Abstract
The stationary-phase sigma factor (RpoS) regulates many cellular responses to environmental stress conditions such as heat, acid, and alkali shocks. On the other hand, mutations at the rpoS locus have frequently been detected among pathogenic as well as commensal strains of Escherichia coli. The objective of this study was to perform a functional analysis of the RpoS-mediated stress responses of enterohemorrhagic E. coli strains from food-borne outbreaks. E. coli strains belonging to serotypes O157:H7, O111:H11, and O26:H11 exhibited polymorphisms for two phenotypes widely used to monitor rpoS mutations, heat tolerance and glycogen synthesis, as well as for two others, alkali tolerance and adherence to Caco-2 cells. However, these strains synthesized the oxidative acid resistance system through an rpoS-dependent pathway. During the transition from mildly acidic growth conditions (pH 5.5) to alkaline stress (pH 10.2), cell survival was dependent on rpoS functionality. Some strains were able to overcome negative regulation by RpoS and induced higher β-galactosidase activity without compromising their acid resistance. There were no major differences in the DNA sequences in the rpoS coding regions among the tested strains. The heterogeneity of rpoS-dependent phenotypes observed for stress-related phenotypes was also evident in the Caco-2 cell adherence assay. Wild-type O157:H7 strains with native rpoS were less adherent than rpoS-complemented counterpart strains, suggesting that rpoS functionality is needed. These results show that some pathogenic E. coli strains can maintain their acid tolerance capability while compromising other RpoS-dependent stress responses. Such adaptation processes may have significant impact on a pathogen's survival in food processing environments, as well in the host's stomach and intestine.
Enteric food-borne pathogens have developed sophisticated stress response mechanisms suited for a variety of ecological conditions encountered in both animal hosts and the external environment. The low infectious dose of enterohemorrhagic Escherichia coli, a food- and water-borne pathogen, is most likely due to a combination of several factors, such as its abilities to produce Shiga-like toxin and to survive gastric acidity and acidic foods, as well as efficient stress resistance mechanisms (19, 25, 36). Under adverse environmental conditions, such as nutrient limitation, osmotic shock, or low pH, cellular RpoS levels increase dramatically and RNA polymerase-containing RpoS transcribes over 70 genes involved in stress resistance and protection (39, 54). RpoS is a key element in the survival of several food-borne human pathogens, including Shigella flexneri, salmonellae, and diarrheagenic E. coli (24, 41, 47). In spite of having a central role in bacterial viability, data from genome sequence projects, as well as other comparative studies with pathogenic E. coli strains, have revealed that the rpoS locus is highly polymorphic (16, 33, 38).
Several recent studies have analyzed the occurrence of rpoS mutants under laboratory chemostatic growth conditions as well as from environmental and clinical isolates of enteric pathogens (27, 28, 38, 56). Under certain circumstances, variation at the rpoS locus has been thought to confer a selective advantage to cells within a bacterial population experiencing nutrient deprivation (17, 28, 57). Several of these studies utilized assays such as temperature tolerance, resistance to hydrogen peroxide, and/or synthesis of glycogen to measure the frequency of rpoS mutation (29, 38, 51). Mutations at the rpoS locus were frequently observed under chemostatic growth when E. coli K-12 cells were fed a suboptimal concentration of glucose (38). On the other hand, certain external stress conditions, such as mildly acidic conditions, significantly attenuated the frequency of rpoS mutations (14, 27). In spite of the diverse virulence characteristics of pathogenic E. coli, one common trait that is very evident is the ability of these strains to withstand gastric acidity (18, 30, 36, 53). In fact, acid tolerance plays a vital role in the survival and virulence of diarrheagenic E. coli strains (9, 42, 46). In S. flexneri and diarrheagenic E. coli strains, the induction of two of three acid resistance pathways under aerobic growth conditions is positively regulated by RpoS (3, 30, 47). The first acid resistance system (AR1) is referred to as the glucose-repressible oxidative pathway, and it protects cells from acidic stress above pH 3.0 (30, 31, 46). The structural components of this acid resistance system (other than RpoS), as well as the mechanisms by which it protects the cells, are still unknown (2, 9). The second acid resistance system (AR2) is glutamate-dependent acid resistance (GDAR), and it can protect cells from acidic stress below pH 3 (22, 46). In spite of the central regulatory role of RpoS, clinical and food-borne isolates of pathogenic E. coli with rpoS mutations have been reported (5, 53), suggesting that the pathogen may rely on rpoS-independent induction of acid resistance systems.
