Abstract
The discovery of an abundant and diverse virus community in oceans and lakes has profoundly reshaped ideas about global carbon and nutrient fluxes, food web dynamics, and maintenance of microbial biodiversity. These roles are exerted through massive viral impact on the population dynamics of heterotrophic bacterioplankton and primary producers. We took advantage of a shallow wetland system with contrasting microhabitats in close proximity to demonstrate that in marked contrast to pelagic systems, viral infection, determined directly by transmission electron microscopy, and consequently mortality of prokaryotes were surprisingly low in benthic habitats in all seasons. This was true even though free viruses were abundant throughout the year and bacterial infection and mortality rates were high in surrounding water. The habitats in which we found this pattern include sediment, decomposing plant litter, and biofilms on aquatic vegetation. Overall, we detected viruses in only 4 of a total of ∼15,000 bacterial cells inspected in these three habitats; for comparison, nearly 300 of ∼5,000 cells suspended in the water column were infected. The strikingly low incidence of impact of phages in the benthos may have important implications, since a major portion of microbial biodiversity and global carbon and nutrient turnover are associated with surfaces. Therefore, if failure to infect benthic bacteria is a widespread phenomenon, then the global role of viruses in controlling microbial diversity, food web dynamics, and biogeochemical cycles would be greatly diminished compared to predictions based on data from planktonic environments.
Viruses are ubiquitous in ecosystems, highly abundant and extraordinarily diverse (7, 20, 32, 37, 38, 39). Shotgun sequencing has yielded rough estimates of between 400 and 7,000 viral genotypes in 200 liters of seawater (5), and total densities in marine and freshwaters typically exceed 1010 per liter (20, 32, 37, 39). Most importantly, due to their massive impact on prokaryotic and microalgal population dynamics, viruses are a dynamic and functionally important component of pelagic ecosystems (20, 32, 37, 39). In particular, virus-induced mortality (VIM) of heterotrophic prokaryotes in open marine and inland waters can be as high as 10 to 50%, even approaching 100%, of prokaryotic cell production (37, 39).
These figures and current concepts on viral functional importance are largely derived from planktonic environments (20, 32, 37, 39). However, large numbers of prokaryotes are embedded in a biofilm matrix which creates habitat conditions that differ vastly from those in the open water (14, 17, 30). A key question, therefore, is to what extent current concepts and data based on planktonic systems can be extrapolated to their benthic counterparts.
A suitable system to address this issue is marshes at land-water interfaces, which comprise multiple, structurally distinct microhabitats in close proximity. Vascular plants in marshes also tend to be highly productive while experiencing small grazing losses during the growing season. As a result, they supply abundant resources to heterotrophic organisms (27), and corresponding microbial activity, productivity, and turnover can be especially high (13). Bacteriophages could be instrumental in this rapid turnover, since viral lysis not only kills host cells but also results in the release of cell fragments, dissolved organic carbon, and nutrients that can be readily used for bacterial growth (20, 37). At a given location and thus under identical macroenvironmental conditions, infection rates should be highest in microhabitats where bacterial productivity is greatest. Conversely, if the spatial arrangement or biofilm structure in benthic habitats provides protection from phage infection, then virus proliferation and VIM should be lower than in adjacent, less productive microhabitats, most notably in the water column. We tested these predictions by simultaneously examining virus-bacterium relationships in four microhabitats of a freshwater marsh whose close proximity ensured identical macroenvironmental conditions.
MATERIALS AND METHODS
Field procedures.
Virus dynamics were studied in four littoral marsh microhabitats of Lake Hallwil, a eutrophic lake in Central Switzerland (47°17′N, 8°14′E) at 449 m above sea level (13). A randomized block sampling design was used to designate six sampling plots (2.8 by 1.3 m each) in the littoral marsh, dominated by the common reed Phragmites australis. The six plots were sampled monthly over the course of a year. Depth-integrated water samples were taken with an acid-washed polyvinylchloride tube and stored in 1-liter acid-washed glass bottles. Epiphytic biofilms were collected by clipping off reed stems just above the sediment surface and cutting the submerged parts into three 10-cm sections, which were inserted in sterile screw-cap glass tubes containing lake water. Sediment samples were collected with a hand-held corer (6.5-cm diameter). The depth of the aerobic surface sediment layer was measured in the field with a calibrated oxygen microelectrode (model O2NAD-1; Toepffer Lab Systems, Göppingen, Germany). The aerobic top sediment layer (2 to 29 mm) was then transferred to an autoclaved glass vial. Plant litter on the sediment surface was collected with a manually operated bilge pump within an area defined by a custom-made Plexiglas cylinder (30-cm diameter). The collected material was passed over a 1-mm mesh screen, rinsed directly in the field with lake water, and placed in a plastic box (8). All samples were returned to the laboratory in a cool box and immediately processed upon arrival. Temperature changes were minimized throughout sample processing.
