Abstract
Extraintestinal growth of fecal bacteria can impair accurate assessment of watershed health. Anaerobic fecal bacteria belonging to the order Bacteroidales are attractive candidates for fecal source tracking because they have host-specific distributions and do not grow well in the presence of high oxygen concentrations. Growth of general and human-specific fecal Bacteroidales marker organisms in environmental samples (sewage) and persistence of the corresponding genetic markers were investigated using bromodeoxyuridine (BrdU) DNA labeling and immunocapture, followed by PCR detection. Background amplification of unlabeled controls occasionally occurred when a high number of PCR cycles was used. By using fluorescent detection of PCR products obtained after 15 cycles, which was determined to be quantitative, we enriched for BrdU-labeled DNA and did not detect unlabeled DNA. By using pure cultures of Bacteroides vulgatus, the ability of Bacteroidales bacteria to take up and incorporate BrdU into nascent DNA was confirmed. Fecal Bacteroidales organisms took up and incorporated BrdU into DNA during growth. In sewage incubated aerobically at the in situ temperature, Bacteroidales genetic marker sequences persisted for at least 24 h and Bacteroidales fecal bacteria grew for up to 24 h as well. Detection by PCR using a low, quantitative cycle number decreased the sensitivity of the assay such that we were unable to detect fecal Bacteroidales human-specific marker sequences in unlabeled or BrdU-labeled fractions, even when fluorescent detection was used. Using 30 PCR cycles with unlabeled fractions, human-specific Bacteroidales sequences were detected, and they persisted for up to 24 h in sewage. These data support the utility of BrdU labeling and immunocapture followed by length heterogeneity PCR or fluorescent detection using low numbers of PCR cycles. However, this method may not be sensitive enough to identify cells that are present at low densities in aquatic environments.
Fecal pollution from failing septic systems, urban and agricultural runoff, and wild animals affects human and environmental health (26) and causes economic losses (13) due to closure of shellfish beds and recreational waters. An ability to accurately identify nonpoint sources of fecal pollution is vital for effective remediation of such pollution. Most United States state agencies monitor water quality and estimate human health risk in accordance with the U.S. Environmental Protection Agency policies (45). The methods used rely on cultivation and enumeration of bacterial indicators, such as Escherichia coli and fecal enterococci. Implementation of these detection methods has substantially reduced the incidence of waterborne disease, but the methods have failed to discriminate between sources of fecal pollution and require laborious and time-consuming laboratory manipulations. In addition, once in the environment, fecal bacteria may rapidly lose the ability to be cultured by traditional methods but retain metabolic functions (28) or pathogenicity (33, 35).
To accurately estimate the human health risk associated with exposure to fecal pathogens, an indicator should not proliferate in the environment, should persist as long as pathogens, and should be present at the same time at concentrations proportional to the concentrations of pathogens (42). E. coli and enterococci are not well correlated with pathogenic Salmonella spp. (25), Campylobacter spp. (6, 19, 25, 27), Cryptosporidium spp. (6, 19, 25, 27), human enteroviruses (19, 25), including adenoviruses (29), or coliphages (20). In addition, E. coli and enterococci can survive and persist in natural environments such as freshwater lakes and streams, algal wrack, beach sand, and tropical soils (8, 17, 34, 46).
We previously proposed PCR-based detection of 16S rRNA gene markers for uncultivated anaerobic fecal bacteria in the order Bacteroidales as a rapid method for diagnosing fecal pollution and discriminating its source(s) (4, 12, 14, 38). Two PCR primers, HF134 and HF183, specifically detect human feces (5). Bacteroidales genes can be used as fecal indicators in a real-time quantitative PCR assay (11), and recently a human-specific quantitative Bacteroidales PCR assay was developed (37) based on the HF183 primer (5). These assays, combined with quantitative assays for fecal pathogens, have the potential to determine fecal pollution sources and support inferences about associated health risks. Several studies have suggested that Bacteroidales genotypes exhibit geographical stability (14, 37), as well as host specificity (4, 12, 37), and may be appropriate for quantitative source tracking on a global scale.
Use of this method for quantitative source tracking requires an understanding of both the persistence and the possible proliferation of these bacteria in the environment. Because Bacteroidales bacteria are obligate anaerobes, their survival in the extraintestinal environment is thought to be limited (21, 22). Reservoirs for Bacteroidales bacteria are believed to be restricted to the body cavities of animals; no strains adapted to aquatic environments are currently known.
We used a cultivation-independent bromodeoxyuridine (BrdU) DNA labeling technique to measure the persistence and proliferation of these bacteria. Under thymidine (TdR) auxotrophy conditions, cells pulse-labeled with BrdU, a thymidine base analog, take up and incorporate BrdU into newly synthesized DNA in place of thymidine. Uptake of BrdU and subsequent incorporation of this molecule into DNA can therefore be used to identify actively growing bacteria (32, 39). Separation of BrdU-labeled DNA using immunocapture, followed by PCR of ribosomal genes in the labeled fraction, allows species-level identification of growing bacteria in mixed communities (2, 7, 44), bypassing traditional culture methods. Following immunocapture, successful PCR amplification of unlabeled (supernatant) DNA demonstrates persistence of targeted cells or DNAs, whereas PCR amplification of labeled (immunocaptured) DNA demonstrates growth. The BrdU labeling and immunocapture method has reportedly been used successfully to identify metabolically active populations of marine bacteria (44), soil fauna (7), and arbuscular mycorrhizae (2).
