Abstract
Geranyl-coenzyme A (CoA)-carboxylase (GCase; AtuC/AtuF) and methylcrotonyl-CoA-carboxylase (MCase; LiuB/LiuD) are characteristic enzymes of the catabolic pathway of acyclic terpenes (citronellol and geraniol) and of saturated methyl-branched compounds, such as leucine or isovalerate, respectively. Proteins encoded by two gene clusters (atuABCDEFGH and liuRABCDE) of Pseudomonas aeruginosa PAO1 were essential for acyclic terpene utilization (Atu) and for leucine and isovalerate utilization (Liu), respectively, as revealed by phenotype analysis of 10 insertion mutants, two-dimensional gel electrophoresis, determination of GCase and MCase activities, and Western blot analysis of wild-type and mutant strains. Analysis of the genome sequences of other pseudomonads (P. putida KT2440 and P. fluorescens Pf-5) revealed candidate genes for Liu proteins for both species and candidate genes for Atu proteins in P. fluorescens. This result concurred with the finding that P. fluorescens, but not P. putida, could grow on acyclic terpenes (citronellol and citronellate), while both species were able to utilize leucine and isovalerate. A regulatory gene, atuR, was identified upstream of atuABCDEFGH and negatively regulated expression of the atu gene cluster.
Acyclic terpenes, such as citronellol and geraniol, are aroma compounds frequently occurring in plants. Citronellol (3,7-dimethyl-6-octen-1-ol) is used in the food and perfume industries but is also used as a natural repellent of mosquitoes (19). Geraniol is an aroma compound typical for plants of the genus Geranium; it is structurally related to citronellol and differs from the latter only by the presence of an additional double bond. Citronellol and geraniol are model compounds of acyclic monoterpenes and belong to the family of acyclic methyl-branched molecules derived from isoprene. Related compounds are carotenoids, steroids, and polyisoprene (rubber). Leucine and isovalerate are examples of saturated molecules with a methyl-branched carbon backbone. Recently, it was found that geraniol and similar acyclic terpenes can have significant effects on mammalian cells and can even induce apoptosis in vitro in pancreatic cancer cells (5, 7, 11, 22). Apparently, the physiological activities and cellular functions of monoterpenes are underestimated. The poor information on the biochemical routes of monoterpenes in organisms might be one reason for this lack of knowledge. Citronellol is the only acyclic monoterpene for which some information exists on its biochemistry in microorganisms (see below).
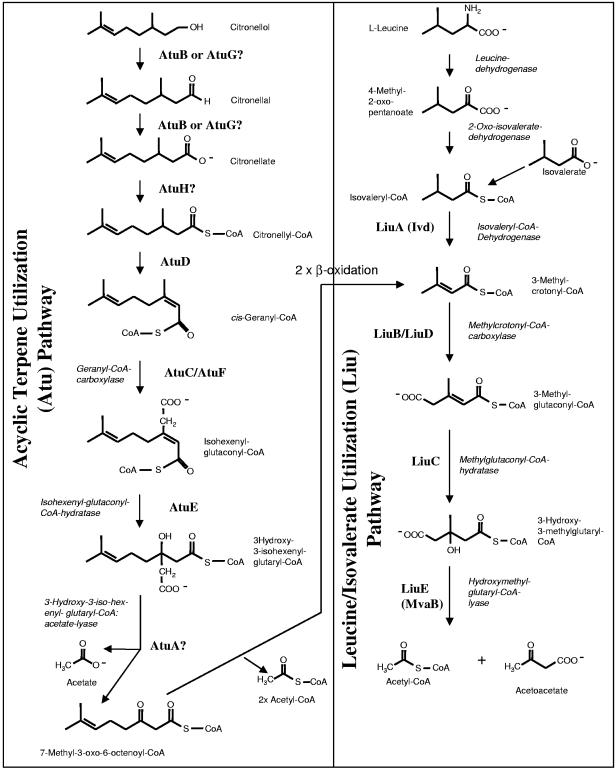
The degradation pathway of acyclic (mono)terpenes, such as citronellol or geraniol, was investigated in Pseudomonas citronellolis by Seubert and coworkers in the 1960s (28-32) and by Fall and coworkers in the 1970s (12, 14, 15). A putative pathway leading from citronellol to acetate, acetyl coenzyme A (acetyl-CoA), and acetoacetate as end products based on the above-mentioned biochemical investigations is shown in Fig. 1. The pathway includes three phases. (i) Citronellol and geraniol are oxidized to citronellate and geranylate, respectively, and activated to the corresponding CoA thioesters. Citronellyl-CoA is oxidized to geranyl-CoA. Geranyl-CoA is subsequently carboxylated by geranyl-CoA carboxylase (GCase), hydrated at the double bond by isohexenylglutaconyl-CoA-hydratase, and the product is cleaved into acetate and 7-methyl-3-oxo-6-octenoyl-CoA. As a result, the branched β-methyl group that would inhibit β-oxidation, is cleaved off as acetate. This biochemical route is named the acyclic terpene utilization (Atu) pathway (Fig. 1). (ii) 7-Methyl-3-oxo-6-octenoyl-CoA can be now oxidized by two rounds of β-oxidation. (iii) The product, 3-methylcrotonyl-CoA, concurs with the leucine/isovalerate utilization (Liu) pathway, which includes a second hydratase and carboxylase step catalyzed by methylglutaconyl-CoA-hydratase and methylcrotonyl-CoA carboxylase (MCase) (Fig. 1). MCase differs from GCase in P. citronellolis in its substrate specificity (15, 18). Studies of Fall and coworkers suggested that utilization of acyclic terpenes might be very similar in Pseudomonas aeruginosa and probably in Pseudomonas mendocina (6, 14).
FIG. 1.
Acyclic terpene utilization (Atu) and leucine/isovalerate utilization (Liu) pathways according to Seubert and Fass (30) as modified by Höschle et al. (20). Proposed functions based on phenotype analysis, enzyme activity determination, Western blot analysis, and/or amino acid alignments of identified gene products are indicated. Question marks indicate speculative assignments.