Intrigued by the frequent occurrence of mutations in rpoS and by the fact that the general stress response is controlled by this sigma factor, we undertook analyses of rpoS alleles from outbreak and clinical isolates of pathogenic E. coli strains. E. coli strains were subjected to an array of stress tests in which survival is primarily considered to be RpoS dependent (10, 39, 43). We observed several strains (26 out of 53) with phenotypic characteristics that were atypical of a functional RpoS allele (i.e., heat sensitivity and/or lack of glycogen synthesis), even though they had a functional rpoS according to their acid resistance phenotype (5). Temperature tolerance and ability to synthesize glycogen have been used previously to monitor the frequency of rpoS mutations (27, 38). Isolates of Shiga toxin-producing E. coli of serotypes O157:H7 (two strains), O111:H11 (one strain), and O26:H11 (one strain) with a wide spectrum of glycogen synthesis patterns were chosen for in-depth phenotypic analysis. The objective of the experiments described here was to evaluate rpoS heterogeneity and its effect on stress responses among pathogenic E. coli strains.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains of E. coli used were strains 52 (ATCC 43895, Nar Rfr) (11), 258 and 251 (serotype O157:H7), 229 (serotype O111:H11), and 205 (serotype O26:H11), all of which have been described previously (5). Strain 55 is an rpoS::Ap mutant of strain 52 (8), and pPS4.4 carries a wild-type rpoS gene on a pACYC184-based low-copy-number plasmid (53). The culture purity of the pathogenic E. coli isolates was ascertained by examining the catalase activity of at least 96 colonies for each strain (38). Frozen stocks maintained at −75°C were streaked onto Luria-Bertani (LB) agar, and, after overnight growth at 37°C, a single colony was inoculated into one of the following media: (i) minimal E medium (52) containing 0.4% glucose at pH 7.0 (EG minimal medium) (31), (ii) LB growth medium or (iii) LB buffered with 100 mM MES (morpholineethanesulfonic acid) (pH 5.5), or (iv) LB buffered with 100 mM MOPS (morpholinepropanesulfonic acid) (pH 8.0). Most diarrheagenic E. coli strains are auxotrophs and require amino acids or vitamins for growth in minimal media (1, 35). The precise requirement for vitamins and amino acids for individual strains was not determined; instead, EG minimal medium was supplemented with 50 μg of yeast extract per ml. Strain 52 is auxotrophic for thiamine, nicotinamide, and riboflavin (1). When grown in EG medium supplemented with vitamins or EG medium supplemented with yeast extract, the strain had identical GDAR phenotypes (4). To obtain stationary-phase cultures, cells were grown in the media on an orbital shaker (220 rpm, 37°C) for 20 to 22 h to an optical density at 600 nm (OD600) of 3.5 or higher.
The strains carrying plasmids (pPS4.4 or pACYC184) were grown in the media described above supplemented with chloramphenicol (35 μg ml−1).
Acid, alkali, and heat shock assays.
To examine the phenotypic expression of the RpoS-mediated acid resistance system (AR1), stationary-phase cells were diluted directly from the growth media (1:200) into EG medium, pH 3.0, and challenged at 37°C for 2 h. For the GDAR system, the cells were diluted directly from the growth media (1:200) into EG medium, pH 2.0, supplemented with 1.5 mM glutamate and challenged at 37°C for 1 h. EG medium was prewarmed to 37°C, and the pH was adjusted with 6 N HCl (3). The population of cells at the beginning of the acid challenge (after 1:200 dilution in the acid shock media) was in the range of ∼0.5 × 107 to 1.5 × 107 cells/ml (final concentration), which was necessary to avoid cell density-dependent artifacts (11). Control acid challenge experiments were performed in EG medium (pH 2.0) without the addition of glutamate. Viable counts were determined after acid challenge by dilution of the cells in phosphate-buffered saline (PBS) (50 mM, pH 7.2) and immediate plating on LB agar.
For heat shock assays, stationary-phase cells from LB-MES broth were diluted directly (1:200) into PBS preequilibrated to 58°C. Viable counts were determined at 10 s and at 1.5, 3, 5, and 7.5 min by dilution of the cells in PBS and immediate plating on LB agar.
For alkali shock assays, stationary-phase cells from LB, LB-MES, or LB-MOPS broth were diluted directly (1:200) into 100 mM CAPS (3-cyclohexylamine-1-propane sulfonic acid), pH 10.2 (adjusted with 100 mM NaOH), preequilibrated to 37°C. Viable counts were determined at 10 s and at 1, 2, and 4 h by dilution of cells in PBS and immediate plating on LB agar.
Experiments involving stress tolerance, as well as β-galactosidase assays, were repeated three to five times, and the results were calculated with consideration of all data points.
Detection of glycogen synthesis.
Glycogen synthesis was monitored as described previously (21) with minor modifications (55). Briefly, overnight cultures were plated onto Kornberg agar medium (55) and, after 24 h of incubation at 37°C, plates were further incubated at 10°C for 24 h and then stained with an iodine solution for 1 to 2 min. Dark brown colonies indicated the synthesis of glycogen, while pale brown or white colonies indicated partial or no synthesis of glycogen.
β-Galactosidase assays.
Enzyme activity was determined from cells grown to the stationary growth phase in EG minimal medium for 22 to 24 h. Cells were centrifuged, resuspended in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 [pH 7.0]), and then permeabilized by treatment with sodium dodecyl sulfate (SDS) and chloroform (37). For all experiments, stationary-phase cultures at 24 h after inoculation with an OD600 of 2.8 or more were used, and the assays were performed with Z buffer containing 50 mM β-mercaptoethanol. In all experiments, β-galactosidase activity (change in OD420 per minute) was normalized to cell density (OD600) and was compared to appropriate controls assayed at the same time.
Western blot analysis.