Here we describe the results of a study in which we used the BrdU immunocapture method to monitor the growth of Bacteroidales organisms in sewage. Because strains containing the human-specific Bacteroidales markers have not been cultivated, we first tested the ability of Bacteroides vulgatus, the closest cultivated phylogenetic relative, to take up and incorporate BrdU into newly synthesized DNA. To circumvent problems arising from background amplification of unlabeled control DNA, we developed a fluorescent detection method employing quantitative length heterogeneity PCR (LH-PCR) (41). LH-PCR is a method that chromatographically separates fluorescently labeled PCR products by length with a DNA automated sequencer such that the relative fluorescence of each fragment is proportional to its abundance. Using this technique, Suzuki and colleagues (40, 41) showed that kinetic biases occur as the number of PCR cycles increases but that the PCR remains quantitative relative to the initial template concentration with a low number of cycles, which can be established empirically. We amplified ribosomal gene fragments from strains with different fragment lengths, which allowed us to establish quantitative PCR conditions (in which the proportion of fragments in PCR products was equal to the proportion of fragments in the template). We then graphically tested the efficiency of the BrdU immunocapture method. Finally, we pulse-labeled sewage influent for 24 h with BrdU to determine whether Bacteroidales fecal bacteria grew under environmental conditions and whether the BrdU method could be used to detect persistence and growth at environmentally relevant concentrations.
MATERIALS AND METHODS
Bacterial strains.
We obtained B. vulgatus ATCC 4245 from the laboratory of Abigail Salyers, University of Illinois, Champaign-Urbana. The control organism Fulvimarina pelagi HTCC 2506 (9) was obtained from Jang-Cheon Cho at Oregon State University.
Sewage influent.
Sewage influent was collected from the Corvallis Wastewater Reclamation Plant, Corvallis, OR.
BrdU labeling.
Sewage influent and/or pure cultures of B. vulgatus and E. coli were supplemented with 33 nM TdR (Sigma, St. Louis, Mo.) and 20 μM BrdU (Sigma); unlabeled negative controls received 33 nM TdR in the absence of BrdU. Amended cultures were incubated at 37°C until the log phase (approximately 4 h). Sewage was incubated at the in situ temperature (21°C) under aerobic conditions for 4, 8, 12, and 24 h. Triplicate tubes of sewage influent were incubated for 4, 8, 12, or 24 h. Supplements were added to sewage samples 4 h prior to harvest (i.e., at 0, 4, 8, and 20 h) so that each sample was incubated with the supplements for only 4 h immediately before DNA extraction.
DNA extraction.
Following BrdU labeling, DNA was extracted from cultures and sewage samples.
(i) Bacterial cultures.
A log-phase culture suspension (4 ml) was pelleted, washed, divided, placed in two 1.7-ml microcentrifuge tubes, and repelleted. We extracted DNA from pure bacterial cultures using a DNeasy tissue kit (QIAGEN, Valencia, Calif.), with the following modifications: two washes with buffer AW2 were used, and the DNA was eluted twice by adding 50 μl of buffer AE to the silica column, warming the preparation to 65°C for 10 min, and centrifuging the preparation at 10,000 rpm for 1 min.
(ii) Sewage samples.
DNA was extracted from sewage influent using a DNeasy tissue extraction kit (QIAGEN), with several modifications. First, 7-ml samples were filtered through 0.2-μm Supor membrane filters (Pall Gelman Laboratory, Ann Arbor, MI) under a vacuum (at least 15 lb/in2). The filters were rolled, placed in sterile 15-ml polypropylene tubes containing 500 μl guanidine isothiocyanate buffer (5 M guanidine thiocyanate, 100 mM EDTA, 0.5% N-lauroyl sarcosine), and then vortexed for 60 s to completely saturate the filters. The filters were stored at −80°C until DNA was extracted. After thawing, the filters were vortexed again for 60 s. Incubation with buffer ATL and proteinase K was omitted. Tubes containing filters with guanidine isothiocyanate buffer received 500 μl buffer AL and were vortexed for 60 s, and then each tube received 500 μl of 100% ethanol and was vortexed for an additional 60 s. Lysates were removed from the tubes and loaded onto individual DNeasy spin columns. The filters were discarded. Instead of the recommended single wash step with buffer AW2, three 500-μl washes were used. DNA was eluted twice by adding 50 μl of buffer AE to the silica column and then warming the preparation to 65°C for 10 min and centrifuging it at 10,000 rpm for 1 min.
Immunocapture.
To separate BrdU-labeled DNA from unlabeled DNA, we used the immunocapture procedure described by Urbach et al. (44). First, 9 μl of herring sperm DNA (1.25 mg/ml; Invitrogen, Carlsbad, CA) per sample was boiled for 5 min and quick-frozen with dry ice and ethanol. Monoclonal mouse anti-BrdU antibody (0.44 mg/ml immunoglobulin G; 1 μl per sample; Sigma) was added to the herring sperm DNA, mixed, and incubated in the dark at room temperature for 30 min with occasional mixing. DNA (1 μg) from pure cultures or DNA recovered from sewage extraction, measured using a PicoGreen assay (Molecular Probes Inc., Eugene, OR), was suspended in 10 μl phosphate-buffered saline (PBS), boiled for 5 min, and quick-frozen on dry ice and ethanol. Each DNA sample received 10 μl of the anti-BrdU antibody-herring sperm DNA mixture. Samples were incubated in the dark at room temperature for 30 min with occasional mixing. Goat anti-mouse immunoglobulin G-coated paramagnetic beads (4 × 108 beads/ml; Dynal Biotech, Oslo, Norway) were washed three times with 1 ml PBS-0.1% nonacetylated bovine serum albumin (BSA) (Invitrogen), and the volume was adjusted to the original volume with PBS-0.1% BSA. Twenty-five microliters of the bead suspension was added to each sample, which was incubated in the dark at room temperature with constant mixing for 30 min. Bead supernatants were removed and saved. The remaining antibody-bound DNA was washed seven times with PBS-0.1% BSA and eluted with 100 μl of 1.7 mM BrdU with constant agitation for 30 min. The DNA recovered was concentrated by ethanol precipitation. Dry pellets were dissolved in 8 μl (immunocaptured fractions) and 20 μl (bead supernatants) molecular grade water with gentle heat and mixing. All washes were performed using a magnetic particle concentrator (MPC-S; Dynal).
PCR detection of immunocaptured and unlabeled fractions.
