Abstract
The utilization of Saccharomyces cerevisiae strains overproducing glycerol and with a reduced ethanol yield is a potentially valuable strategy for producing wine with decreased ethanol content. However, glycerol overproduction is accompanied by acetate accumulation. In this study, we evaluated the effects of the overexpression of GPD1, coding for glycerol-3-phosphate dehydrogenase, in three commercial wine yeast strains in which the two copies of ALD6 encoding the NADP+-dependent Mg2+-activated cytosolic acetaldehyde dehydrogenase have been deleted. Under wine fermentation conditions, the engineered industrial strains exhibit fermentation performance and growth properties similar to those of the wild type. Acetate was produced at concentrations similar to that of the wild-type strains, whereas sugar was efficiently diverted to glycerol. The ethanol yield of the GPD1 ald6 industrial strains was 15 to 20% lower than that in the controls. However, these strains accumulated acetoin at considerable levels due to inefficient reduction to 2,3-butanediol. Due to the low taste and odor thresholds of acetoin and its negative sensorial impact on wine, novel engineering strategies will be required for a proper adjustment of the metabolites at the acetaldehyde branch point.
Glycerol is the most abundant by-product of alcoholic fermentation after ethanol and carbon dioxide (19, 22, 25) and contributes to the sensory characteristics of wine, particularly smoothness and overall body (8, 18, 19). Consequently, many attempts to increase the glycerol yield during fermentation have been made (34, 37). Furthermore, rerouting of the carbon flux towards glycerol leads to a decreased ethanol yield as a result of carbon diversion and decreased NADH availability. This is of great importance, since a large number of quality wines produced by modern winemaking practices, which favor harvesting fully ripened grapes, frequently contain an excessive ethanol content. Consequently, in recent years, there has been an increasing demand for wines with reduced ethanol content, which has been driven by both health and economic concerns. The utilization of strains with a low ethanol yield to produce wines with reduced alcohol content may be an alternative approach to physical methods, which are expensive and detrimental for aroma compounds.
In Saccharomyces cerevisiae, glycerol is synthesized by the reduction of dihydroxyacetone phosphate followed by a subsequent dephosphorylation catalyzed by glycerol-3-phosphate dehydrogenase and glycerol-3-phosphatase, respectively. Glycerol overproduction has been obtained by overexpression of either GPD1 or GPD2, encoding glycerol-3-phosphate dehydrogenase isoforms (5, 15, 16, 17, 24, 27). Wine yeast strains overexpressing GPD1 produce between 12 and 18 g/liter of glycerol, and the final ethanol concentration is 1% to 1.5% (vol/vol) lower (23, 24). As a result of carbon shift and increased utilization of NADH through the glycerol pathway, GPD1 strains exhibit major alterations in central metabolism, such as increased formation of succinate, acetaldehyde, acetoin, 2,3-butanediol, and acetate.
Excessive production of acetate (above 1 g/liter) is a major side effect, since the maximum amount desirable in wine is around 0.6 g/liter. Acetate is produced mainly during fermentation by oxidation of acetaldehyde catalyzed by the cytosolic Mg2+-activated NADPH-dependent acetaldehyde dehydrogenase (ACDH) Ald6p (14, 26, 31). Since a deletion of ALD6 results in a substantially lower acetate yield (26), this strategy was applied to laboratory- and wine-derived strains overexpressing GPD1 (6, 7, 23). However, industrial strains exhibit major differences compared to laboratory strains (24). In addition, the environmental conditions for yeast cells during grape fermentation must significantly differ from those in laboratory media. Therefore, the consequences of GPD1 overexpression and ALD6 deletion on yeast physiology and technological performance need to be appraised in commercial wine yeast strains under enologically relevant conditions. We constructed three industrial strains from which the two copies of ALD6 were deleted and in which GPD1 is overexpressed. The potentialities of these strains for the wine industry were evaluated by analyzing the impact of these modifications on growth, fermentation performance, and metabolite profiles during fermentation under winemaking conditions. The results of this evaluation will serve as a strong basis for further improvements of these strains.
MATERIALS AND METHODS
Strains and plasmids.
Escherichia coli DH5α was used for cloning experiments. E. coli cultivation and media were described previously (32). All the yeast strains used in this study were S. cerevisiae strains and are described in Table 1. Yeast strains were maintained and grown in YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose). The multicopy vector pVT100U-GPD1 carrying GPD1 under the control of the ADH1 promoter and terminator and its derivative, pVT100U-ZEO-GPD1, have been described previously (15, 24). Transformants were selected on SD minimal medium (6.7 g/liter yeast nitrogen base without amino acids, 2% glucose) or on YPD medium supplemented with 150 μg/ml of geneticin or 100 μg/ml of phleomycin.
TABLE 1.