Cultures were incubated for 18 to 22 h at 37°C to a stationary growth phase (OD600 of 3.5 or higher) in LB-MES medium. Cells of each strain were collected by centrifugation (12,500 × g, 4 min), washed twice in saline, resuspended at 1 OD600 unit/ml in loading buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 2.5% β-mercaptoethanol, 0.01% Na-azide, 0.1% bromophenol blue), and placed in a boiling water bath for 5 min to make a whole-cell protein sample. Each protein extract was fractionated on SDS-polyacrylamide gradient gels (4 to 20%). After electrophoretic transfer of proteins onto nitrocellulose membranes, the RpoS protein was revealed by using a mouse monoclonal antibody (NeoClone Biotechnology International, Madison, WI) raised against a 38-kDa (RpoS) subunit of E. coli RNA polymerase, followed by anti-mouse antibody raised in a goat coupled to peroxidase as a secondary antibody. A SuperSignal chemiluminescence kit (Pierce Biotechnology, Rockford, IL) was used to detect the primary antibody, and the signal was captured on preflashed Kodak X-ray film.
Caco-2 cell adherence assays.
The human intestinal cell line Caco-2 was obtained from the ATCC (Manassas, VA). Frozen stock cultures were maintained in GIBCO (Invitrogen, Gaithersburg, MD) cell freezing medium held at −140°C. The tissue culture cells were cultivated at 37°C in a 94% air-6% CO2 atmosphere in minimal essential medium supplemented with Earle's salts, 10% fetal bovine serum, 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 1 mM sodium pyruvate. All cell culture media and supplements were obtained from GIBCO.
The adherence assays were performed essentially as described previously (5). In each well of a 24-well cell culture cluster, approximately 2 × 107 mid-log-phase bacteria (OD650, 0.3 to 0.4) were added to confluent monolayers of about 5 × 105 epithelial cells per well. Adherence was allowed to occur for 1 h at 37°C in a 94% air-6% CO2 atmosphere. The monolayers were washed three times with Hanks' balanced salt solution and then lysed with 0.1% Triton X-100 in 0.9% saline, and adherent bacteria were enumerated by spread plate counting on Trypticase soy agar (BBL Becton Dickinson Microbiology Systems, Cockeysville, MD). Adherence was expressed as the percentage of the inoculum surviving the washing treatment (i.e., percent recovery). All assays were conducted in quadruplicate and independently repeated at least twice. Results are expressed as averages of the replicate experiments. Recovery data percentages were calculated and analyzed as described previously by using the Student t test and one-factor analysis of variance followed by Dunns' or Student-Newman-Kuels multiple comparisons of means when significant differences (P < 0.05) were found (49).
DNA manipulations and cloning procedures.
Standard molecular biology methods were used for chromosomal and plasmid DNA isolation, restriction enzyme digestion and ligation, electroporation, and transformations (44). For cloning of the rpoS gene, PCR was performed with a 1:9 mixture of Vent and Taq DNA polymerase enzymes, as previously described (13, 51). PCR products were cloned in pGEM-T Easy vector (Promega Corporation, Madison, WI), and DNA sequencing was performed with double-stranded plasmid DNA templates and SP6 and T7 primers at the Iowa State University Sequencing Facility (Ames, IA), using the PCR-based dideoxynucleotide terminator protocol (Applied Biosystems, Foster City, CA).
Nucleotide sequence accession numbers.
The rpoS sequences of the isolates have been deposited in the GenBank database under accession numbers DQ287967 (52), DQ287968 (229), DQ287969 (258), DQ287970 (205), and DQ287971 (251).
RESULTS
Acid-induced synthesis of AR1 and cross-protection to heat tolerance.
We determined whether strains were capable of inducing higher levels of AR1 when grown to the stationary growth phase in LB-MES, pH 5.5, versus LB-MOPS, pH 8.0. All strains exhibited higher survival rates during acid shock (pH 3.0, 2 h) when grown in LB-MES (pH 5.5) than when the cells were grown in LB-MOPS (pH 8.0) (Table 1). Strain 55 is an rpoS::Ap mutant and lacked the ability to induce either AR1 or GDAR under aerobic growth conditions (9, 11). All of the strains (except strain 55) synthesized abundant RpoS when analyzed by Western blotting (Fig. 1) and were also able to induce GDAR (data not shown).
TABLE 1.
RpoS-dependent expression of the acid resistance system AR1a
| Strain (serotype) | Mean % survival ± SD after acid shock (pH 3.0, 2 h) for cells grown in:
|
|
|---|---|---|
| LB-MOPS (pH 8.0) | LB-MES (pH 5.5) | |
| 52 (O157:H7) | 3.4 ± 0.9 | 47.3 ± 4.9 |
| 55 (O157:H7) | <0.01 | <0.01 |
| 55-1 (O157:H7) | 2.4 ± 1.9 | 37.3 ± 6.3 |
| 251 (O157:H7) | 1.4 ± 0.8 | 41.3 ± 11.1 |
| 258 (O157:H7) | 4.4 ± 2.9 | 57.3 ± 8.1 |
| 205 (O26:H11) | 1.4 ± 0.7 | 37.3 ± 9.4 |
| 229 (O111:H11) | 1.3 ± 1.4 | 33.1 ± 6.7 |
Acid resistance was determined by diluting stationary-phase cells directly from the growth medium into EG medium (1:200), pH 3.0, with challenge at 37°C for 2 h. The initial cell density was in the range of ∼0.5 × 107 to 1.5 × 107 cells/ml.