Bead supernatants and immunocaptured fractions were added to PCR mixtures (2 μl per mixture). Each 25-μl reaction mixture consisted of 1× PCR buffer containing 2.0 mM MgCl2 (TaKaRa BIO Inc., Otsu, Shiga, Japan), each deoxynucleoside triphosphate at a concentration of 0.2 mM, each primer at a concentration of 0.2 μM, 0.08% BSA, and 0.025 U Ex Taq DNA polymerase (TaKaRa BIO Inc.). The amplification conditions included an initial denaturation step of 94°C for 3 min, followed by 94°C for 1 min, the annealing temperature specific for each primer pair for 45 s, 72°C for 45 s for the numbers of cycles described below, and then a final extension step at 72°C for 7 min. The annealing temperature for bacterial primer 27F (AGAGTTTGATCMTGGCTCAG) (40) paired with universal primer 338R (GCTGCCTCCCGTAGGAGT) (40) was 55°C. The annealing temperature for the Bacteroidales-specific primers Bac32F (AACGCTAGCTACAGGCTT) (4) and 6-carboxyfluorescein (6-FAM)-labeled Bac708R (CAATCGGAGTTCTTCGTG) (4) was 53°C. For human-specific Bacteroidales primers HF134 (GCCGTCTACTCTTGGCC) (5) and HF183 (ATCATGAGTTCACATGTCCG) (5) paired with 6-FAM-labeled 708R, the annealing temperature was 63°C. PCR products were visualized on 1.5% agarose gels stained with ethidium bromide (Sigma), using a UVP gel imager (UVP, Upland, CA), or by fluorescent fragment (GeneScan) analysis.
GeneScan analysis.
6-FAM-labeled PCR products were diluted by an empirically determined amount to avoid saturating the fluorescent detector, as follows: 15 PCR cycles, no dilution; 20 PCR cycles, 1:2 dilution; 25 PCR cycles, 1:10 dilution; and 30 PCR cycles, 1:10 dilution. Aliquots (2 μl) of diluted PCR products were submitted to the Central Services Laboratory at Oregon State University for resolution with a model ABI 3100 capillary sequencer and GeneScan software (ABI: Applied Biosystems Inc., Freemont, Calif.). GENESCAN500-ROX (ABI) was added as an internal size standard to each sample when the expected fragment length was less than 400 bp; MAPMARKER1000 was added as an internal size standard to samples when fragments were expected to be more than 400 bp long. Fragment sizes were estimated using the Local Southern Calling method provided in GeneScan, version 3.1 (ABI).
BrdU labeling of B. vulgatus.
To determine the ability of bacteria in the order Bacteroidales to take up and incorporate BrdU, we used pure cultures of B. vulgatus and labeled them with BrdU as described above. To detect B. vulgatus BrdU-labeled DNA, we used 30 PCR cycles with 16S rRNA gene primers 27F and 338R and visualized the products by gel electrophoresis as described above.
Establishing conditions under which immunocapture separates BrdU-labeled DNA from unlabeled DNA.
Because we observed background amplification of unlabeled control DNA, we determined the ability of the immunocapture method to accurately separate BrdU-labeled DNAs from unlabeled DNAs. Mixtures of F. pelagi and B. vulgatus DNAs at different proportions were used as templates, resulting in fragments of different lengths that could be distinguished and quantified by LH-PCR.
(i) Quantitative cycle number.
To determine PCR conditions that yielded the same proportion of PCR fragments in the product as in the template, we combined F. pelagi and B. vulgatus genomic DNAs at ratios of 1:1, 1:10, and 10:1. Then PCRs with 15, 20, or 25 cycles were carried out as described above using 2-μl portions (5 ng/μl) of the mixtures as templates with 6-FAM-labeled primer 27F and primer 338R. The PCR products were submitted for GeneScan analysis within 24 h. The quantitative cycle number was determined by comparing the initial template ratios to the recovered fragment abundance values calculated using ratios of relative fluorescence units (RFU). The number of cycles that resulted in fragment abundance values with ratios equal to the initial template ratios was considered quantitative.
(ii) Determining immunocapture using LH-PCR detection.
We combined unlabeled F. pelagi and BrdU-labeled B. vulgatus genomic DNAs at ratios of 1:1, 1:10, and 10:1. Preimmunocapture LH-PCR was carried out using 2-μl portions (5 ng/μl) of these mixtures; the remaining DNA was used in immunocapture reactions as described above. For detection we performed LH-PCR with 15, 20, and 25 cycles, using 6-FAM-labeled primer 27F with primer 338R.
Growth in sewage samples.
To determine whether Bacteroidales cells grew in sewage, incubation mixtures were labeled with BrdU, DNA was extracted, the immunocapture protocol was carried out as described above, and results were detected by PCR with general or human-specific Bacteroidales primers using numbers of cycles determined to be quantitative.
(i) General Bacteroidales quantitative cycle number.
To determine a quantitative PCR cycle number using Bacteroidales-specific primers Bac32F and 6-FAM-labeled Bac708R (4), we used different amounts of B. vulgatus template DNA and quantified PCR products by using GeneScan after 15, 20, 25, and 30 PCR cycles. The following amounts of template DNA were added to PCR mixtures, as determined by the PicoGreen assay (Molecular Probes Inc.): 0.5 ng, 1 ng, 5 ng, and 10 ng. The PCR products were submitted for GeneScan analysis within 24 h. The quantitative cycle number was determined as described above.
(ii) Human-specific Bacteroidales quantitative cycle number.
Similarly, we determined the quantitative PCR cycle number for Bacteroidales human-specific primers HF134 and HF183 (5) in combination with 6-FAM-labeled Bac708R (4). The following amounts of cloned plasmid DNA were added to PCR mixtures: 23 ng, 11.65 ng, 2.33 ng, and 1.17 ng. The PCR products were quantified by GeneScan after 15, 20, 25, and 30 cycles. The quantitative cycle number was determined as described above.
(iii) Detection of growth and persistence in sewage samples.