Knowledge of the structural genes involved in catabolism of methyl-branched compounds was poor until recently. A cluster of six genes (gnyRDBHAL [open reading frames PA2011 to PA2016], renamed the liuRABCDE gene cluster) was reported to be necessary for degradation of linear terpenes in P. aeruginosa (10). However, studies in our lab suggested that the liu gene cluster is only indirectly involved in terpene utilization (Fig. 1) and that another gene cluster (atu gene cluster, atuABCDEFGH [Fig. 1]) is more likely to encode proteins specific for acyclic terpene utilization (20). Recent results of Aguilar et al. (1) were in agreement with this assumption. In this study we identified most of the putative gene products of the atu and liu gene clusters by two-dimensional (2D) gel electrophoresis. The importance of individual atu and liu genes for functionality of the two combined pathways was investigated by GCase and MCase activity determination, insertion mutagenesis, and Western blot analysis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Growth of bacteria in liquid culture was performed as described elsewhere (20). Growth on solid media with liquid carbon sources was performed in separate incubators to avoid cross contamination by vapors. Liquid cultures contained 0.5% glucose or 0.075% glucose and 0.1% of sodium citronellate or 0.1% sodium isovalerate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli JM109 | Cloning strain | 36 |
| E. coli S17-1 | Mobilizing strain | 33 |
| Pseudomonas aeruginosa PAO1 | Wild type; utilizes citronellol and isovalerate | ATCC (ATCC 15692) |
| Pseudomonas fluorescens Pf-5 | Wild type; utilizes citronellol and isovalerate | 25 |
| Pseudomonas citronellolis | Wild type; utilizes citronellol and isovalerate | 28 |
| Pseudomonas putida KT2440 | Wild type; utilizes isovalerate | ATCC (ATCC 47054) |
| P. aeruginosa strainsa | ||
| PAO1 Smr | Spontaneous streptomycin-resistant mutant of PAO1 (≥500 μg/ml); Smr | 21 |
| PAO1 Smr 22B/1/1 (atuE::mini-Tn5) | Transposon mutant of PAO1; Tcr transposon inserted in atuE | This study |
| PAO1 Smr ins-atuR | pKnockout-G::atuR; Smr Gmr | This study |
| PAO1 Smr ins-atuA | pKnockout-G::atuA; Smr Gmr | This study |
| PAO1 Smr ins-atuB | pKnockout-G::atuB; Smr Gmr | This study |
| PAO1 Smr ins-atuC | pKnockout-G::atuC; Smr Gmr | This study |
| PAO1 Smr ins-atuD | pKnockout-G::atuD; Smr Gmr | This study |
| PAO1 Smr ins-atuE | pKnockout-G::atuE; Smr Gmr | This study |
| PAO1 Smr ins-atuF | pKnockout-G::atuF; Smr Gmr | 20 |
| PAO1 Smr ins-atuG | pKnockout-G::atuG; Smr Gmr | This study |
| PAO1 Smr ins-atuH | pKnockout-G::atuH; Smr Gmr | This study |
| PAO1 Smr ins-liuD | pKnockout-G::liuD; Smr Gmr | This study |
| PAO1 Smr ins-liuC | pKnockout-G::liuC; Smr Gmr | This study |
| Plasmids | ||
| pUT miniTn5-Tc | Mutagenesis plasmid; Tcr | 9 |
| pKnockout-G | Suicide vector for rapid gene inactivation in P. aeruginosa | 35 |
| pKnockout-G::atuR | Including an 3′ and 5′ truncated fragment of atuR; Ampr Gmr | This study |
| pKnockout-G::atuA | Including an 3′ and 5′ truncated fragment of atuA; Ampr Gmr | This study |
| pKnockout-G::atuB | Including an 3′ and 5′ truncated fragment of atuB; Ampr Gmr | This study |
| pKnockout-G::atuC | Including an 3′ and 5′ truncated fragment of atuC; Ampr Gmr | This study |
| pKnockout-G::atuD | Including an 3′ and 5′ truncated fragment of atuD; Ampr Gmr | This study |
| pKnockout-G::atuE | Including an 3′ and 5′ truncated fragment of atuE; Ampr Gmr | This study |
| pKnockout-G::atuF | Including an 3′ and 5′ truncated fragment of atuF; Ampr Gmr | 20 |
| pKnockout-G::atuG | Including an 3′ and 5′ truncated fragment of atuG; Ampr, Gmr | This study |
| pKnockout-G::atuH | Including an 3′ and 5′ truncated fragment of atuH; Ampr Gmr | This study |
| pKnockout-G::liuD | Including an 3′ and 5′ truncated fragment of liuD; Ampr Gmr | This study |
| pKnockout-G::liuC | Including an 3′ and 5′ truncated fragment of liuC; Ampr Gmr | This study |
Strains with insertion mutations in genes are indicated by “ins” preceding the gene.
Insertion mutagenesis.
Transposon mutagenesis with pUTminiTn5-Tc and identification and sequencing of transposon insertion fragments were performed as described previously (21). Gene disruptions were carried out using pKnockout-G (35) as described previously (20). Correctness of the respective insertion event was verified by PCR using one gene-specific and one pKnockout-specific primer (data not shown). Polar downstream effects were avoided by selection of those mutants in which the lac promoter of pKnockout (constitutively expressed in P. aeruginosa) was oriented colinearly to the respective gene cluster, resulting in constitutive transcription of the genes. Insertion mutagenesis of open reading frame PA2885 (atuR) was performed as described later in the text.
Synthesis of geranyl-CoA and HPLC-(ESI)MS determination of CoA compounds.
Synthesis of geranyl-CoA was done by the mixed-anhydride method described elsewhere (17) with some modifications. Geranic acid (770 μmol) was dissolved in 5.1 ml tetrahydrofurane and neutralized by adding an equimolar amount of triethylamine. Ethylchloroformate (770 μmol) was added, and the mixture was stirred for 30 min at room temperature and filtered (4 μm pore size). The filtrate containing the anhydride was added dropwise to 58 μmol coenzyme A that had been dissolved in a 3:2 mixture (pH 8.0) of 12 ml water to tetrahydrofurane (with solid NaHCO3). After the mixture was stirred for 25 min, 4 ml of water was added and the pH was adjusted to 3.0 with 2N HCl. The solution was extracted three times with diethyl ether. The aqueous phase was lyophilized. Liquid chromatography coupled to mass spectrometry with an electrospray interface [(ESI)MS] was run on an HP1100 high-performance liquid chromatography (HPLC) system (Agilent, Waldbronn, Germany) coupled with a Micromass VG platform II quadrupole mass spectrometer and an electrospray interface. Chromatographic conditions were as follows: column, Hypersil gold C18 (1.9 μm; 50 × 2.1 mm); 25°C; flow rate, 0.2 ml/min; eluent mixture A (10 mM ammonium formiate [98%]-methanol [2%] [vol/vol]) and eluent mixture B (acetonitrile gradient; percent acetonitrile 3% [at 0 to 3 min] to 100% [at 20 to 24 min]). Geranyl-CoA synthesized as mentioned above was identified by detection of the expected quasimolecular ion ([M-H]−) m/z 916 and the corresponding Na adduct ([M-2H+Na]−) m/z 938 in the major HPLC peak (not shown).