[3H]leucine incorporation.
Biomass production of heterotrophic prokaryotes was determined by measuring rates of [3H]leucine incorporation with a protocol specifically adapted to samples rich in organic matter (10). Samples were incubated for 30 min in a [3H]leucine solution prepared in 0.2-μm-filtered lake water. Incubations were stopped by adding trichloroacetic acid (5% final concentration). Macromolecules were precipitated on ice, and proteins were extracted in hot alkaline solution (0.5 M NaOH, 25 mM EDTA, 0.1% sodium dodecyl sulfate) and radioassayed (11). A scintillation cocktail (Hionic-Fluor; Packard Bioscience, Meriden, Conn.) with a high uptake capacity for alkaline solutions and effective suppression of chemiluminescence was used.
Microscopy.
Water samples (20 ml) for microscopy were preserved with buffered formaldehyde (0.1% sodium pyrophosphate buffer, 2% final concentration) and kept at 4°C in the dark for viral and bacterial counts. Biofilms were carefully scraped off the reed stems. The volume of the slurry was adjusted to 50 ml with filtered lake water in a graduated tube, the tube was vortexed, and a 5-ml aliquot was preserved with 5 ml of pyrophosphate-buffered formalin. A 0.5-ml aliquot of the intact surface sediment sample (i.e., including pore water) was taken with a sterile 1-ml disposable syringe with the Luer-lock end cut off and preserved with 10 ml of pyrophosphate-buffered formalin. Subsamples of the collected plant litter (∼500 mg [wet weight]) also were preserved in 10 ml of pyrophosphate-buffered formalin.
The preserved plant litter, sediment, biofilm, and water samples were treated with an ultrasonic probe (Branson Sonifier 250, 80-W output, 76-μm amplitude [9]), and the bacteria and viruses in the resulting suspension were stained for epifluorescence counts (28) using SYBR green II (11, 35). Briefly, appropriate aliquots of the suspensions were filtered (0.02-μm-pore-size Anodisc filters; Whatman), and the trapped cells and viruses were stained for 15 min and viewed under a Zeiss Axioskop 2 epifluorescence microscope (100-W high-pressure bulb, Chroma light filter set no. 41001 for SYBR green II, with excitation at 480 nm, beam splitter at 505 nm, and emission at 530 nm). Bright green virus-like particles (VLPs) were clearly visible against a dark black background. Bacterial numbers and biovolumes and numbers of VLPs were determined with an image analysis system (Visitron, Puchheim, Germany) (11, 13). All images were processed separately for viruses and prokaryotic cells to optimize contrast for either small VLPs or sharp definition of cell boundaries. To account for the halo around particles stained by the fluorescent dye, particles of <0.3 μm in diameter were considered viruses. Setting the threshold to slightly smaller diameters (e.g., 0.2 μm) had little effect on virus counts.
On four sampling occasions corresponding to the meteorological middles of winter, spring, summer, and fall, viruses were analyzed by transmission electron microscopy (TEM). The ultrasonicated biofilm, sediment, and litter samples were diluted three- to eightfold and filtered through 3-μm membrane filters at ≤10 kPa to remove large particles. Litter samples were then sonicated for 1 min, and sediment samples were vortexed (80 s at maximum speed) before filtration. Samples were harvested by ultracentrifugation with a swing-out rotor onto 400-mesh electron microscope copper grids with Formvar carbon support films (Science Service, Munich, Germany) (30). Samples were centrifuged at 70,000 × g for 20 min at 15°C. The supernatants were discarded, and the grids were stained for 60 s with uranyl acetate (2%, wt/wt) and then washed twice for 20 s with distilled water. Bacteria and viruses were counted with a JEOL model 1200EX TEM operated at 80 kV at a magnification of 30,000. The high voltage facilitated identification of bacterial cells containing mature phages (34). At least 350 bacterial cells were inspected in each sample to determine the frequency of visibly infected cells (FVIC) and the number of viruses released upon host cell lysis (burst size). This resulted in a total of 30,000 inspected cells, half of them in water samples and about 5,000 in each of the three benthic habitats. Phage sizes and morphologies of virus particles outside and inside bacterial cells were determined in 18 photographs from selected water samples.