Recovered bead supernatants and immunocaptured fractions from sewage incubation mixtures from each time (4, 8, 12, and 24 h) were added to PCR mixtures (2 μl per reaction mixture). Analyses were performed with all bead supernatants and immunocaptured DNA for both BrdU-labeled samples and unlabeled controls. PCRs were performed as described above. Samples were subjected to LH-PCR analysis using products obtained after 15 PCR cycles with primer pairs Bac32F-Bac708R, HF134-Bac708R, and HF183-Bac708R; PCR with primers 27F and 338R was included as a labeling control.
Limits of detection at the quantitative cycle number.
We determined the limit of detection of target sequences with each of the primer pairs at its quantitative cycle number. PCR mixtures primed with Bac32F and 6-FAM-labled Bac708R received the following amounts of genomic B. vulgatus DNA: 0.5 pg, 1 pg, 5 pg, 0.01 ng, 0.05 ng, 0.1 ng, 0.5 ng, 1 ng, and 5 ng. PCR mixtures primed with either HF134 or HF183 in combination with 6-FAM-labeled Bac708R received the following amounts of plasmid DNA containing the human-specific sequences: 5 pg, 0.01 ng, 0.05 gn, 0.1 ng, 0.5 ng, 1 ng, and 5 ng, quantified by spectrophotometry (BioSpec-1601; Shimadzu Corp., Kyoto, Japan). Amplification was performed as described above. Detection was by fluorescent fragment analysis as described above.
Immunocapture with fluorescent PCR: reproducibility.
We used B. vulgatus and E. coli to determine reproducibility and to establish the lower limits of detection of the assay with mixed populations.
(i) BrdU labeling.
Pure cultures of B. vulgatus and E. coli were subjected to BrdU labeling as described above. DNA was extracted using the DNeasy (QIAGEN) protocol (see above) and was quantified using PicoGreen (Molecular Probes Inc.). DNAs were diluted to obtain a concentration of 10 ng/μl.
(ii) Immunocapture.
Labeled B. vulgatus and E. coli DNAs were combined at the following ratios: 1:100, 1:10, 1:1, 10:1, and 100:1 (1 ng). Immunocapture was performed with three sets of replicate samples as previously described. Each set of replicate samples was processed independent of the other replicate samples. DNA pellets were dissolved in 8 μl (immunocaptured fraction) and 20 μl (supernatant) of molecular biology grade water.
(iii) Fluorescent PCR and GeneScan analysis.
Fifteen cycles of PCR with primers Bac32F and 6-FAM-labeled Bac708R were performed using preimmunocapture DNA mixtures, bead supernatants, and immunocaptured fractions obtained after the labeling and immunocapture procedure for all three replicates. Each fluorescently labeled PCR product (2 μl) was subjected to GeneScan analysis.
RESULTS
To determine whether Bacteroidales fecal bacteria can grow under environmental conditions, we labeled cultures of B. vulgatus and sewage influent with BrdU and used the immunocapture technique (2, 7, 44) to recover labeled DNA. Labeled (immunocaptured) or unlabeled (supernatant) DNA was used as the template in PCRs performed with primers specific for the order Bacteroidales or human-specific Bacteroidales fecal bacteria. Amplification of unlabeled DNA demonstrated that there was marker persistence, whereas amplification of BrdU-labeled DNA demonstrated that cell growth occurred.
BrdU immunocapture and 30-cycle PCR.
We successfully amplified B. vulgatus genes from immunocaptured DNA, indicating that B. vulgatus took up BrdU during growth and incorporated it into newly synthesized DNA. However, we obtained inconsistent results when we used 30 PCR cycles to detect BrdU-labeled DNA. Frequently, amplification occurred in the immunocaptured fraction of the unlabeled control (Fig. 1); if the antibody technique isolated only BrdU-labeled DNA, the immunocaptured unlabeled control should not have been amplified. Occasional amplification of the immunocaptured fraction of the unlabeled controls occurred even when a variety of blocking agents, including herring sperm DNA, Roche blocking agent, Tween 20, and Denhardt's reagent, were evaluated alone and in combination (data not shown).
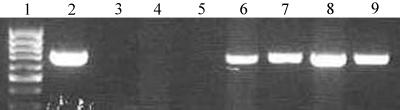
FIG. 1.
Agarose gel electrophoresis following 30 cycles of PCR performed with primers 27F and 338R and with immunocaptured fractions of B. vulgatus BrdU-labeled and unlabeled DNAs, showing robust amplification of the unlabeled control (lane 9). Lanes from left: 1, 100-bp ladder (size standard); 2, PCR positive control; 3, PCR negative control; 4, no-DNA bead supernatant; 5, no-DNA immunocaptured fraction; 6, BrdU-labeled bead supernatant; 7, BrdU-labeled immunocaptured DNA; 8, unlabeled bead supernatant; 9, unlabeled immunocaptured DNA.
Establishing conditions under which immunocapture separates BrdU-labeled DNA from unlabeled DNA.
The amplification observed in immunocaptured fractions of unlabeled controls could have been caused by the high sensitivity of PCR amplifying background-level fragments of unlabeled DNA or by the lack of specificity of the assay. In order to use the BrdU assay to detect growth, it was necessary to distinguish between these hypotheses. To determine the ability of the antibody assay to separate BrdU-labeled DNA from unlabeled DNA, we used a more sensitive technique, LH-PCR (41). Following PCR with 16S rRNA gene primers 27F and 338R, LH-PCR revealed a naturally occurring 37-bp difference between the length of amplicons derived from pure cultures of B. vulgatus (352 bp) and the length of amplicons derived from pure cultures of F. pelagi (315 bp), which was caused by insertions and deletions in rRNA genes (41).
First, we empirically established PCR conditions that were quantitative; that is, the ratios of amplified PCR products were the same as the ratios of the products in the initial template mixtures. When we added known proportions of F. pelagi and B. vulgatus unlabeled DNAs to PCR mixtures and stopped the reactions after 15, 20, and 25 cycles, amplification bias occurred as the cycle number increased. With 15 PCR cycles the ratio of fragments in the products corresponded to the original template proportions (Table 1); when more than 20 cycles were used (Table 1), this relationship diminished.