2D gel electrophoresis.
The cells of interest (2 to 3 g) were resuspended in 1 ml of 0.1 M HEPES buffer (pH 7.4) per g of cells before 100 μl DNase I (100 μg/ml), 50 μl RNase A (10 mg/ml), and 150 μl 10 mM MgSO4 were added. The suspension was passed two times through a precooled French press cell at 800 lb/in2 and centrifuged at 80,000 × g for 1 h at 4°C. 2D electrophoresis was performed using 18-cm-long immobilized pH gradient strips (pH 3 to 10 or pH 4 to 7) that had been rehydrated under mineral oil at room temperature overnight {340 μl rehydration solution contained 7 M urea, 2 M thiourea, 2% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 0.002% (wt/vol) bromphenol blue, 10 mM dithiothreitol (DTT), 0.5% Pharmalyte pH 3 to 10 or pH 5 to 8, and 500 μg soluble protein}. Isoelectric focusing was performed at 500 V for 1 min, 500 to 3,500 V for 1 h 30 min and at 3,500 V for 6 h 20 min in a Multiphor II apparatus under mineral oil at 20°C. The focused strips were equilibrated in buffer I containing 4% (wt/vol) sodium dodecyl sulfate (SDS), 50 mM Tris, 6 M urea, 30% (vol/vol) glycerol, 0.002% (wt/vol) bromphenol blue, and 2% (wt/vol) DTT for 15 min and then in buffer II containing 2.5% (wt/vol) iodoacetamide instead of DTT for another 15 min at room temperature. Each strip was run into a 10% (wt/vol) SDS-polyacrylamide gel for the molecular mass dimension using a Hoefer isodalt apparatus (Amersham Biosciences) at 100 V overnight. Protein spots were detected by overnight colloidal Coomassie blue staining in 8% (wt/vol) ammonium sulfate, 2% (vol/vol) phosphoric acid, 5% (wt/vol) Coomassie blue G250, and 20% (vol/vol) methanol. Equipment was from GE Healthcare.
Determination of GCase and MCase activity.
Late exponential cells (carbon sources as indicated in the text) were collected by centrifugation at 4°C and washed with mineral salts medium without a carbon source. The pellet was resuspended in 1 ml of 25 mM Tris-HCl, pH 7.5, per g of cells. The supernatant obtained after the cells were ruptured by French press treatment (twice) and centrifuged (80,000 × g, 1 h) was used. GCase and MCase activities were measured at 340 nm (30°C) as described by Fall (13). The assay mixture contained the following (in 1 ml of 0.1 M Tris-HCl, pH 8.0): 10 mM MgCl2, 0.5 mM ATP, 10 mM KHCO3, 0.2 mM phosphoenolpyruvate, 0.1 mg ml−1 NADH, 0.5 mg ml−1 bovine serum albumin, 6.3 U ml−1 pyruvate kinase, 13.0 U ml−1 lactate dehydrogenase, and 10 to 20 μl of soluble crude extract. The assay was started by adding 150 μM (final concentration) methylcrotonyl-CoA or geranyl-CoA. One unit of activity is defined as 1 μmol of product formed per minute. Isolation of biotin-containing proteins from soluble cell extracts was done with immobilized monomeric avidin as described previously (20). Protein determination was performed by the Bradford procedure (4).
Protein identification by peptide mass fingerprinting.
For peptide mass fingerprinting, the protein spots of interest were excised from Coomassie blue-stained gels and subjected to in-gel digestion with trypsin as described previously (26). Peptides were extracted by sequential addition of 12 μl water and 10 μl 0.1% (vol/vol) trifluoroacetic acid in 30% (vol/vol) acetonitrile. The resulting peptide solution (0.5 μl) was mixed on a stainless steel sample plate with 0.5 μl of a saturated cyano-4-hydroxy-trans-cinnamic acid solution in 50% (vol/vol) acetonitrile and 0.1% (vol/vol) trifluoroacetic acid. Samples were analyzed manually with an Applied Biosystems (Weiterstadt, Germany) Voyager STR matrix-assisted laser desorption ionization-time of flight mass spectrometer in the positive reflector mode at 20 kV and 63% grid voltage, and the delay time was set at 125 ns. External calibration was performed using calibration mixtures 1 and 2 of the Sequazyme peptide mass standard kit. Data analysis was performed using Voyager Control Panel 5.0 and Voyager Data Explorer 3.5 software. The generated mass lists were used to search a local digestion product database of 5,567 P. aeruginosa PAO1 proteins (34) using ProteinProspector MS-Fit (8) available at http://prospector.ucsf.edu/.