FIG. 1.
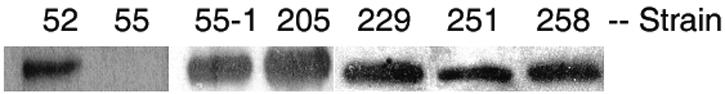
Western blot analysis of RpoS expression in pathogenic E. coli strains. Wild-type E. coli strains, rpoS::Ap mutant strain 55, and strain 55-1 carrying pPS4.4 (rpoS) were aerobically grown to the stationary phase in LB-MES medium, and equivalent amounts of protein were loaded into each lane and probed with anti-RpoS antibody to detect RpoS (38 kDa). Individual strains used are indicated above each lane.
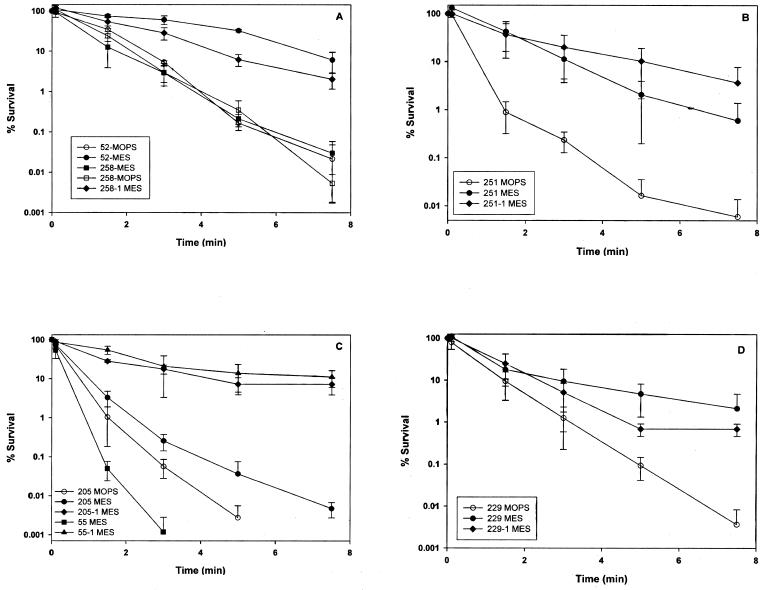
We then determined whether strains exhibited acid-induced cross-protection against heat stress. Strain 258 failed to exhibit acid-induced cross-protection to heat stress (Fig. 2A), but it was observed in strains 52, 229, and 251 (Fig. 2A, B, and D).
FIG. 2.
E. coli strains were grown to the stationary growth phase in LB-MES medium (pH 5.5) and diluted 1:200 in PBS preequilibrated at 58°C. The surviving population was determined at the indicated time points by withdrawing samples and plating appropriate dilutions immediately on LB agar.
When grown in LB-MES (pH 5.5), strain 205 displayed increased heat resistance compared to growth in LB-MOPS (pH 8.0) (Fig. 2C). In contrast, strain 55, grown in LB-MES, did not exhibit acid-induced heat resistance (Fig. 2C). Overall, when grown in LB-MES (pH 5.5), strain 229 displayed greater acid-induced heat tolerance (Fig. 2D) than did strains 55, 205, 251, and 258 (Fig. 2A, B, and C).
Glycogen synthesis and stress tolerance analyses after mobilization of the wild-type rpoS allele.
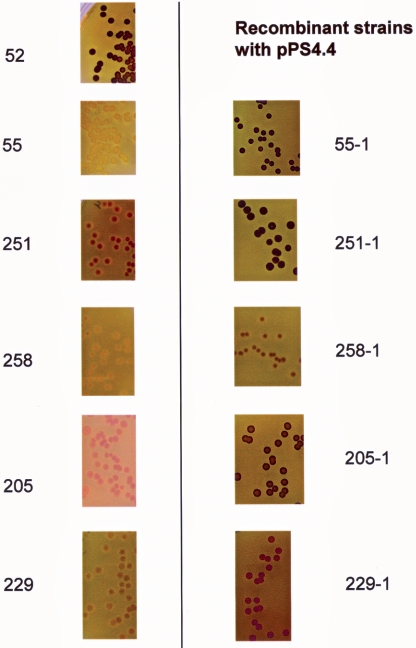
Like heat tolerance, the ability to synthesize glycogen has also been used as an indicator of the rpoS status of cell cultures in the chemostat as well as in clinical isolates of E. coli (27, 38). Strain 52, and to a lesser extent strain 251, showed dark brown colonies upon staining with iodine, indicating abundant glycogen synthesis. All other strains failed to synthesize glycogen when grown on Kornberg agar medium (Fig. 3).
FIG. 3.
Glycogen synthesis patterns of pathogenic E. coli strains. The wild-type strains and recombinant strains carrying pPS4.4 were plated on Kornberg agar (containing 50 μg/ml chloramphenicol for recombinant strains) and stained with iodine solution. Dark brown colonies indicate abundant glycogen synthesis, while pale yellow colonies are devoid of glycogen.
