensitive pneumococci in a model biofilm (5). Understanding the factors that give rise to IP in polymicrobial biofilms could facilitate narrow-spectrum antibiotic therapies and help reduce incidences of treatment failure.
Acknowledgments
We thank Michelle Sanders, Stefka Taskaev, and Brian Lovell for their assistance with experiments, Robert Simmons for help with CLSM, and Dan Rozen and Don Ahearn for their comments. The vector pUCSpec was kindly provided by June Scott, Emory University. S. Gilbert was supported by the University Scholar program, Georgia State University. We thank the Biology Department, Georgia State University, for supporting B. A. Stubblefield.
This research was supported by the American Heart Association Southeast Affiliate (grant no. 0160196B).
REFERENCES
- 1.Adair, C. G., S. P. Gorman, B. M. Feron, L. M. Byers, D. S. Jones, C. E. Goldsmith, J. E. Moore, J. R. Kerr, M. D. Curran, G. Hogg, C. H. Webb, G. J. McCarthy, and K. R. Milligan. 1999. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 25:1072-1076. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M., and R. Bartha. 1997. Microbial ecology: fundamentals and applications, 4th ed. Addison-Wesley Publishing Co., New York, N.Y.
- 3.Brook, I. 1994. Indirect pathogenicity. Infect. Dis. Clin. Pract. 3:S21-S27. [Google Scholar]
- 4.Brook, I. 2002. Microbiology of polymicrobial abscesses and implications for therapy. J. Antimicrob. Chemother. 50:805-810. [DOI] [PubMed] [Google Scholar]
- 5.Budhani, R., and J. Struthers. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 42:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, D. L., C. J. Jakielaszek, L. A. Miller, and J. A. Poupard. 1999. Escherichia coli ATCC 35218 as a quality control isolate for susceptibility testing of Haemophilus influenzae with haemophilus test medium. Antimicrob. Agents Chemother. 43:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceri, H., M. Olson, C. Stremick, R. Read, D. Morck, and A. Buret. 1999. The Calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlan, R., and J. Costerton. 2001. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, R., and K. Harding. 2004. Bacteria and wound healing. Curr. Opin. Infect. Dis. 17:91-96. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, E., and J. Keasling. 2004. Bench scale flow cell for nondestructive imaging of biofilms. In J. Spencer and A. Ragout de Spencer (ed.), Environmental microbiology methods and protocols. Humana Press, Totowa, N.J.
- 11.Hansen, M., R. J. Palmer, C. Udsen, D. White, and S. Molin. 2001. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology 147:1383-1391. [DOI] [PubMed] [Google Scholar]
- 12.Heydorn, A., A. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. Ersboell, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 13.Husmann, L. K., D. L. Yung, S. K. Hollingshead, and J. R. Scott. 1997. Role of putative virulence factors of Streptococcus pyogenes in mouse models of long-term throat colonization and pneumonia. Infect. Immun. 65:1422-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorgensen, J. H., and J. D. Turnidge. 2003. Susceptibility test methods: dilution and disk diffusion methods, p. 1115-1116. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 15.Kaieda, S., H. Yano, N. Okitsu, Y. Hosaka, R. Okamoto, M. Inoue, and H. Takahashi. 2005. In vitro investigation of the indirect pathogenicity of beta-lactamase-producing microorganisms in the nasopharyngeal microflora. Int. J. Pediatr. Otorhinolaryngol. 69:479-485. [DOI] [PubMed] [Google Scholar]
- 16.Kataoka, D., H. Fujiwara, T. Kawakami, Y. Tanaka, A. Tanimoto, and S. Ikawa. 2003. The indirect pathogenicity of Stenotrophomonas maltophilia. Int. J. Antimicrob. Agents 22:601-606. [DOI] [PubMed] [Google Scholar]
- 17.Kreft, J. U. 2004. Biofilms promote altruism. Microbiology 150:2751-2760. [DOI] [PubMed] [Google Scholar]
- 18.Kucers, A. 1979. The use of antibiotics: a comprehensive review with clinical emphasis, 3rd. ed. William Heinerman Medical Books Ltd., London, United Kingdom.
- 19.LeBlanc, D., L. Lee, and J. Inamine. 1991. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1804-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livermore, D., F. Moosdeen, M. Lindridge, P. Khog, and J. Williams. 1986. Behaviour of TEM-1 beta-lactamase as a resistance mechanism to ampicillin, mezlocillin and azlocillin in Escherichia coli. J. Antimicrob. Chemother. 17:139-146. [DOI] [PubMed] [Google Scholar]
- 21.Lorian, V. (ed.). 1996. Antibiotics in laboratory medicine, 4th. ed. Williams and Wilkins, Baltimore, Md.
- 22.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson, G., S. Knobel, W. Sharif, S. Kain, and D. Piston. 1997. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73:2782-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickering, S., R. Bayston, and B. Scammell. 2003. Electromagnetic augmentation of antibiotic efficacy in infection of orthopaedic implants. J. Bone Joint Surg. Br. 85:588-593. [DOI] [PubMed] [Google Scholar]
- 25.Power, E. G. M. 1998. Bacterial resistance to antibiotics, p. 181-200. In W. B. Hugo and A. D. Russell (ed.), Pharmaceutical microbiology. Blackwell Science, London, United Kingdom.
- 26.Shen, H., and Y.-T. Wang. 1993. Characterization of enzymatic reduction of hexavalent chromium by Escherichia coli ATCC 33456. Appl. Environ. Microbiol. 59:3771-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsien, R. 1998. The green fluorescent protein. Annu. Rev. Biochem. 67:509-544. [DOI] [PubMed] [Google Scholar]
- 28.Wright, G. D. 2005. Bacterial resistance to antibiotics: enzymatic degradation and modification. Adv. Drug Deliv. Rev. 57:1451-1470. [DOI] [PubMed] [Google Scholar]