RESULTS
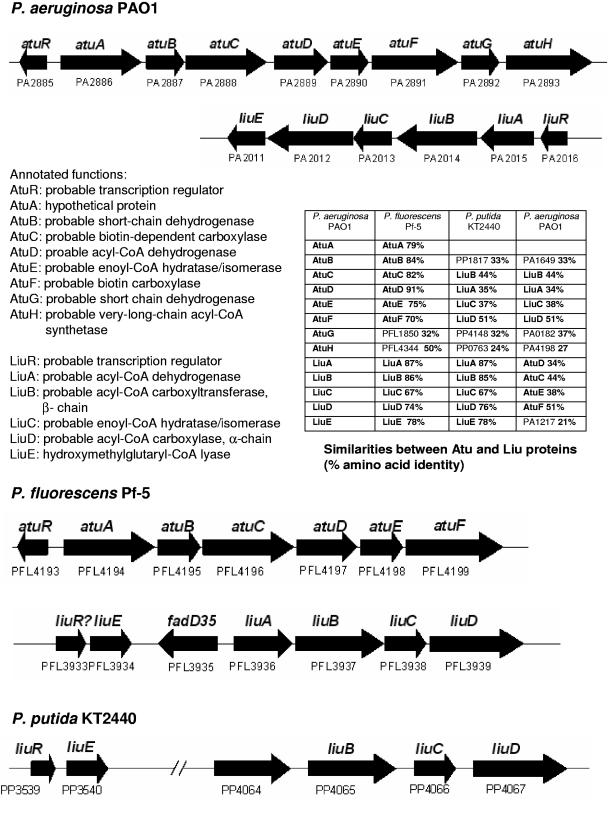
Screening of >8,000 mini-Tn5-induced mutants for defects in acyclic terpene utilization revealed one mutant (22B-1-1) that was unable to utilize any of the four monoterpenes tested as the sole source of carbon and energy (citronellol, citronellate, geraniol, and geranylate). Growth of this mutant on mineral salts medium (containing leucine [0.2%], isovalerate [0.2%], acetate [0.3%], succinate [0.6%], or glucose [0.5%]) or NB medium was not impaired and indicated that the mutation was specific for metabolism of acyclic terpenes. The site of transposon insertion was identified at position 3243626 of the P. aeruginosa genome. This position is located in the coding region of open reading frame PA2890 (atuE) that is part of a gene cluster containing eight genes with open reading frames PA2886 to PA2893 (atuABCDEFGH) and that had been annotated as a putative enoyl-CoA hydratase/isomerase gene (Fig. 1 and 2). Comparison of the putative gene products of the atuABCDEFGH gene cluster with the database revealed high amino acid similarities (70 to 96%) to gene products of similar gene clusters of genome-sequenced Pseudomonas fluorescens Pf-5 and to gene products of a recently cloned atu gene cluster of P. citronellolis (16), but not to gene products of other sequenced pseudomonads (Pseudomonas putida) (Fig. 2). The similarity values of the Atu proteins to respective Liu proteins of the liuABCDE gene cluster of P. aeruginosa and related pseudomonads were between 34 and 51%. Finding amino acid similarities between Atu and Liu proteins is not surprising, because both pathways have several similar reactions (Fig. 1). As shown in Table 2, all three bacterial species with an atu gene cluster (P. aeruginosa, P. citronellolis, and P. fluorescens Pf-5) were able to utilize acyclic terpenes, but species without an atu gene cluster (P. putida) were not. All mentioned species have a liu gene cluster and are able to utilize leucine and isovalerate as the sole source of carbon and energy. This finding is in agreement with the assumption that the atuABCDEFGH gene cluster encodes proteins of the acyclic terpene-utilizing pathway (Fig. 1) and that the liu gene cluster is necessary for leucine/isovalerate utilization (Fig. 1).
FIG. 2.
Comparison of the relative positions of atu and liu genes in P. aeruginosa PAO1, P. fluorescens Pf-5, and P. putida KT2440. Annotated functions and amino acid similarity values of P. aeruginosa atu and liu gene products are also given (percent identity).
TABLE 2.
Phenotypes of selected wild-type and mutant strains
| Strainb | Growth ona:
|
||||||
|---|---|---|---|---|---|---|---|
| CL | GL | CA | GA | Leu | IsoV | Succ | |
| PAO1 | ++ | + | ++ | ++ | + | ++ | ++ |
| PAO1 22B/1/1 (atuE::mini-Tn5) | − | − | − | −' | + | ++ | ++ |
| PAO1 ins-atuA | −* | − | −* | − | + | ++ | ++ |
| PAO1 ins-atuB | −* | − | −* | − | + | ++ | ++ |
| PAO1 ins-atuC | − | − | − | − | + | ++ | ++ |
| PAO1 ins-atuD | −* | −* | −* | − | + | ++ | ++ |
| PAO1 ins-atuE | + | + | + | + | + | ++ | ++ |
| PAO1 ins-atuF | −* | − | −* | − | + | ++ | ++ |
| PAO1 ins-atuG | ++ | + | ++ | ++ | + | ++ | ++ |
| PAO1 ins-atuH | ++ | + | ++ | ++ | + | ++ | ++ |
| PAO1 ins-liuC | −* | − | − | − | − | −* | ++ |
| PAO1 ins-liuD | −* | − | −* | − | − | −* | ++ |
| PAO1 ins-atuR | ++ | + | ++ | ++ | + | ++ | ++ |
| P. citronellolis | ++ | + | ++ | ++ | ++ | ++ | ++ |
| P. fluorescens Pf-5 | ++ | − | ++ | − | ++ | + | ++ |
| P. putida KT2440 | − | − | − | − | ++ | ++ | ++ |
Bacteria were incubated on solid mineral salts medium with the following carbon sources at 30°C for 4 days: citronellol (CL), geraniol (GL), citronellate (CA), geranylate (GA), leucine (Leu), isovalerate (IsoV), and succinate (Succ). Good growth (++), growth (+), poor growth (±), and no growth (−) is indicated. An asterisk indicates that single colonies came up spontaneously during incubation and probably represent suppression mutants.
Strains with insertion mutations in genes are indicated by “ins” preceding the gene.
2D gel electrophoresis.
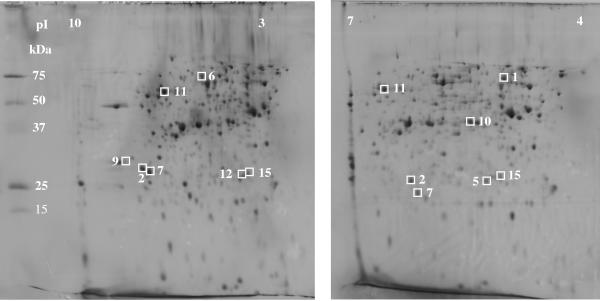
To investigate which proteins of the two pathways (Atu versus Liu) were specifically induced during growth on acyclic terpenes and on saturated methyl-branched compounds, 2D gel electrophoresis was performed. We compared cell extracts of succinate-grown cells (control) with citronellol-, citronellate-, and isovalerate-grown cells. Each sample was separated repeatedly in a wide (pH 3 to 10) and narrow (pH 4 to 7) pH range during isoelectric focusing to obtain maximal resolution. Several spots that were present only in citronellate-, citronellol-, and/or isovalerate-grown cells but that were absent or were present in reduced intensity in succinate-grown cells were identified (examples shown in Fig. 3; for details, see Table 3). The respective spots were isolated after Coomassie blue staining and subjected to trypsin peptide analysis, and the resulting masses were compared with theoretical values deduced from the P. aeruginosa genome database. Table 3 shows the results: five of the eight potential Atu proteins (AtuA, AtuB, AtuE, AtuF, and AtuG) were identified only in citronellol- and citronellate-grown cells and were not present in isovalerate- or succinate-grown cells. Interestingly, one other spot corresponding to the gene product of open reading frame PA1342, which is part of a four-gene cluster (PA1339 to PA1342) with putative function as an ABC transporter, was also specifically expressed in cells grown on acyclic terpenes. This protein might be involved in uptake of acyclic terpenes. Three of the six predicted liu gene products (LiuA, LiuB, and LiuC) and several other proteins were identified in terpene-grown and isovalerate-grown cells (Table 3) but were absent or present at a significantly lower spot intensity in succinate-grown cells. The latter include proteins with putative functions related to the tricarboxylic acid cycle (isocitrate lyase and glutamate synthase) and to C2 carbon metabolism (acetyl-CoA-acetyltransferase and acetyl-CoA-acetoacetate transferase). This finding is not surprising, as both isovalerate and acyclic terpenes are metabolized to acetate, acetyl-CoA, and acetoacetate and enter the tricarboxylic acid cycle and the glyoxylate shunt.