Data analysis.
Frequencies of infected cells (FIC) in water samples were calculated from FVIC by using an empirical relationship (36). An empirical relationship was also used to calculate virus-induced mortality (VIM) of prokaryotic cells in water samples (3). Virus production was calculated by multiplying burst size by the number of cells lysed by viruses per unit of time (37), where the number of lysed prokaryotes equals the product of VIM and cell production. Cell production, in turn, was calculated from biomass production of prokaryotes (i.e., [3H]leucine incorporation), assuming that 1 μm3 of prokaryotic biomass equaled 7.8 fg of C and that the volume of the average cell in water samples was 0.02 μm3 (8). Biomass production was calculated from leucine incorporation rates using a conversion factor established for freshwater sediment bacteria (12). Isotope dilution has been found to be minor (1.17) (10) and was therefore not taken into account. For comparison, FIC, VIM, and virus production were also calculated for epiphytic biofilms, sediment, and plant litter, but meaningful interpretation of the results in strict quantitative terms is limited by the very small FVIC values.
RESULTS
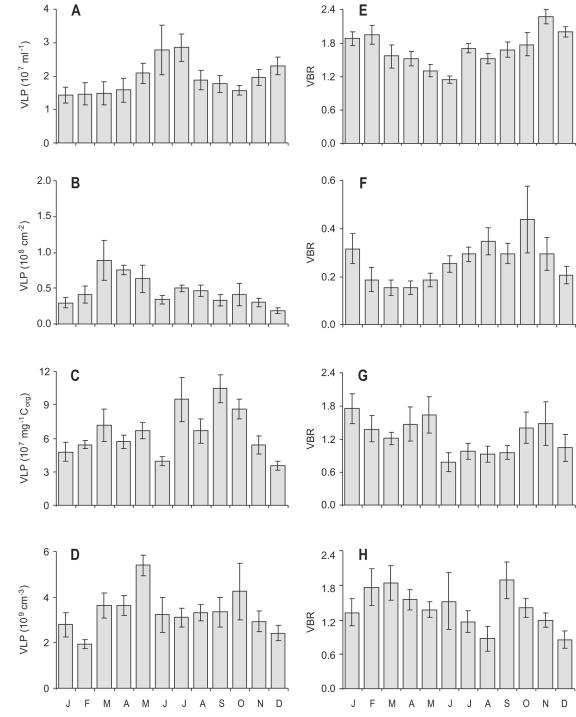
Epifluorescence counts indicated large numbers of free VLPs in all aquatic microhabitats investigated in this study: the water column of a littoral freshwater marsh dominated by common reed (Phragmites australis), biofilms on the reed stems, decaying plant litter, and the aerobic top sediment layer (Fig. 1). The abundance of VLPs was positively correlated with prokaryotic cell abundance (Pearson correlation on log-transformed data; r > 0.32, P < 0.01), the strongest correlations occurring in water and plant litter (P < 0.0001; r = 0.88 and r = 0.76, respectively). Although virus-to-bacterium ratios (VBRs) were at the low end of literature values (15, 26, 37), VLPs were consistently more abundant than prokaryotes, except in biofilms (Fig. 1). Comparisons of virus counts by epifluorescence microscopy and TEM of 16 randomly selected samples comprising the four microhabitats yielded similar abundances when the contrast between the fluorescently stained virus particles and the black filter background was optimized (paired t test on ln-transformed counts; P = 0.70). TEM revealed viruses mostly with hexagonal or nearly spherical heads, either with or without tails. Capsid diameters ranged from 25 to 100 nm, but most (88%) were between 30 and 70 nm, very similar to viruses in pelagic environments (2, 37).
FIG. 1.