TABLE 1.
Ratios of relative fluorescence units resulting from LH-PCR products following 15 and 20 cycles of PCR in which unlabeled F. pelagi and BrdU-labeled B. vulgatus DNA templates at known ratios were amplified using primers 27F and 338R
| F. pelagi/B. vulgatus ratio in template |
F. pelagi/B. vulgatus ratio in PCR products
|
|
|---|---|---|
| 15 cycles | 20 cycles | |
| 1:10 | 0.0597 | 0.2540 |
| 1:1 | 0.5660 | 0.7162 |
| 10:1 | 5.060 | 6.261 |
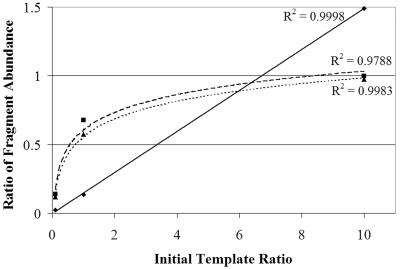
We subjected mixtures containing known proportions of unlabeled F. pelagi DNA and BrdU-labeled B. vulgatus DNA to immunocapture. We analyzed bead supernatants, immunocaptured eluates, and mixtures prior to immunocapture using LH-PCR with 15, 20, and 25 cycles. Fifteen LH-PCR cycles resulted in fragment abundances proportional to the fragment abundances in the original template mixtures. We observed amplification bias with 20 cycles; amplification shifted from linear to logarithmic, approaching a 1:1 final amplicon composition, consistent with previously published data (40) (Fig. 2).
FIG. 2.
As the number of cycles increased, the ratio of the initial template concentrations (F. pelagi/B. vulgatus) relative to the ratio of fragment abundances shifted from linear to logarithmic. Solid line, 15 cycles (linear); dashed line, 20 cycles (logarithmic); dotted line, 25 cycles (logarithmic). Fragments were amplified using primers 27F and 338R.
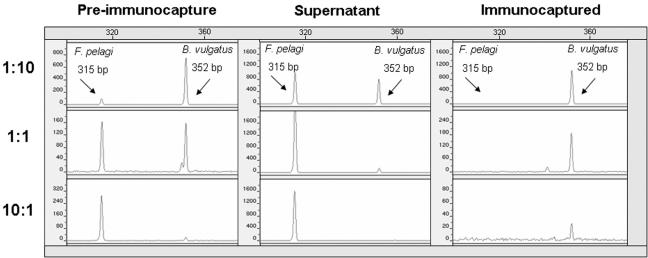
Immunocapture followed by a 15-cycle LH-PCR with bead supernatants, as well as immunocaptured fractions, revealed that immunomagnetic separation enriched for BrdU-labeled DNA (Fig. 3). In repeated experiments, unlabeled F. pelagi DNA was never identified in the immunocaptured fraction when a 15-cycle PCR was used for detection (data not shown). These results demonstrated that immunocapture was specific for BrdU-labeled DNA; however, with a high number of cycles unlabeled “background” DNA was occasionally amplified as a result of blocker leakage or PCR sensitivity. We eliminated amplification of background DNA by using fluorescent detection of PCR amplicons following 15 cycles.
FIG. 3.
LH-PCR electropherograms after 15 PCR cycles, showing the proportions of unlabeled F. pelagi and BrdU-labeled B. vulgatus before and after immunocapture when primers 27F and 338R were used.
BrdU labeling and immunocapture of sewage influent.
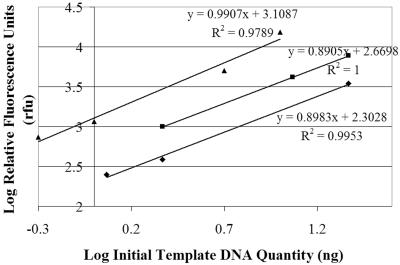
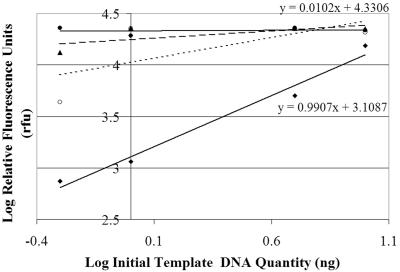
We labeled sewage with BrdU and incubated it for up to 24 h to measure the growth of Bacteroidales cells and the persistence of Bacteroidales molecular markers in sewage, using immunocapture followed by fluorescent PCR detection. To detect Bacteroidales with general and human-specific Bacteroidales primers, we first established conditions under which the primer pairs were quantitative by varying the template concentrations and establishing the numbers of cycles for which the product concentration was proportional to the template concentration. Plotting the log of fragment abundance (RFU) versus the log of the initial template concentrations indicated that for all Bacteroidales-specific primer pairs used in this study, PCR was quantitative when 15 cycles of amplification were used (Fig. 4). With 20 or more cycles we observed bias in the sample results from LH-PCR fragment analysis, and the slopes approached zero as the number of cycles increased (Fig. 5).
FIG. 4.
PCR for 15 cycles revealed an increase in amplicon fluorescence that was directly proportional to an increase in the initial template concentration. Symbols: ⧫, general Bacteroidales; ▪, HF134; ▴, HF183. The equations for the lines are in point slope form.
FIG. 5.
PCRs using the general Bacteroidales primers (32F and 708R) for 15, 20, 25, and 30 cycles. With more than 15 cycles, amplification bias was observed as amplicon fluorescence reached constant values and the slope values approached zero. Symbols: ⧫, 15 cycles; ○, 20 cycles; ▴, 25 cycles; •, 30 cycles. Line equations are given for 15 and 30 cycles.
As a control for the BrdU labeling process we also amplified bacterial 16S rRNA gene sequences in case we could not detect Bacteroidales DNA in the immunocaptured fractions. Bacterial 16S rRNA genes were detected throughout the 24-h period. We detected BrdU-labeled Bacteroidales DNA at 4, 8, 12, and 24 h, indicating that growth of fecal Bacteroidales cells occurred for at least 24 h. The Bacteroidales molecular signal also persisted in the unlabeled DNA fraction for the entire 24-h time course.