FIG. 3.
Two-dimensional gel electrophoresis of soluble P. aeruginosa proteins after growth on citronellol. The positions of marker proteins and the direction of isoelectric focusing are indicated. Two different pH gradients (pH 3 to 10 NL [left] and pH 4 to 7 [right]) were used. Proteins spots not visible in succinate-grown cells are indicated. The numbering of the spots is the same as in Table 3.
TABLE 3.
Proteins, genes, and assigned functions in catabolism of acyclic terpenes and isovaleratea
| Spot no. | PA no. | Proposed name | No. of peptides that matched | % Coverage | Expression of proteins in cells grown onb:
|
Proposed function | |||
|---|---|---|---|---|---|---|---|---|---|
| Succ | IsoV | CL | CA | ||||||
| 1 | 2886 | AtuA | 12 | 32 | − | − | + | + | 3-Hydroxy-3-isohexenylglutaryl-CoA:acetate-lyase? |
| 2 | 2887 | AtuB | 6 | 32 | − | − | + | + | Citronellol/citronellal dehydrogenase |
| 3 | 2888 | AtuC | WB | − | − | + | + | GCase, β-subunit | |
| 4 | 2889 | AtuD | ND | Citronellyl-CoA dehydrogenase | |||||
| 5 | 2890 | AtuE | 8 | 34 | − | − | + | + | Isohexenylglutaconyl-CoA hydratase |
| 6 | 2891 | AtuF | 20/WB | 44 | − | − | + | + | GCase, α-subunit (biotin containing) |
| 7 | 2892 | AtuG | 20 | 68 | − | − | + | + | Citronellol/citronellal dehydrogenase |
| 8 | 2893 | AtuH | ND | Citronellyl-CoA synthetase | |||||
| 9 | 1342 | 15 | 42 | − | − | + | + | Periplasmic binding protein (ABC transporter) | |
| 10 | 2015 | LiuA | 15 | 37 | − | + | + | + | Isovaleryl-CoA Dehydrogenase |
| 11 | 2014 | LiuB | 19/WB | 45 | − | + | + | + | MCase, β-subunit |
| 12 | 2013 | LiuC | 9 | 28 | − | + | + | + | Methylglutaconyl-CoA hydratase |
| 13 | 2012 | LiuD | WB | − | + | + | + | MCase, α-subunit (biotin containing) | |
| 14 | 2011 | LiuE | ND | 3-Hydroxymethylglutaryl-CoA lyase | |||||
| 15 | 1999 | AtoD | 6 | 41 | − | + | + | + | Acetyl-CoA:acetoacetate-CoA transferase |
| 16 | 2001 | AtoB | 15 | 75 | ± | + | + | + | Acetyl-CoA acetyltransferase |
| 17 | 2634 | Icl | 16 | 33 | ± | + | + | + | Isocitrate lyase |
| 18 | 5035 | GltD | 16 | 40 | ± | ± | ± | ± | Glutamate synthase |
Hypothetical proteins are given in normal roman type. Proteins with experimentally verified function are given in bold type. Expression of proteins was shown in cells grown on succinate (Succ), isovalerte (IsoV), citronellol (CL), and citronellate (CA) by 2D gel electrophoresis or by Western blotting (WB) and trypsin peptide analysis or not determined (ND).
Symbols: −, no expression; +, significant expression; ±, poor or uncertain expression.
Insertion mutations in atu and liu gene cluster.
To investigate the importance of the individual genes of the atu and liu gene clusters, all eight atu genes and two of the liu genes were mutated by insertion mutagenesis (Tables 1 and 2). pKnockout-G was used as an insertion vector throughout. This vector has a constitutive (in P. aeruginosa) lac promoter, and care was taken that this lac promoter was oriented colinearly to the direction of transcription of the target gene. By doing this, we avoided downstream effects of the insertion. Other methods of gene inactivation, such as site-directed deletion of a gene, led to a downstream effect, as revealed by the inability of an atuA deletion mutant to express biotin-containing carboxylase subunit (AtuF) (data not shown). In contrast to this, insertion of pKnockout in atuA resulted in constitutive expression of AtuF and confirmed the functionality of the pKnockout-encoded lac promoter. Table 2 shows the phenotypes of the respective insertion mutants in comparison to the wild type. Insertion in atuA, atuB, atuC, atuD, or atuF resulted in inability of the strains to utilize acyclic terpenes, while growth on leucine, isovalerate, and unrelated carbon sources was not impaired. Insertions in atuG or atuH had no detectable effects. Mutants with insertion in liuC or liuD could not utilize either acyclic terpenes or leucine or isovalerate. An interesting observation was made for two different atuE insertion mutants: while the mini-Tn5-induced atuE mutant was completely unable to utilize acyclic terpenes, the pKnockout-derived atuE insertion mutant showed reduced but nevertheless significant growth on acyclic terpenes (Table 2). Apparently, the insertions had polar downstream effects, but the constitutively expressed lac promoter compensated for these effects in the pKnockout-derived mutant. The result indicated that AtuE (putative isohexenyl-glutaconyl-CoA hydratase [see below]) is important but is obviously not essential for growth on acyclic terpenes and can be partially replaced by other (hydratase) isoenzymes. This conclusion is different from results recently reported on the effect of atuE insertion (1).
MCase and GCase activities.