Seasonal changes in the abundance of free virus-like particles (VLPs) and ratios of viruses to bacteria (VBRs) in four microhabitats of a littoral freshwater marsh. (A and E) Marsh water column. (B and F) Biofilm on reed stems. (C and G) Decaying plant litter. (D and H) Aerobic top sediment layer. Error bars depict ±1 SE; n = 6 (each histogram).
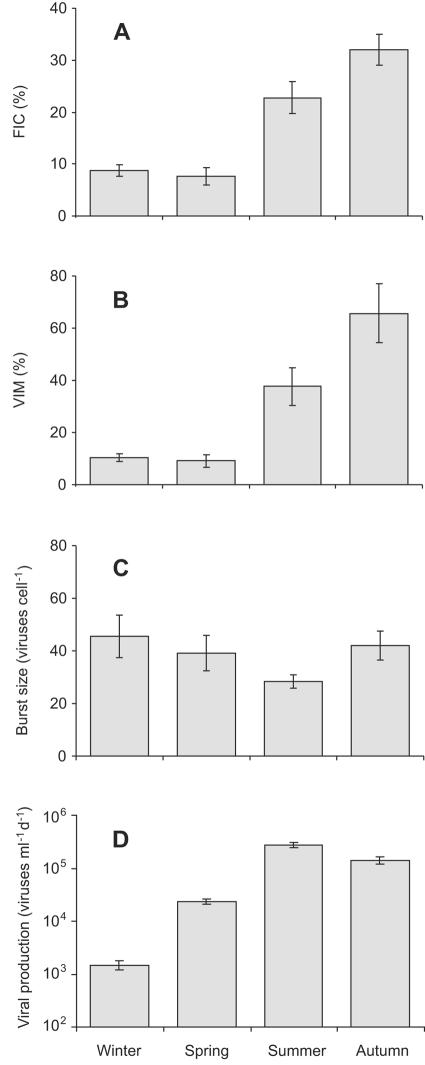
Prokaryotic cells infected by viruses were observed in all samples from the marsh water column. Mature phages were found in 292 of the 15,443 inspected cells (1.9%), corresponding to FIC between 8 and 32% of the total numbers (Fig. 2A). VIM in the marsh water was estimated to remove up to 66% of prokaryote production, with significant differences among seasons (Fig. 2B). The burst size of individual cells varied from 7 to 200, with a mean of 38 ± 3 (mean ± standard error [SE]) mature phages per cell, very similar to literature data from pelagic environments (37, 39). This resulted in a production of 0.18 × 104 to 32 × 104 viruses ml−1 of marsh water day−1 (Fig. 2D).
FIG. 2.
Seasonal changes in parameters characterizing virus-bacterium interactions in the water column of a freshwater marsh. (A) FIC. (B) VIM. (C) Number of mature viruses per bacterial cell (burst size). (D) Virus production rate (per milliliter of water per day). Error bars depict ±1 SE; n = 6 (each histogram).
In marked contrast to high infection rates in the marsh water column, none of the 4,970 cells inspected in biofilms on submerged plant surfaces was infected by phages. Likewise, only a single infected cell was found in aerobic sediment, although 4,269 bacteria were viewed, and only 3 out of 5,145 cells associated with decaying plant litter contained visible phages. Thus, based on inspection by TEM of nearly 15,000 cells of benthic prokaryotes, the FVIC for the three benthic habitats was consistently <0.1% of the total cell numbers. This is roughly 2 orders of magnitude lower than observed in the surrounding water column. As a consequence, estimated FIC never exceeded 0.4% in any of the benthic habitats, and virus production and VIM were invariably small (VIM < 1%).
DISCUSSION
Our discovery that the tight interactions between virioplankton and pelagic heterotrophic prokaryotes—shown more than a decade ago (2, 29) and since substantiated in both marine and freshwaters (20, 31, 37, 39)—may not invariably apply to benthic habitats is a striking finding with potentially far-reaching implications. Most remarkable is the observation that phage-infected cells were virtually absent from all three benthic microhabitats examined, even though (i) free viruses were abundant in those microhabitats and (ii) infection and VIM of prokaryotes was high in the surrounding water body. This exceedingly low incidence of infection implies that the critical roles of phages in regulating pelagic microbial population and food web dynamics, carbon and nutrient cycling, and likely the exchange of genetic material (20, 37, 39) may not universally apply to benthic habitats.