Fluorescent fragment analysis of the PCR products obtained following 15 cycles did not detect the Bacteroidales human-specific markers in either supernatants or immunocaptured fractions, even when we used 10 times the concentration of labeled sewage DNA in the immunocapture procedure. Either the human-specific markers did not persist or grow or the fluorescent detection was not sensitive enough when 15 PCR cycles were used. We estimated the limits of detection for fluorescent fragment analysis following 15 PCR cycles using decreasing concentrations of template DNA. We detected as little as 0.01 ng of B. vulgatus genomic DNA, corresponding to 1.24 × 104 copies of the 16S rRNA gene, and 0.1 ng of human-specific Bacteroidales plasmid DNA, corresponding to 2 × 107 copies of the gene.
To examine the reproducibility of the assay, as well as to establish the lowest level of BrdU-labeled target DNA that could be detected in a mixed population of BrdU-labeled DNAs, we conducted three replicate immunocapture experiments using five different proportions of BrdU-labeled B. vulgatus and E. coli DNAs. We reliably detected amounts down to 0.5 ng and occasionally detected as little as 0.01 ng of BrdU-labeled template DNA (Table 2) using the general Bacteroidales primers.
TABLE 2.
Fluorescent PCR efficiency with 15 cycles when known quantities of BrdU-labeled target DNA were immunocaptured in a community of BrdU-labeled DNA
| Starting proportion (ng)
|
Detection of B. vulgatus before immunocapture
|
Detection of B. vulgatus following immunocapture
|
|||||
|---|---|---|---|---|---|---|---|
| E. coli | B. vulgatus | Replicate 1 | Replicate 2 | Replicate 3 | Replicate 1 | Replicate 2 | Replicate 3 |
| 0.99 | 0.01 | +a | + | ± | − | − | + |
| 0.9 | 0.1 | + | + | + | − | + | + |
| 0.5 | 0.5 | + | + | + | + | + | + |
| 0.1 | 0.9 | + | + | + | + | + | + |
| 0.01 | 0.99 | + | + | + | + | + | + |
+, positive; −, negative; ±, weak positive (RFU values less than values that have statistical significance).
DISCUSSION
A major limitation in studies of the ecology of allochthonous organisms is the inability to cultivate the majority of these organisms under laboratory conditions. BrdU labeling of environmental samples or natural assemblages, followed by immunocytochemical separation, can allow detection of actively growing members of a population and, in the case of pollution source tracking, allows a distinction to be made between the persistence of a molecular marker and the growth of the indicator organism(s).
Our data demonstrate that analysis of growth by BrdU immunocapture and PCR detection with 30 cycles is not reliable due to the innate background commonly observed in immunological assays, which can subsequently be amplified by PCR. Previous studies in which 30 PCR cycles were used to detect BrdU-labeled sequences did not show PCR results with unlabeled controls (2, 7). Unlabeled Roseobacter and lake water DNAs were observed in LH-PCR electropherograms (44) in a study in which PCR was used to detect BrdU-labeled Alteromonas mixed with unlabeled Roseobacter DNA following immunocapture. In order to eliminate PCR detection of unlabeled DNA, we used LH-PCR to verify that there was enrichment for BrdU-labeled DNA in a mixed community and PCR with fluorescent detection to determine a quantitative cycle number that reliably amplified only BrdU-labeled DNA. By comparing known template proportions to fragment abundances following PCR with the quantitative cycle number, we showed that the BrdU immunocapture technique enriched for BrdU-labeled DNA and did not amplify unlabeled DNA. We optimized our analyses by analyzing dilutions of PCR products from each sample. Thus, the observed bias was not due to overloading the sequencer with excessive PCR product. Peak amplitudes between 100 and 5,000 RFU are considered the most reliable peak amplitudes when the ABI 3100 sequencer is used; therefore, we retained data for peaks in this range.
Although highly reproducible, using PCR with fluorescent detection with the low number of cycles required for quantitative results decreased the sensitivity of this assay; we could not detect the human-specific Bacteroidales group in bead supernatants or immunocaptured fractions from BrdU-labeled sewage. This may have been due to low cell densities. The smallest amount of BrdU-labeled DNA that could be detected consistently was 0.5 ng, and detection of 0.1 ng and 0.01 ng of BrdU-labeled template was erratic (in contrast, a standard PCR with 30 cycles routinely detects 100 gene copies using these primers [38]). Because the human-specific clades are rare in sewage compared to Bacteroidales in general, it is likely that the number of cycles for detection of these targets could be increased without compromising the quantitative results. We do not believe that the inability to detect was due to inhibition. It is possible that impurities resulting from extraction of DNA from sewage interfered with antibody binding; however, we routinely detected growth of general Bacteroidales cells in sewage, suggesting that interference with antibody binding was negligible in these experiments. A third possibility is that we did not detect cells belonging to the Bacteroidales human marker clade because they did not grow, although other Bacteroidales cells did; this would imply that different Bacteroidales marker types may exhibit differential survival.
We demonstrated that Bacteroidales fecal bacteria grew for up to 24 h in sewage when preparations were incubated aerobically at the in situ temperature of sewage and that the marker(s) persisted for at least 24 h under the same conditions. This is in agreement with the findings of Kreader (22), who showed that the molecular signal for Bacteroides distasonis persisted for up to 14, 5, and 2 days at 4°C, 14°C, and 24°C, respectively, in unfiltered river water. Additionally, Seurinck and colleagues (37) found that the human-specific marker (HF183) persisted for up to 24 days at 4°C and 12°C and for up to 8 days at 28°C in fresh river water (37).