Comparison of the amino acid sequences of AtuC/AtuF and LiuB/LiuD showed high similarities to the two subunits of biotin-containing carboxylases. Since the Atu and Liu pathway each contain one carboxylase step (GCase and MCase) and since the AtuC and AtuF insertion mutants were unable to grow on acyclic terpenes but still utilized isovalerate as a carbon source, we assumed that AtuC/AtuF and LiuB/LiuD represented GCase and MCase subunits, respectively. To find direct experimental evidence for this assumption, we assayed GCase and MCase activity. P. aeruginosa PAO1 wild type and two insertion mutants (atuF and liuD) were grown on glucose, citronellate, and isovalerate. Isovalerate- and citronellate-grown cultures (each 0.1%) additionally contained 0.075% glucose in order to enable growth of the mutants. None of the strains contained significant MCase or GCase activity after growth on glucose (<3 mU/mg). Soluble cell extracts of citronellate-grown wild-type cells contained 33 mU/mg and 15 mU/mg MCase and GCase activity, respectively, indicating that the Atu and Liu pathways were both operating (Table 4). MCase activity (45 mU/mg) but no detectable GCase activity was determined for isovalerate-grown wild-type cells, confirming that the Liu pathway is operating in isovalerate-grown cells, while GCase, a key enzyme of the Atu pathway, is not expressed. Mutant liuD (putative MCase subunit) contained no or very low MCase or GCase activity after growth in the presence of isovalerate but showed significant GCase activity (19 mU/mg) in citronellate-exposed cells. GCase activity in the atuF mutant (putative GCase subunit) was not detected, but high MCase specific activity was determined in isovalerate (39 mU/mg)- and citronellate-grown cells (17 mU). These results confirm that liuD and atuF code for MCase and GCase, respectively.
TABLE 4.
Specific GCase and MCase activities of soluble cell extractsa
| P. aeruginosa strainb and carbon source | Sp act (mU/mg)
|
|
|---|---|---|
| GCase | MCase | |
| WT, glucose | ≤3 | ≤3 |
| WT, citronellate | 15 | 33 |
| WT, isovalerate | ≤3 | 45 |
| ins-liuD, glucose | ≤3 | ≤3 |
| ins-liuD, citronellate | 19 | ≤3 |
| ins-liuD, isovalerate | 3.3 | ≤3 |
| ins-atuF, glucose | ≤3 | ≤3 |
| ins-atuF, citronellate | ≤3 | 17 |
| ins-atuF, isovalerate | ≤3 | 39 |
Because of the presence of NADH oxidase activities in P. aeruginosa, significant carboxylase activities could be determined only above 3 mU/mg.
Wild-type strain (WT) and strains with insertion mutations (indicated by “ins” before a gene) were used.
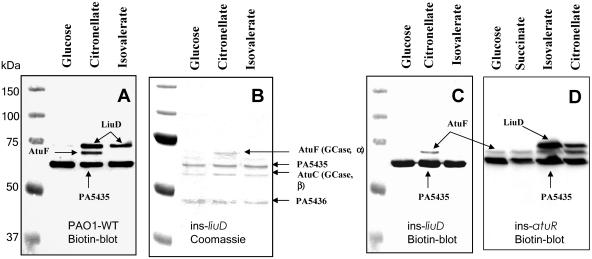
Isolation of carboxylases and Western blot analysis.
Biotin-containing proteins of wild-type cells and of the liuD insertion mutant were purified by avidin affinity chromatography and separated into subunits by reducing SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 4A to C). The identities of isolated subunits with LiuB/LiuD and AtuC/AtuF in the wild type were shown by trypsin peptide mass spectrometry (data not shown). As expected from MCase and GCase activities (Table 4), LiuD (MCase biotin subunit) was present in both isovalerate- and citronellol-grown wild-type cells, while AtuF (GCase biotin subunit) was detected only in citronellol-grown cells. No LiuD but significant AtuF signal appeared in the liuD mutant. In addition, a constitutively expressed carboxylase (PA5435 and PA5436 gene products) was also found, which is in agreement with earlier findings (1, 10, 20).
FIG. 4.
Western blot analysis (A, C, and D) of P. aeruginosa PAO1 proteins after SDS-PAGE for biotin-containing proteins. Soluble cell extracts after avidin purification were separated on 10% SDS-polyacrylamide gels and stained with Coomassie brilliant blue (B) or transferred to polyvinylidene difluoride membranes and screened for biotin-containing proteins (A and C). Gene products identified by trypsin peptide analysis of isolated protein bands from panel B are indicated. Western blot analysis of P. aeruginosa PAO1 ins-atuR proteins after SDS-PAGE for biotin-containing proteins (D). Note the constitutive expression of AtuF. PAO1-WT, PAO1 (wild type); ins-liuD, insertion liuD mutant; ins-atuR, insertion atuR mutant.
Identification of atuR as a repressor of the atu gene cluster.
Regulation of atu gene cluster expression has not yet been investigated. Upstream of the atu gene cluster and in the direction opposite of transcription, an open reading frame (PA2885) with similarity to transcriptional regulators is located (Fig. 2). An insertion mutant in PA2885 was constructed. The direction of the pKnockout-encoded lac promoter was oriented in the same direction as that of PA2885, i.e., opposite that of the atu gene cluster so that constitutive expression of the atu gene cluster from the pKnockout-encoded lac promoter was avoided. The insertion mutant obtained had no detectable effect on growth on any of the substrates tested (Table 2). To test whether the PA2885 gene product could act as a repressor of the atu gene cluster, we performed Western blotting for biotin proteins of soluble cell extracts after growth on unrelated (glucose and succinate) and related (isovalerate and citronellate) compounds and used detection of biotin-containing subunit AtuF as a marker for atu gene cluster expression. As shown in Fig. 4D, AtuF was constitutively expressed in the PA2885 insertion mutant on all tested substrates including succinate and glucose, while the wild type expressed AtuF only in cells grown on acyclic terpenes (Fig. 4A). LiuD (MCase biotin-containing subunit) was still regulated as in the wild type and was expressed only in isovalerate- and citronellate-grown cells. Apparently, PA2885 is necessary to repress atu gene expression on unrelated carbon sources and does not influence expression of the liu genes. We assume that PA2885 represents a repressor of atu gene cluster expression and named it atuR.
DISCUSSION
In this study we identified six out of eight potential Atu proteins to be specifically expressed in cells that had been grown on linear terpenes, such as citronellol or citronellate, by 2D gel electrophoresis and/or trypsin peptide analysis of isolated biotin proteins (Fig. 3 and 4). AtuD and AtuH were the only gene products of the gene cluster that were not yet identified. We assume that AtuD and AtuH were also expressed but possibly may have been overlaid by a constitutively expressed protein in the 2D picture of hundreds of spots. There was generally no visible differences in the proteome profiles of citronellol- and citronellate-grown cells; the above-mentioned Atu proteins were present in both citronellol-grown cells and citronellate-grown cells but not in isovalerate- or succinate-grown cells (Table 3). These results confirmed that the Atu proteins are specific for utilization of acyclic terpenes and revealed that genes responsible for proteins that oxidize citronellol to citronellate and genes coding for catabolic steps downstream of citronellate are simultaneously expressed. A differentiation in upper and lower pathway (1, 10) appears artificially. Annotation data suggest that AtuB and/or AtuG (both probably short-chain dehydrogenases) could encode citronellol dehydrogenase and citronellal dehydrogenase and AtuH (probable acyl-CoA synthetase) citronellyl-CoA synthetase. AtuD (probable acyl-CoA dehydrogenase) could catalyze the oxidation of citronellyl-CoA to geranyl-CoA (Fig. 1). The finding that insertions in atuG or atuH or in atuE have no or only poor effects does not exclude the participation of the gene products in the Atu pathway. Since the P. aeruginosa genome contains many genes encoding proteins with similarities to short-chain dehydrogenases, acyl-CoA synthetases, and hydratases, inactivation of atuG, atuH, and atuE may be compensated for by expression of isogenes resulting in suppression of a detectable phenotype. Specific expression on acyclic terpenes was shown in the proteomics experiment at least for AtuE and AtuG (Table 3).