In order to examine how much phenotypic diversity may be attributed to native rpoS, we cloned and sequenced the chromosomal copies of rpoS from strains 52, 205, 229, 251, and 258 (see below). Additionally, we mobilized the wild-type rpoS allele cloned on the low-copy-number plasmid pPS4.4 (53) to all strains used in this study. (The wild type and the corresponding recombinant strains carrying pPS4.4 are referred to without and with the suffix “-1,” e.g., strains 205 and 205-1, respectively.) The recombinant strain 55-1, as well as strains 258-1, 251-1, and 205-1, gained heat resistance (Fig. 2; data not shown for 229-1) and the ability to synthesize glycogen (Fig. 3). Strain 229 was heat tolerant but defective in glycogen synthesis, and it gained the ability to synthesize glycogen upon mobilization of pPS4.4 (Fig. 3).
The role of RpoS in alkali shock survival.
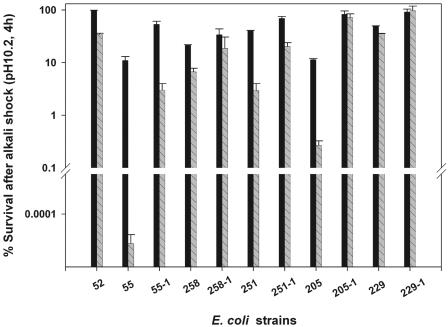
We further examined whether pathogenic strains with diverse sets of rpoS alleles would show polymorphic phenotypes of alkali stress tolerance. When cells were grown in LB broth, the final pH of the growth medium, after 20 to 22 h of growth, was between 8.5 and 8.8. Strain 52, when grown to stationary phase in LB broth, showed 99% survival after alkali shock in 100 mM CAPS (pH 10.2) for 4 h, compared to 12% survival by strain 55 (rpoS::Ap) (Fig. 4). Other strains tested were also able to withstand alkali shock, and their percent survival was in the range of 11% (strain 205) to 49% (strain 229). However, when grown in LB-MES, pH 5.5, alkali tolerance was strain dependent, and strain 55 (used as a negative control) was sensitive to alkali, with less than 0.001% of cells surviving after a 4-h exposure to 100 mM CAPS (pH 10.2) (Fig. 4). In comparison, strains 251 and 258 were much more resistant; 3.5 and 8.4% of the cells survived the alkali challenge, respectively. Strain 205 was the most sensitive among this group, with only 0.1% of cells surviving alkali shock, whereas strain 229 was the most alkali resistant, with 44% of cells surviving alkali shock. The mobilization of the wild-type allele of rpoS on pPS4.4 slightly improved the alkali tolerance of cells grown in LB (or LB-MOPS); when grown in LB-MES, the recombinant strains 55-1, 258-1, 251-1, 205-1, and 229-1 were able to withstand alkali shock as well as strain 52 (Fig. 4).
FIG. 4.
Phenotypic differences in alkali tolerance among various E. coli strains. E. coli strains were grown to the stationary growth phase in LB (filled bars) or LB-MES (striped bars) medium and diluted 1:200 in 100 mM CAPS buffer (pH 10.2) preequilibrated at 37°C. The surviving population was determined after 4 h by withdrawing samples and plating appropriate dilutions immediately on LB agar.
Starvation-induced β-galactosidase activity.
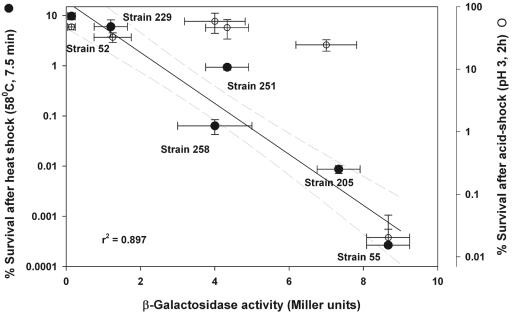
It has been reported that rpoS mutations lead to a significantly higher expression of the glucose scavenging system, especially during starvation (16, 27, 38). We examined the β-galactosidase activities of various strains in the stationary growth phase and compared them to the heat and acid tolerance phenotypes (Fig. 5). Strain 52 had the lowest β-galactosidase activity but the highest heat and alkaline tolerance, while the reverse was true for the rpoS::Ap strain 55. In general, the strains exhibited an inverse relationship (r2 = 0.897) for heat tolerance and β-galactosidase activity but not with reference to acid tolerance (Fig. 5).
FIG. 5.
Comparison of β-galactosidase activity with heat and acid survival of various E. coli strains. Heat tolerance data (filled circles) were obtained from Fig. 2, and data on acid resistance by AR1 (open circles) were obtained from Table 1. The regression line (solid) (r2 = 0.897) and 99% confidence intervals (broken lines) for heat tolerance and β-galactosidase activity were calculated with SigmaPlot, version 8.0.
Influence of different rpoS alleles on Caco-2 cell adherence.