This conclusion seems at odds with recent reports assigning roles to viruses in marine sediments similar to those in open-water environments (23, 24). However, previous inferences about viral importance in benthic habitats have been based mainly on viral abundance and VBRs (1, 16, 37, 39), on positive correlations between viral and bacterial numbers and activity (24, 25), or on virus production in sediments as determined with dilution and incubation techniques, the last approach including indirect estimates of VIM (21, 23, 26). Importantly, direct evidence of virus infection and VIM based on microscopic observation of viruses within host cells has been lacking for sediments and other benthic habitats. Nevertheless, estimates of VIM in a freshwater sediment that were derived from virus decay experiments were more than three times lower (average of 6%) than in the overlying water (average of 20%), prompting the conclusion that sediment of the investigated cut-off river meander was an unfavorable environment for virus proliferation (18). This conclusion was also reached based on results from sediment analyses in a series of West African lakes where virus infection of benthic bacteria was low, as in the present study (2a). Data from marine sediments suggested that virus-induced cell lysis of prokaryotes also was a relatively minor process in the benthic carbon cycle (21, 26); however, this conclusion was based primarily on the disproportionately high external carbon supply, whereas estimates of VIM were 20% and 7 to 48%, respectively. Clearly, the current information on the impact of benthic viruses needs to be augmented by thorough analyses from a range of environments before a general assessment of viral impact on benthic prokaryotes can emerge. However, our near-zero infection and mortality rates indicate that inferences about benthic viral importance can result in serious errors when solely derived from viral abundance or VBR or when indiscriminately extrapolated from relationships established in planktonic microbial communities.
How can the apparent discrepancy between the high abundance of viruses on the one hand and the low infection rate (FIC) and impact on prokaryotes (VIM) on the other hand be reconciled? Explanations relate primarily to the complex spatial structure of benthic bacterial microhabitats, where microorganisms tend to be associated with surfaces. In particular, (i) the exopolymer matrix in which surface-associated microbes are embedded may provide physical protection against virus encounter (17), unless viruses carry enzymes on their surface that are capable of degrading biofilm exopolymers (31); (ii) viruses may be scavenged by adsorption to mineral or organic particles and thus be rendered inactive (23); (iii) viruses may be entrapped in biofilms and concentrated 100-fold, as observed for virus-sized particles in biofilms on reed stems (19); (iv) receptors for viral attachment on host cell surfaces may be masked by components of the exopolymer matrix (37) or cell aggregation (31); (v) proteases concentrated within biofilms may catalyze the digestion of capsid proteins (37); and (vi) greater resistance to infection mediated by rapid modification of viral receptors on the host membrane or by intracellular defense responses (4) may occur. The rationale behind the last argument is that short generation times promote rapid evolution of phage resistance on absolute time scales. It is in line with these ideas that biofilm bacteria in simple model systems show notable resistance to bacteriophage infection (14). Thus, although the exact mechanisms responsible for the lack of infection in our benthic marsh habitats are unknown, a large number of plausible reasons could account for the phenomenon.
The difficulty of infecting a bacterial host in benthic habitats could be exacerbated by the greater diversity of benthic microbial communities. Since phages are host specific (20, 38), the likelihood of finding a suitable host declines as microbial communities become more diverse. Sequencing of metagenomic libraries has shown that local viral diversity can indeed be high, with even the most abundant viral genome constituting only a small fraction (<5%) of the total virus community (5, 7). Given that prokaryotic diversity in sediments is orders of magnitude greater than in pelagic environments (33) and that theory predicts viral and bacterial diversity to covary, with viral genotypes typically 10 times more abundant (38), viral diversity in sediments is expected to be very large. This prediction is consistent with the finding that 1 kg of marine sediment can contain more than 104 and possibly up to 106 viral genotypes (6, 7). Therefore, chances for viral infection in the benthos might be even lower than in less diverse planktonic microbial communities, even though the high densities of prokaryotes and viruses in benthic habitats (24) foster host-virus encounter.