Since atuC/atuF and atuE code for the two subunits of GCase and for isohexenylglutaconyl-CoA hydratase (see below), atuA is the only remaining gene of the gene cluster whose function remains unknown. AtuA (unknown hypothetical protein) was specifically expressed only in citronellol- and citronellate-grown cells. AtuA could be involved in transport of the substrates into the cell. However, a gene outside of the atu gene cluster (PA1342, putative ABC transporter together with adjacent genes, PA1339 to PA1342 [Table 3]) is more likely to be involved in transport because the PA1342 gene product was also specifically expressed in citronellol- and citronellate-grown cells but was not visible in 2D gels of isovalerate-grown cells. The only remaining function of the Atu pathway (Fig. 1) is 3-hydroxy-3-isohexenylglutaryl-CoA:acetate-lyase (HHG lyase), and we speculate that atuA could encode the missing lyase. Insertion mutagenesis of atuA showed that AtuA is essential for a functional Atu pathway. There is no significant similarity of the AtuA amino acid sequence to any other known protein, not even to the hydroxymethylglutaryl-CoA lyase (HMG lyase, LiuE/MvaB). This may not be a surprise, because a HHG lyase gene has not been described so far and HHG lyase differs from HMG lyase in the reaction products (2, 23, 24, 27). While the former splits off an acetate molecule, the latter cleaves another carbon bond, releasing acetyl-CoA instead of acetate (29, 30). This difference might be caused by different structures of the two lyase proteins. Substrate specificity analysis of partially purified HHG lyase had shown that it cannot fulfill the function of HMG lyase in vitro (29, 30). It is therefore unlikely that HMG-CoA lyase (LiuE [see below]) has a dual function in the Liu and Atu pathway and can catalyze both lyase reactions as proposed by others (1).
Expression of four out of the five Liu proteins (LiuA, LiuB, LiuC, and LiuD) of the liuABCDE gene cluster was identified by 2D gel electrophoresis in cells that been grown on isovalerate, citronellol, and/or citronellate (Fig. 3 and 4 and Table 3). This indicated that the Liu proteins are essential for isovalerate utilization and are indirectly necessary for utilization of acyclic terpenes, because the Atu pathway concurs the Liu pathway at methylcrotonyl-CoA (Fig. 1). LiuB and LiuD have been definitively shown to encode the two subunits of MCase (Fig. 4). Comparison with the database shows that LiuC and AtuE both have strong similarities to many enoyl-CoA hydratases. Since the Atu and Liu pathway each contain one hydratase step, namely, isohexenylglutaconyl-CoA-hydratase and methylglutaconyl-CoA-hydratase (Fig. 1), and were specifically expressed during growth on citronellate (AtuE and LiuC) and on leucine/isovalerate (only LiuC), it is very likely that atuE and liuD encode isohexenylglutaconyl-CoA-hydratase and methylglutaconyl-CoA-hydratase, respectively.
LiuE is the only protein of the Liu gene cluster whose expression could not be shown directly in 2D gels. The LiuE amino acid sequence shows high similarities to HMG lyase of Pseudomonas mevaloni and other species (2). For Rhodospirillum rubrum, the involvement of HMG lyase in metabolism of leucine has been described elsewhere (3). On the basis of these findings, we assume that liuE encodes HMG lyase in P. aeruginosa. Only bacteria that have both the atu and liu gene clusters, such as P. aeruginosa, P. fluorescens Pf-5, and P. citronellolis, are able to utilize citronellol. We predict that the citronellol- and isovalerate-degrading species P. mendocina will also have functional atu and liu gene clusters. Inspection of the database revealed that gene clusters highly similar to the atu gene cluster of P. aeruginosa are present in Marinobacter aquaeolei VT8 and in Hahella chejuensis KCTC 2396. The functions of these hypothetical proteins remain to be identified.
Acknowledgments
We thank R. Schäfer for assistance during transposon mutagenesis and S. Drescher and M. Panas for assistance during 2D gel electrophoresis. We also thank A. Rooney for providing P. fluorescens Pf-5.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to D.J.