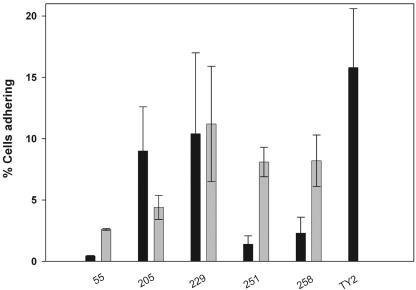
Previously we reported the positive influence of the two acid resistance pathway regulators rpoS and gadE on adherence of E. coli O157:H7 to Caco-2 cells (5). We examined strains that showed rpoS-mediated functional heterogeneity for stress tolerance phenotypes for their adherence properties (Fig. 6). As shown in Fig. 6, both O157:H7 wild-type strains 251 and 258, each with a native rpoS allele, were less adherent to Caco-2 cells than their recombinant rpoS-complemented counterparts (251-1 and 258-1). However, strains 205 (O26:H11) and 229 (O111:H11) either adhered better than or equally as well as their rpoS-complemented mates, 205-1 and 229-1, respectively. Salmonella enterica serovar Typhi strain TY2 and enterohemorrhagic E. coli strain 55, an rpoS::Ap mutant, were used as positive and negative controls, respectively.
FIG. 6.
Adherence phenotypes of pathogenic E. coli strains. Bacteria grown to mid-log phase were used to infect Caco-2 cell monolayers and were allowed to adhere for 1 h at 37°C in a 94% air-6% CO2 atmosphere. The monolayers were washed three times with Hanks' balanced salt solution and then lysed with 0.1% Triton X-100 in saline. Bacterial adherence (enumerated by spread plate counting) is expressed as the percentage of the inoculum surviving the washing treatment (percent recovery). All assays were conducted in quadruplicate and independently repeated at least twice. Black bars, wild-type E. coli strains; gray bars, strains carrying pPS4.4 (rpoS). The observed differences with 251 versus 251-1, 258 versus 258-1, and 205 versus 205-1 are significant (P < 0.001). Error bars indicate standard deviations.
Analysis of upstream and structural gene sequences of rpoS alleles.
Barring one amino acid substitution, there was very little variation observed in the DNA sequences of the rpoS coding regions from various strains (Table 2). The rpoS sequence of strain 205 contains one difference at nucleotide (nt) 482, resulting in a change from glutamine to proline at position 161. The remaining differences in DNA sequence did not alter the amino acid sequence. The 24-nt upstream region containing the ribosome-binding site (AGGAG), which is essential for the stationary-phase induction of rpoS translation (23), was conserved in all strains (Table 2). There was no change in the DNA sequence at the putative antisense element, nt −88 to −107 (12), presumably involved in pairing at the ribosome-binding site (data not shown). DNA sequence at the predicted DsrA-binding site (20, 34), nt −97 to −122, was also conserved in all strains (data not shown).
TABLE 2.
DNA sequence at the rpoS locus in various E. coli strains
| Strain | Nucleotide sequence (amino acid) at positiona:
|
||||
|---|---|---|---|---|---|
| −24 | 385-387 | 481-483 | 541-543 | 817-819 | |
| 52 | GGGATCACGGGTAGGAGCCACCTTATG | CGT (Arg) | CAA (Gln) | ACA (Thr) | GTA (Val) |
| 229 | GGGATCACGGGTAGGAGCCACCTTATG | CGC (Arg) | CAA (Gln) | ACC (Thr) | GTG (Val) |
| 258 | GGGATCACGGGTAGGAGCCACCTTATG | CGT (Arg) | CAA (Gln) | ACC (Thr) | GTA (Val) |
| 251 | GGGATCACGGGTAGGAGCCACCTTATG | CGT (Arg) | CAA (Gln) | ACA (Thr) | GTA (Val) |
| 205 | GGGATCACGGGTAGGAGCCACCTTATG | CGC (Arg) | CCA (Pro) | ACC (Thr) | GTG (Val) |
The ribosome-binding site is shown in italics, and the translational start codon is shown in bold. Nucleotide changes at specific positions are underlined.
DISCUSSION
We observed functional heterogeneity among alleles of rpoS in food-borne and clinical isolates of pathogenic strains of E. coli from serotypes O157:H7, O111:H11, and O26:H11. It appears unlikely that these nondomesticated strains could be carrying major gene deletions or multiple defects specifically with reference to host adherence, acid and heat tolerance, or glycogen synthesis pathways, since the mere expression of RpoS from a low-copy-number plasmid restored heat tolerance (strains 258, 251, and 205) (Fig. 2), host adherence (strains 251 and 258) (Fig. 6), and glycogen synthesis (strains 258, 229, and 205) (Fig. 3). Moreover, all strains were able to synthesize RpoS (Fig. 1) and induce AR1 when grown in aerobic shake cultures in LB-MES, pH 5.5 (Table 1), further supporting the fact that they do possess fully operational acid resistance systems as well as a functional rpoS gene.
There are a number of different measurable phenotypes that are used to characterize mutation frequencies in E. coli (6, 7, 32). Glycogen synthesis and heat tolerance profiles have been used recently to monitor mutations at the rpoS locus (16, 27, 38). Although glycogen production or accumulation is not required for growth under laboratory conditions, the presence of glycogen may increase the viability of E. coli under adverse conditions or in specific ecological niches (40). However from the number of outbreak strains that we examined, several strains had heat resistance profiles and glycogen synthesis abilities that were inconsistent with their RpoS status. For example, strain 251 synthesized significant quantities of glycogen but was heat sensitive, while strain 229 was heat tolerant but failed to synthesize glycogen. When pPS4.4 was mobilized in strains 229 and 251, the recombinant strain 229-1 was able to synthesize glycogen (Fig. 3), while strain 251-1 gained heat tolerance (Fig. 2B).