These arguments do not yet account fully for the mismatch we observed between high viral abundance and low infection rates, a phenomenon that has been termed the “infection paradox” (37). However, several solutions could reconcile the striking discrepancy. They include (i) physical import of viruses (22, 23), conceivably together with high loads of plankton carried into the marsh from the open lake in a conveyer belt-like fashion (see reference 13); (ii) possibly a low decay rate of viruses (e.g., references 16, 18, and 31; but see reference 23); (iii) a prevalence of temperate phages, which could be more abundant in sediment than in water (6; but see reference 24) and not only remain invisible inside cells but also be effective at conferring resistance to new infections; and (iv) infection by chronic phages, since large filamentous forms have been found to be a conspicuous component of a viral sediment community (25) and, like temperate phages, would hardly be detected inside cells by the TEM technique we used. These points illustrate the range of possible mechanisms that may resolve the infection paradox; well-designed experiments are needed to determine which are the most important.
Methodological bias is highly unlikely to account for our unexpected results of virtually nonexistent virus infection in benthic habitats. The key argument is that parameters characterizing viral dynamics in the marsh water displayed patterns and relationships with bacteria akin to those commonly observed in pelagic environments. In particular, the frequency of infected cells and bacterial mortality rates showed VIMs as high as 66%, with clear seasonal changes, demonstrating that in the marsh water viral cell lysis was a major cause of bacterial mortality. Our methods used to determine infection rates in the marsh water and benthic habitats were identical. This includes ultrasonication of water samples. Consequently, the good match with published accounts of our results on virus-prokaryote interactions in the water column bolsters confidence that the weak virus infections in the benthic habitats were real—not a methodological artifact.
If the low virus infection rates found here in three benthic microhabitats of a marsh turn out in the future to be widespread in freshwater, and possibly other benthic environments, then the implications could be far-reaching. The current paradigm is that bacteriophages are instrumental in controlling prokaryotic population dynamics, microbial biodiversity, and carbon and nutrient cycles in aquatic ecosystems (20, 31, 37, 38). Are these roles of phages and the well-established patterns for the pelagic zone of lakes and oceans universal? The present results suggest they are not. Do freshwater benthic habitats differ fundamentally from marine sediments (cf. references 1 and 18), and if so, why? Or are freshwater marshes featuring high organic matter content just an exceptionally unsuitable habitat for benthic viruses? Given the potential key role of viruses in controlling microbial diversity, food web dynamics, and biogeochemical cycles, it is critically important to understand the reasons for viral prevalence and the underlying mechanisms, as well their variations across major ecosystem types.
Acknowledgments
We thank O. Fries, C. Hoyle, K. Saukkonen, and D. Steiner for field and laboratory assistance; R. Danovaro, I. Hewson, M. Middelboe, and M. G. Weinbauer for constructive comments on the manuscript; and J. A. Fuhrman for discussion. The Canton Aargau kindly granted us access to the field site.
This work benefited from support received from the Swiss National Science Foundation (grant 3100-050439.97) and the Research Commission of ETH Zurich (grant 0-23010-00).
REFERENCES
- 1.Baker, P. W., and L. G. Leff. 2004. Seasonal patterns of abundance of viruses and bacteria in a Northeast Ohio (USA) stream. Arch. Hydrobiol. 161:225-233. [Google Scholar]
- 2.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 2a.Bettarel, Y., M. Bouvy, C. Dumont, and T. Sime-Ngando. Virus-bacterium interactions in water and sediment of West African inland aquatic systems. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 3.Binder, B. 1999. Reconsidering the relationship between virally induced bacterial mortality and frequency of infected cells. Aquat. Microb. Ecol. 18:207-215. [Google Scholar]
- 4.Bohannan, B. J. M., and R. E. Lenski. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol. Lett. 3:362-377. [Google Scholar]
- 5.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart, M., B. Felts, S. Kelley, J. M. Mahaffy, J. Nulton, P. Salamon, and F. Rohwer. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. R. Soc. Lond. B 271:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitbart, M., and F. Rohwer. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278-284. [DOI] [PubMed] [Google Scholar]
- 8.Buesing, N. 2002. Microbial productivity and organic matter flow in a littoral reed stand. Dissertation no. 14667. Ph.D. thesis. ETH Zurich, Switzerland.