REFERENCES
- 1.Aguilar, J. A., A. N. Zavala, C. Diaz-Perez, C. Cervantes, A. L. Diaz-Perez, and J. Campos-Garcia. 2006. The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:2070-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D. H., and V. W. Rodwell. 1989. Nucleotide sequence and expression in Escherichia coli of the 3-hydroxy-3-methylglutaryl coenzyme A lyase gene of Pseudomonas mevalonii. J. Bacteriol. 171:6468-6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltscheffsky, M., M. Brosche, T. Hultman, L. Lundvik, P. Nyren, Y. Sakai-Nore, A. Severin, and A. Strid. 1997. A 3-hydroxy-3-methylglutaryl-CoA lyase gene in the photosynthetic bacterium Rhodospirillum rubrum. Biochim. Biophys. Acta 1337:113-122. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Burke, Y. D., M. J. Stark, S. L. Roach, S. E. Sen, and P. L. Crowell. 1997. Inhibition of pancreatic cancer growth by the dietary isoprenoids farnesol and geraniol. Lipids 32:151-156. [DOI] [PubMed] [Google Scholar]
- 6.Cantwell, S. G., E. P. Lau, D. S. Watt, and R. R. Fall. 1978. Biodegradation of acyclic isoprenoids by Pseudomonas species. J. Bacteriol. 135:324-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carnesecchi, S., R. Bras-Goncalves, A. Bradaia, M. Zeisel, F. Gosse, M. F. Poupon, and F. Raul. 2004. Geraniol, a component of plant essential oils, modulates DNA synthesis and potentiates 5-fluorouracil efficacy on human colon tumor xenografts. Cancer Lett. 215:53-59. [DOI] [PubMed] [Google Scholar]
- 8.Clauser, K. R., P. Baker, and A. L. Burlingame. 1999. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871-2882. [DOI] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaz-Perez, A. L., A. N. Zavala-Hernandez, C. Cervantes, and J. Campos-Garcia. 2004. The gnyRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 70:5102-5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, R. E., D. Lau, A. El-Sohemy, and M. C. Archer. 2004. Geraniol and beta-ionone inhibit proliferation, cell cycle progression, and cyclin-dependent kinase 2 activity in MCF-7 breast cancer cells independent of effects on HMG-CoA reductase activity. Biochem. Pharmacol. 68:1739-1747. [DOI] [PubMed] [Google Scholar]
- 12.Fall, R. R. 1981. 3-Methylcrotonyl-CoA and geranyl-CoA carboxylases from Pseudomonas citronellolis. Methods Enzymol. 71(Part C):791-799. [DOI] [PubMed] [Google Scholar]
- 13.Fall, R. R. 1976. Stabilization of an acetyl-coenzyme A carboxylase complex from Pseudomonas citronellolis. Biochim. Biophys. Acta 450:475-480. [DOI] [PubMed] [Google Scholar]
- 14.Fall, R. R., J. L. Brown, and T. L. Schaeffer. 1979. Enzyme recruitment allows the biodegradation of recalcitrant branched hydrocarbons by Pseudomonas citronellolis. Appl. Environ. Microbiol. 38:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fall, R. R., and M. L. Hector. 1977. Acyl-coenzyme A carboxylases. Homologous 3-methylcrotonyl-CoA and geranyl-CoA carboxylases from Pseudomonas citronellolis. Biochemistry 16:4000-4005. [DOI] [PubMed] [Google Scholar]
- 16.Förster-Fromme, K., and D. Jendrossek. 2005. Unpublished results.
- 17.Guan, X., T. Diez, T. K. Prasad, B. J. Nikolau, and E. S. Wurtele. 1999. Geranoyl-CoA carboxylase: a novel biotin-containing enzyme in plants. Arch. Biochem. Biophys. 362:12-21. [DOI] [PubMed] [Google Scholar]
- 18.Hector, M. L., and R. R. Fall. 1976. Multiple acyl-coenzyme A carboxylases in Pseudomonas citronellolis. Biochemistry 15:3465-3472. [DOI] [PubMed] [Google Scholar]
- 19.Hierro, I., A. Valero, P. Perez, P. Gonzalez, M. M. Cabo, M. P. Montilla, and M. C. Navarro. 2004. Action of different monoterpenic compounds against Anisakis simplex s.l. L3 larvae. Phytomedicine 11:77-82. [DOI] [PubMed] [Google Scholar]
- 20.Höschle, B., V. Gnau, and D. Jendrossek. 2005. Methylcrotonyl-CoA carboxylase and geranyl-CoA carboxylase are involved in leucine/isovalerate utilisation (Liu) and in acyclic terpenes utilisation (Atu) and are encoded by liuB/liuD and atuC/atuF in Pseudomonas aeruginosa. Microbiology 151:3649-3656. [DOI] [PubMed] [Google Scholar]
- 21.Höschle, B., and D. Jendrossek. 2005. Utilization of geraniol is dependent on molybdenum in Pseudomonas aeruginosa: evidence for different metabolic routes for oxidation of geraniol and citronellol. Microbiology 151:2277-2283. [DOI] [PubMed] [Google Scholar]
- 22.Izumi, S., O. Takashima, and T. Hirata. 1999. Geraniol is a potent inducer of apoptosis-like cell death in the cultured shoot primordia of Matricaria chamomilla. Biochem. Biophys. Res. Commun. 259:519-522. [DOI] [PubMed] [Google Scholar]
- 23.Miziorko, H. M., and C. Narasimhan. 2000. Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA lyase. Methods Enzymol. 324:139-149. [DOI] [PubMed] [Google Scholar]
- 24.Narasimhan, C., and H. M. Miziorko. 1992. Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl-CoA lyase: characterization of the isolated recombinant protein and investigation of the enzyme's cation requirements. Biochemistry 31:11224-11230. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. Myers, D. V. Mavrodi, R. T. DeBoy, R. Seshadri, Q. Ren, R. Madupu, R. J. Dodson, A. S. Durkin, L. M. Brinkac, S. C. Daugherty, S. A. Sullivan, M. J. Rosovitz, M. L. Gwinn, L. Zhou, D. J. Schneider, S. W. Cartinhour, W. C. Nelson, J. Weidman, K. Watkins, K. Tran, H. Khouri, E. A. Pierson, L. S. Pierson III, L. S. Thomashow, and J. E. Loper. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23:873-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaffer, S., B. Weil, V. D. Nguyen, G. Dongmann, K. Gunther, M. Nickolaus, T. Hermann, and M. Bott. 2001. A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22:4404-4422. [DOI] [PubMed] [Google Scholar]
- 27.Scher, D. S., and V. W. Rodwell. 1989. 3-Hydroxy-3-methylglutaryl coenzyme A lyase from Pseudomonas mevalonii. Biochim. Biophys. Acta 1003:321-326. [DOI] [PubMed] [Google Scholar]
- 28.Seubert, W. 1960. Degradation of isoprenoid compounds by microorganisms. I. Isolation and characterization of an isoprenoid-degrading bacterium, Pseudomonas citronellolis n. sp. J. Bacteriol. 79:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seubert, W., and E. Fass. 1964. Studies on the bacterial degradation of isoprenoids. IV. The purification and properties of beta-isohexenylglutaconyl-CoA-hydratase and beta-hydroxy-beta-isohexenylglutaryl-CoA-lyase. Biochem. Z. 341:23-34. [PubMed] [Google Scholar]
- 30.Seubert, W., and E. Fass. 1964. Studies on the bacterial degradation of isoprenoids. V. The mechanism of isoprenoid degradation. Biochem. Z. 341:35-44. [PubMed] [Google Scholar]
- 31.Seubert, W., E. Fass, and U. Remberger. 1963. Studies on the bacterial degradation of isoprenoids. III. Purification and properties of geranyl carboxylase. Biochem. Z. 338:265-275. [PubMed] [Google Scholar]
- 32.Seubert, W., and U. Remberger. 1963. Studies on the bacterial degradation of isoprenoids. II. The role of carbon dioxide. Biochem. Z. 338:245-264. [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Windgassen, M., A. Urban, and K. E. Jaeger. 2000. Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193:201-205. [DOI] [PubMed] [Google Scholar]
- 36.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]