It is known that at least some bacteria in the Bacteroidales group are not obligately anaerobic. Bacteroides fragilis, which was previously considered an obligate anaerobe, requires nanomolar concentrations of oxygen for growth and possesses an O2-dependent cytochrome (3). These findings are consistent with the observation that Bacteroidales cells exhibit some degree of oxygen tolerance when they are manipulated in the laboratory. In addition, sewage influent often contains flocculent material. Bacteria colonize small particles, creating anaerobic microniches in an overall aerobic environment (43), such as surface water. Finally, we did not shake or otherwise aerate our sewage incubation mixtures. When we added the oxygen-sensitive, colorimetric indicator resazurin to 10-ml sewage samples and incubated them aerobically for 4 h, the mixtures stratified such that the top one-third of a tube was pink, indicating the presence of oxygen, but the lower two-thirds was clear, indicating anaerobiosis (data not shown). This observation, combined with our data demonstrating that there was growth of Bacteroidales cells during aerobic incubation of sewage influent, suggests that Bacteroidales fecal bacteria may be able to persist and grow in low-oxygen refugia in streams, lakes, estuaries, and bays.
When commensal bacteria are used as source markers for fecal pollution, a reliable means of quantifying marker abundance and growth is necessary in order to implement policies involving human health risk assessment and diagnosis of watershed quality. Studies exploring the growth of indicator organisms should employ quantitative methods at the time of sampling in order to examine the extent of growth in the environmental matrix. Factors that influence the growth of allochthonous organisms include predation, ambient water temperature, UV radiation, and sediments as refugia (1, 15, 18, 28).
Our findings may have a significant impact on microbial ecology studies in which PCR of immunoseparated, BrdU-labeled DNA is used to detect active populations. Because of kinetic biases (40), an unequal ratio of amplicon concentration to initial template concentration can result from high-cycle-number PCR. Unlabeled background DNA amplicons may reach a detectable concentration and appear to be active members of a population or community when in fact they are not, leading to inaccurate conclusions about metabolic or biogeochemical processes in a population or community. The sensitivity of PCR is so great that in order to overcome the problem of amplifying unlabeled background DNA, a quantitative assay should be incorporated into this protocol. Here we demonstrate that PCR performed for a low number of cycles, followed by fluorescent fragment analysis, can overcome the problem of background amplification in these assays.
BrdU labeling and immunocapture followed by a low number of PCR cycles and fluorescent detection is an attractive method for identification of growing cells based on phylotype because it circumvents the need for direct cultivation and isolation, is nonradioactive, and is relatively simple and inexpensive. However, when the growth of uncommon or rare species in aquatic environments is studied, this method is probably not sensitive enough to verify the growth of slowly growing organisms or organisms present at low cell densities. Thus, using PCR to detect growing populations with a low, quantitative cycle number could cause ecologically significant populations that have longer generation times and comprise a small proportion of the overall community to be overlooked.
Other methods for detection of viable or active cells belonging to specific bacterial groups include rRNA-targeted hybridization using specific oligonucleotide probes and microautoradiography. Detection methods that target RNA sequences can be used quantitatively, and the results are reasonable predictors of cell viability because of rapid cellular RNA degradation following cell death. Fluorescence in situ hybridization (FISH) is a nonradioactive method that uses phylogenetic group-specific probes targeting rRNAs to selectively visualize cells, preserving cell size, morphology, and bacterial aggregate composition. Like the BrdU assay, FISH may miss slowly growing or starving cells due to a low rRNA content. In addition, the power of FISH is limited by available sequence data and sequence specificity. However, the sensitivity of FISH assays can be increased by using multiple probes that are specific for the same target organism(s) (24), by using helper probes (16), or by including catalyzed reporter deposition (31). Bacteroidales fecal marker organisms exhibit nearly 100% sequence identity (10), which makes it difficult to design a probe or probe set specific for each host-specific marker and limits the applicability of the FISH technique in our system. Microautoradiography is used to visualize active microbial cells metabolizing a radioactive substrate. When used with 16S rRNA-targeted hybridization, microautoradiography offers the advantage of phylogenetically detecting active cells and deciphering group-specific substrate utilization by the bacteria (23, 30). However, microautoradiography depends on cells' ability to both grow in culture and take up the supplemented substrate. To follow the studies reported here, we are using hybridization and quantitative PCR techniques to quantitatively survey the persistence and growth of host-specific Bacteroidales bacteria in mesocosms that simulate a variety of environmental parameters.
In microbial ecology there are often three questions: who is there, how many, and what are they doing? When used alone, FISH provides information about the identity and abundance of microbial cells but does not determine the function of microbial populations in a community or provide information about growth. FISH paired with BrdU labeling can determine cell growth and the viability of specific microbial species in communities (32). Combining methods that employ 16S rRNA-specific probe hybridization with microautoradiography allows inferences about the viability and function of environmental populations (23, 36) without direct cultivation. Recent advances have broadened our ability to use multiple detection techniques simultaneously. It appears that no single method is best for examining the activity and viability of aquatic microorganisms; thus, to gain maximum insight about the growth, viability, and function of microbial communities, the experimental approach should include multiple methodologies.
Acknowledgments
This work was partially supported by a Charles E. and Clara Marie Eckelman Graduate Assistantship, by Oregon Sea Grant 04-016, and by the Nucleic Acids and Protein Service Core of the Environmental Health Sciences Center, Oregon State University (grant P30 ES00210 from the National Institute of Environmental Health Sciences, National Institutes of Health).
We thank Jakob Pernthaler, Stephen Giovannoni, Linda Dick, and Malcolm Lowry for thoughtful discussions and valuable input.