- 9.Buesing, N., and M. O. Gessner. 2002. Comparison of detachment procedures for direct counts of bacteria associated with sediment particles, plant litter and epiphytic biofilms. Aquat. Microb. Ecol. 27:29-36. [Google Scholar]
- 10.Buesing, N., and M. O. Gessner. 2003. Incorporation of radiolabeled leucine into protein to estimate bacterial production in plant litter, sediment, epiphytic biofilms, and water samples. Microb. Ecol. 45:291-301. [DOI] [PubMed] [Google Scholar]
- 11.Buesing, N. 2005. Bacterial counts and biomass determination by epifluorescence microscopy, p. 199-204. In M. A. S. Graça, F. Bärlocher, and M. O. Gessner, (ed.). Methods to study litter decomposition: a practical guide. Springer, Dordrecht, The Netherlands.
- 12.Buesing, N., and J. Marxsen. 2005. Theoretical and empirical conversion factors for determining bacterial production in freshwater sediments via leucine incorporation. Limnol. Oceanogr. Methods 3:101-107. [Google Scholar]
- 13.Buesing, N., and M. O. Gessner. 2006. Benthic bacterial and fungal productivity and carbon turnover in a freshwater marsh. Appl. Environ. Microbiol. 72:596-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 15.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danovaro, R., C. Corinaldesi, A. Dell'Anno, M. Fabiano, and C. Corselli. 2005. Viruses, prokaryotes and DNA in the sediments of a deep-hypersaline anoxic basin (DHAB) of the Mediterranean Sea. Environ. Microbiol. 7:586-592. [DOI] [PubMed] [Google Scholar]
- 17.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer, U. R., C. Wieltschnig, A. K. T. Kirschner, and B. Velimirov. 2003. Does virus-induced lysis contribute significantly to bacterial mortality in the oxygenated sediment layer of shallow oxbow lakes? Appl. Environ. Microbiol. 69:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flood, J. A., and N. J. Ashby. 2000. Virus-sized particles can be entrapped and concentrated one hundred fold within wetland biofilms. Adv. Environ. Res. 3:403-411. [Google Scholar]
- 20.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 21.Glud, R. N., and M. Middelboe. 2004. Virus and bacteria dynamics of a coastal sediment: implication for benthic carbon cycling. Limnol. Oceanogr. 49:2073-2081. [Google Scholar]
- 22.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 23.Hewson, I., and J. A. Fuhrman. 2003. Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic water. Microb. Ecol. 46:337-347. [DOI] [PubMed] [Google Scholar]
- 24.Mei, M. L., and R. Danovaro. 2004. Virus production and life strategies in aquatic sediments. Limnol. Oceanogr. 49:459-470. [Google Scholar]
- 25.Middelboe, M., R. N. Glud, and K. Finster. 2003. Distribution of viruses and bacteria in relation to diagenetic activity in an estuarine sediment. Limnol. Oceanogr. 48:1447-1456. [Google Scholar]
- 26.Middelboe, M., R. N. Glud, F. Wenzhöfer, K. Ogurib, and H. Kitazato. 2006. Spatial distribution and activity of viruses in the deep-sea sediments of Sagami Bay, Japan. Deep Sea Res. I 53:1-13. [Google Scholar]
- 27.Mitsch, W. J., and J. G. Gosselink. 2000. Wetlands, 3rd ed. Van Nostrand Reinhold, New York, N.Y.
- 28.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 29.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 30.Sime-Ngando, T., J.-P. Mignot, C. Amblard, G. Bourdier, C. Desvilettes, and C. Quiblier-Lloberas. 1996. Caractérisation des particules virales planctoniques dans un lac du Massif Central français: aspects méthodologiques et premiers résultats. Ann. Limnol. 32:259-263. [Google Scholar]
- 31.Sutherland, I. W., K. A. Hughes, L. C. Skillman, and K. Tait. 2004. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232:1-6. [DOI] [PubMed] [Google Scholar]
- 32.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 33.Torsvik, V., L. Øvreås, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 34.Weinbauer, M. G., D. Fuks, and P. Peduzzi. 1993. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl. Environ. Microbiol. 59:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weinbauer, M. G., C. Beckmann, and M. G. Höfle. 1998. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl. Environ. Microbiol. 64:5000-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinbauer, M. G., C. Winter, and M. G. Höfle. 2002. Reconsidering transmission electron microscopy based estimates of viral infection of bacterio-plankton using conversion factors derived from natural communities. Aquat. Microb. Ecol. 27:103-110. [Google Scholar]
- 37.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 38.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 39.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]