Growth to stationary phase or exposure to mild acidity has been shown to induce RpoS-mediated cross-protection against elevated temperatures (10) and was apparent in all strains except 258 (Fig. 2A). With reference to the ability to elicit enhanced levels of AR1 in LB-MES broth (as a gauge of RpoS status), all strains appeared to have a functional RpoS.
There are conflicting reports in the literature on the involvement of RpoS in alkali tolerance of E. coli (45, 46). We observed that with prior exposure to alkaline pH (i.e., growth in either LB or LB-MOPS), cells induce rpoS-independent mechanisms which seem to play a major role in protecting cells from alkali shock. The presence of functional RpoS did aid cells in surviving alkali shock, as evidenced by the poor survival of rpoS mutant strain 55 in comparison with other strains (Fig. 4). On the other hand, during the transition from acidic conditions to alkali shock, cells appear to rely heavily on rpoS-dependent pathways to overcome the alkali shock. Accordingly, RpoS-mediated functional heterogeneity for alkali tolerance was observed when cells were grown in LB-MES, pH 5.5, prior to alkali shock but not when grown in LB (Fig. 4) or in LB-MOPS, pH 8.0 (data not shown).
Studying adherence to Caco-2 cells by gadE and rpoS mutants of pathogenic E. coli strains, we observed a synergistic effect of the two regulators (5). In the work reported here, strains 205, 229, 251, and 258 had “functional” chromosomal rpoS genes and accordingly expressed the RpoS-dependent acid resistance pathways AR1 (Table 1) and GDAR (data not shown). It appears that the chromosomal copy of rpoS from strains 251 and 258 was inefficient in supporting host cell adherence functions (in addition to defects in temperature tolerance and/or glycogen synthesis), since each of these strains adhered better upon mobilization of pPS4.4. Although adherence studies centered on only five strains, the O157:H7 strains showed the greatest need for a functional rpoS allele to regulate adherence. The other strains, 205 (O26:H11) and 229 (O111:H11), adhered at higher populations than the O157:H7 strains, suggesting that there may be differences in the rpoS regulation of adherence mechanisms among serotypes and that in non-O157:H7 strains, adherence may be controlled by other regulators and may be rpoS independent and not directly linked to a stress response (48, 50). An alternative explanation for the infectivity of rpoS mutants is that the patients may be immunocompromised or have reduced gastric acidity.
Nutritional limitations and environmental stress conditions impose conflicting choices on E. coli, where the number of substrates metabolized is negatively regulated by RpoS (27, 38). Likewise, a potential growth advantage of cells in stationary phase has been reported (28, 56, 57) and appears to be related to dysfunctional RpoS activity. The RpoS-containing RNA polymerase holoenzyme has the ability to co-orchestrate with additional regulatory factors. Recent microarray data have indicated that the rpoS regulon is a large cascade-like network that includes expression of secondary regulators, which may result in special regulatory (in)activation of subsets of RpoS-dependent genes (54). The data from this study further revealed that some acid resistance genes can switch RpoS and/or σ70 dependence, depending on specific environmental or stress conditions. We observed that the pathogenic E. coli strains examined here were able to overcome RpoS-mediated negative control over the expression of the sugar uptake system without compromising the induction of acid resistance systems (and the ability to adhere to Caco-2 cells in non-O157:H7 isolates).
Induction of the acid resistance system AR1 in complex growth media appears to accurately report a “minimal required functionality” of RpoS in E. coli strains. Since some temperature-sensitive and glycogen-defective strains were able to launch RpoS-mediated acid resistance, it is imperative that our understanding of RpoS mutation frequencies be reevaluated. It appears from both laboratory studies (26, 38) and the occurrence of rpoS mutations in food-borne populations (5, 53) that regulatory divergence may be more common and that natural regulatory settings may not be uniform even within a species (15, 27). The ability of pathogenic E. coli strains to balance sigma factor-dependent functions without affecting their acid resistance may have significant implications in microbiological food safety and in understanding of the survival strategies of the pathogen in facing multiple stresses in the gastrointestinal environment of the host.
Acknowledgments
This study was supported in part by the Overseas Industrial Attachment Program of the School of Life Sciences and Chemical Technology, Ngee Ann Polytechnic, Singapore.
We thank Ingrid Berlanger, Cheryl Mudd, and Frances Trouth for excellent technical assistance.