REFERENCES
- 1.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artursson, V., and J. K. Jansson. 2003. Use of bromodeoxyuridine immunocapture to identify active bacteria associated with arbuscular mycorrhizal hyphae. Appl. Environ. Microbiol. 69:6208-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonadonna, L., R. Briancesco, M. Ottaviani, and E. Veschetti. 2002. Occurrence of Cryptosporidium oocysts in sewage effluents and correlation with microbial, chemical, and physical water variables. Environ. Monit. Assess. 75:241-252. [DOI] [PubMed] [Google Scholar]
- 7.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byappanahalli, M., M. Fowler, D. Shively, and R. Whitman. 2003. Ubiquity and persistence of Escherichia coli in a midwestern coastal stream. Appl. Environ. Microbiol. 69:4549-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, J.-C., and S. J. Giovannoni. 2003. Fulvimarina pelagi gen. nov., sp. nov., a marine bacterium that forms a deep evolutionary lineage of descent in the order ‘Rhizobiales.’ Int. J. Syst. Evol. Microbiol. 53:1853-1859. [DOI] [PubMed] [Google Scholar]
- 10.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick, L. K., and K. G. Field. 2004. Rapid estimation of number of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick, L. K., M. T. Simonich, and K. G. Field. 2005. Microplate subtractive hybridization to enrich for Bacteroidales genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3179-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfman, M., N. Stoner, and M. Merkel. 2004. Swimming in sewage. Natural Resources Defense Council and the Environmental Integrity Project, New York, N.Y. [Online.] http://www.nrdc.org/water/pollution/sewage/sewage.pdf.
- 14.Field, K. G., E. C. Chern, L. K. Dick, J. Fuhrman, J. Griffith, P. A. Holden, M. G. LaMontagne, J. Le, B. Olson, and M. T. Simonich. 2003. A comparative study of culture-independent, library-independent genotypic methods of fecal source tracking. J. Water Health 1:181-194. [PubMed] [Google Scholar]
- 15.Fish, J. T., and G. W. Pettibone. 1995. Influence of freshwater sediment on the survival of Escherichia coli and Salmonella sp. as measured by three methods of enumeration. Lett. Appl. Microbiol. 20:277-281. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, B. M., F. O. Glockner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujioka, R., C. Dian-Denton, J. Castro, and K. Morphew. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 85:83S-89S. [DOI] [PubMed] [Google Scholar]
- 18.Hood, M. A., and G. E. Ness. 1982. Survival of Vibrio cholerae and Escherichia coli in estuarine waters and sediments. Appl. Environ. Microbiol. 43:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horman, A., R. Rimhanen-Finne, L. Maunula, C.-H. von Bonsdorff, N. Torvela, A. Heikinheimo, and M.-L. Hanninen. 2004. Campylobacter spp., Giardia spp., Cryptosporidium spp., noroviruses, and indicator organisms in surface water in southwestern Finland, 2000-2001. Appl. Environ. Microbiol. 70:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreader, C. A. 1998. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl. Environ. Microbiol. 64:4103-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K. H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. H., C. Malone, and P. F. Kemp. 1993. Use of multiple 16S ribosomal rRNA-targeted fluorescent-probes to increase signal strength and measure cellular rRNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 101:193-201. [Google Scholar]
- 25.Lemarchand, K., and P. Lebaron. 2003. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: relationship with fecal indicators. FEMS Microbiol. Lett. 218:203-209. [DOI] [PubMed] [Google Scholar]
- 26.Lipp, E. K., J. L. Jarrell, D. W. Griffin, J. Lukasik, J. Jacukiewicz, and J. B. Rose. 2002. Preliminary evidence for human fecal contamination in corals of the Florida Keys, USA. Mar. Pollut. Bull. 44:666-670. [DOI] [PubMed] [Google Scholar]
- 27.Lund, V. 1996. Evaluation of E. coli as an indicator for the presence of Campylobacter jejuni and Yersinia enterocolitica in chlorinated and untreated oligotrophic lake water. Water Res. 30:1528-1534. [Google Scholar]
- 28.Menon, P., G. Billen, and P. Servais. 2003. Mortality rates of autochthonous and fecal bacteria in natural aquatic ecosystems. Water Res. 37:4151-4158. [DOI] [PubMed] [Google Scholar]
- 29.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 30.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernthaler, A., J. Pernthaler, M. Schattenhofer, and R. I. Amann. 2002. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl. Environ. Microbiol. 68:5728-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pommepuy, M., M. Butin, A. Derrien, M. Gourmelon, R. R. Colwell, and M. Cormier. 1996. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl. Environ. Microbiol. 62:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Power, M. L., J. Littlefield-Wyer, D. M. Gordon, D. A. Veal, and M. B. Slade. 2005. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 7:631-640. [DOI] [PubMed] [Google Scholar]
- 35.Rahman, I., M. Shahamat, M. A. R. Chowdhury, and R. R. Colwell. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossello-Mora, R., N. Lee, J. Anton, and M. Wagner. 2003. Substrate uptake in extremely halophilic microbial communities revealed by microautoradiography and fluorescence in situ hybridization. Extremophiles 7:409-413. [DOI] [PubMed] [Google Scholar]
- 37.Seurinck, S., T. Defoirdt, W. Verstraete, and S. D. Siciliano. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ. Microbiol. 7:249-259. [DOI] [PubMed] [Google Scholar]
- 38.Shanks, O. C., C. Nietch, M. T. Simonich, M. Younger, D. Reynolds, and K. G. Field. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamooh Bay, Oregon. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 39.Steward, G., and F. Azam. 1999. Bromodeoxyuridine as an alternative to 3H-thymidine for measuring bacterial productivity in aquatic samples. Aquat. Microb. Ecol. 19:57-66. [Google Scholar]
- 40.Suzuki, M., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamplin, M. L. 2003. The application and suitability of microbiological tests for fecal bacteria in pulp mill effluents: a review. Water Qual. Res. Can. 38:221-225. [Google Scholar]
- 43.Tay, S. T.-L., S. Ivanov, S. Yi, W.-Q. Zhuang, and J.-H. Tay. 2002. Presence of anaerobic Bacteroides in aerobically grown microbial granules. Microb. Ecol. 44:278-285. [DOI] [PubMed] [Google Scholar]
- 44.Urbach, E., K. L. Vergin, and S. J. Giovannoni. 1999. Immunochemical detection and isolation of DNA from metabolically active bacteria. Appl. Environ. Microbiol. 65:1207-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.U.S. Environmental Protection Agency. 2001. National coastal condition report EPA-620-R-01-005. U.S. Environmental Protection Agency, Washington, D.C.
- 46.Whitman, R. L., D. A. Shively, H. Pawlick, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]