REFERENCES
- 1.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 3.Bhagwat, A. A. 2003. Regulation of the glutamate-dependent acid-resistant system of diarrheagenic Escherichia coli strains. FEMS Microbiol. Lett. 227:39-45. [DOI] [PubMed] [Google Scholar]
- 4.Bhagwat, A. A., and M. A. Bhagwat. 2004. Comparative analysis of transcriptional regulatory elements of glutamate-dependent acid-resistance systems of Shigella flexneri and Escherichia coli O157:H7. FEMS Microbiol. Lett. 234:139-147. [DOI] [PubMed] [Google Scholar]
- 5.Bhagwat, A. A., L. Chan, R. Han, J. Tan, M. Kothary, J. Jean-Gilles, and B. D. Tall. 2005. Characterization of enterohemorrhagic Escherichia coli strains based on acid resistance phenotypes. Infect. Immun. 73:4993-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 7.Bjorkman, J., I. Nagaev, O. G. Berg, D. Hughes, and D. I. Andersson. 2000. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science 287:1479-1482. [DOI] [PubMed] [Google Scholar]
- 8.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 9.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheville, A. M., K. W. Arnold, C. Buchrieser, C. M. Cheng, and C. W. Kaspar. 1996. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 62:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, S., J. Meng, and A. A. Bhagwat. 2001. Availability of glutamate and arginine during acid challenge determines cell density-dependent survival phenotype of Escherichia coli strains. Appl. Environ. Microbiol. 67:4914-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunning, C., L. Brown, and T. Elliott. 1998. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J. Bacteriol. 180:4564-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell, M. J., and S. E. Finkel. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J. Bacteriol. 185:7044-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferenci, T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1-8. [DOI] [PubMed] [Google Scholar]
- 16.Ferenci, T. 2003. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 11:457-461. [DOI] [PubMed] [Google Scholar]
- 17.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 19.Foster, J. W., and M. Moreno. 1999. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found. Symp. 221:55-69. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399-404. [DOI] [PubMed] [Google Scholar]
- 21.Govons, S., R. Vinopal, J. Ingraham, and J. Preiss. 1969. Isolation of mutants of Escherichia coli B altered in their ability to synthesize glycogen. J. Bacteriol. 97:970-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenhorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirsch, M., and T. Elliott. 2005. Stationary-phase regulation of RpoS translation in Escherichia coli. J. Bacteriol. 187:7204-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibanez-Ruiz, M., V. Robbe-Saule, D. Hermant, S. Labrude, and F. Norel. 2000. Identification of RpoS (σS)-regulated genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:5749-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 26.King, T., and T. Ferenci. 2005. Divergent roles of RpoS in Escherichia coli under aerobic and anaerobic conditions. FEMS Microbiol. Lett. 244:323-327. [DOI] [PubMed] [Google Scholar]
- 27.King, T., A. Ishihama, A. Kori, and T. Ferenci. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolter, R. 1999. Growth in studying the cessation of growth. J. Bacteriol. 181:697-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Large, T. M., S. T. Walk, and T. S. Whittam. 2005. Variation in acid resistance among Shiga toxin-producing clones of pathogenic Escherichia coli. Appl. Environ. Microbiol. 71:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loewe, L., V. Textor, and S. Scherer. 2003. High deleterious genomic mutation rate in stationary phase of Escherichia coli. Science 302:1558-1560. [DOI] [PubMed] [Google Scholar]
- 33.Lombardo, M., I. Aponyi, and S. M. Rosenberg. 2004. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics 166:669-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng, J., M. P. Dolye, T. Zhao, and S. Zhao. 2001. Enterohemorrhagic Escherichia coli, p. 193-213. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 36.Merrell, D. S., and A. Camilli. 2002. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 5:51-55. [DOI] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nystrom, T. 2004. Stationary-phase physiology. Annu. Rev. Microbiol. 58:161-181. [DOI] [PubMed] [Google Scholar]
- 40.Preiss, J., and T. Romeo. 1994. Molecular biology and regulatory aspects of glycogen biosynthesis in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 47:329-399. [DOI] [PubMed] [Google Scholar]
- 41.Price, S. B., C. M. Cheng, C. W. Kaspar, J. C. Wright, F. J. DeGraves, T. A. Penfound, M. P. Castanie-Cornet, and J. W. Foster. 2000. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl. Environ. Microbiol. 66:632-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, S. B., J. C. Wright, F. J. DeGraves, M. P. Castanie-Cornet, and J. W. Foster. 2004. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl. Environ. Microbiol. 70:4792-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robey, M., A. Benito, R. H. Hutson, C. Pascual, S. F. Park, and B. M. Mackey. 2001. Variation in resistance to high hydrostatic pressure and rpoS heterogeneity in natural isolates of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:4901-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sharma, M., and L. R. Beuchat. 2004. Sensitivity of Escherichia coli O157:H7 to commercially available alkaline cleaners and subsequent resistance to heat and sanitizers. Appl. Environ. Microbiol. 70:1795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at acidic pH, p. 1539-1549. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 47.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens, M. P., A. J. Roe, I. Vlisidou, P. M. van Diemen, R. M. La Ragione, A. Best, M. J. Woodward, D. L. Gally, and T. S. Wallis. 2004. Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect. Immun. 72:5402-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tall, B. D., J. F. La Peyre, J. W. Bier, M. D. Miliotis, D. E. Hanes, M. H. Kothary, D. B. Shah, and M. Faisal. 1999. Perkinsus marinus extracellular protease modulates survival of Vibrio vulnificus in eastern oyster hemocytes. Appl. Environ. Microbiol. 65:4261-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torres, A. G., and J. B. Kaper. 2003. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 71:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visick, J., and S. Clarke. 1997. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J. Bacteriol. 179:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 53.Waterman, S. R., and P. L. C. Small. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei, B., S. Shin, D. LaPorte, A. J. Wolfe, and T. Romeo. 2000. Global regulatory mutations in csrA and rpoS cause severe central carbon stress in Escherichia coli in the presence of acetate. J. Bacteriol. 182:1632-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]
- 57.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]