S. cerevisiae strains used and constructed in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| V5 | MATaura3 | INRA UMR SPO, derivative from Champagne wine strain |
| V5 GPD1 | MATaura3/pVT100U-GPD1 | 24 |
| V5 ald6 | MATaura3 ald6::loxP | 31 |
| V5 ald6 GPD1 | MATaura3 ald6::loxP/pVT100U-GPD1 | This study |
| K1M (K1 marquée) | Commercial wine yeast, ICV-INRA, Montpellier | |
| K1M GPD1 | pVT100U-ZEO-GPD1 | This study |
| K1M ald6 | ald6::loxP ald6::kanMX | This study |
| K1M ald6 GPD1 | ald6::loxP ald6::kanMX/pVT100U-ZEO-GPD1 | This study |
| VL1 (Zymaflore VL1) | Commercial wine yeast, Institut d'Oenologie, Bordeaux | |
| VL1 GPD1 | pVT100U-ZEO-GPD1 | This study |
| VL1 ald6 | ald6::loxP ald6::kanMX | This study |
| VL1 ald6 GPD1 | ald6::loxP ald6::kanMX/pVT100U-ZEO-GPD1 | This study |
| BC (Uvaferm BC) | Commercial wine yeast, Davis University, Pasteur no. 1877 | |
| BC GPD1 | pVT100U-ZEO-GPD1 | This study |
| BC ald6 | ald6::loxP ald6::kanMX | This study |
| BC ald6 GPD1 | ald6::loxP ald6::kanMX/pVT100U-ZEO-GPD1 | This study |
Fermentation conditions.
Batch fermentation experiments were carried out in MS synthetic medium that simulates standard grape juice as described previously (1). The MS medium contains 200 g/liter glucose, 6 g/liter malic acid, 6 g/liter citric acid, and a nitrogen source composed of 120 mg/liter nitrogen from ammonium and 340 mg/liter from amino acids. The medium was supplemented with uracil (50 mg/liter) and methionine (115 mg/liter) for growth of V5. During wine fermentation, yeast uses grape phytosterols and unsaturated fatty acids. To fulfill the lipid requirement of yeast cells during anaerobic growth, MS medium was supplemented with 7.5 mg/liter ergosterol, 0.21 g/liter Tween 80, and 2.5 mg/liter oleic acid. The pH of the medium is 3.3. Cells were precultured for 36 h at 28°C in 50-ml flasks filled with MS medium without agitation. The main fermentation culture was inoculated at a density of 1 × 106 cells per ml and carried out at 24°C or 28°C with continuous stirring (350 rpm) in fermentors of 1.1 liter (working volume) equipped with fermentation locks. The CO2 release was determined by an automatic measurement of fermentor weight loss every 20 min. The CO2 production rate dCO2/dt is the first derivate of the amount of CO2 produced with respect to time and was automatically calculated by polynomial smoothing of the CO2 produced (30). Fermentation experiments were performed in triplicate. Since G418 is inefficient at low pHs, fermentations were realized in the absence of selection pressure. Consequently, plasmids were progressively lost during the culture as previously observed (24). Due to this rather uncontrolled process, independent cultures showed significant variation in the production of glycerol and consequently other metabolites, but the same evolution in metabolite profiles was verified in all the experiments.
Chromosomal mapping.
Yeast chromosomal DNA was prepared in plugs as previously described (35), washed once in Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) at 50°C for 30 min, and then washed three times in the same buffer at room temperature for 30 min. Each plug analyzed contained DNA isolated from about 5 × 107 cells. Electrophoresis was performed in a 1% (wt/vol) agarose (Seakem Gold) gel using a transverse alternating-field electrophoresis system (Geneline, Beckman) under the following conditions: a constant voltage of 250 V for a 6-h run time with a 35-s pulse time, followed by 20 h at 275 V with a 55-s pulse time at a constant temperature (14°C). DNA was transferred onto a nylon membrane (Hybond N; Amersham) as previously described (32). Chromosomes were hybridized with a 32P-labeled ALD6-specific oligonucleotide probe (TCTCTTGGGTCTTGGGTAGC) as previously described (4). Southern blots were analyzed with a PhosphorImager (Molecular Dynamics).
Deletion of ALD6 in industrial strains.
The strain V5 ald6::kanMX was constructed as previously described (31) using the short flanking homology PCR-based gene disruption method and the Cre-lox recombination system (12). To delete ALD6 from the industrial strains, a fragment carrying the ald6::kanMX allele of V5 ald6 was amplified using primers CGAGGTCAAGCCTGGCGTGTT and GCAGTTGTTGTACACTAGCTTA, corresponding to regions upstream (positions −169 to −147) and downstream (positions 2178 to 2200) from the ALD6 open reading frame. The primers were designed in such a way as to generate large homologous flanking regions to increase integration efficiency in industrial strains. This fragment, carrying the KanMX marker, was used to transform the industrial strains K1M, VL1, and BC by using the LiAc procedure (33). The transformants that were resistant to G418 were analyzed by PCR with total DNA using primers upstream and downstream from the deleted region, followed by digestion of the PCR product by XhoI, which cleaves the KanMX4 module but is absent from ALD6. Analysis of 10 transformants for each transformed strain led to the selection of several strains lacking one ALD6 copy and carrying at least one wild-type ALD6 gene. The similar band intensity for each copy (deleted or not) suggested that these strains were diploid for ALD6. To delete the remaining wild-type copy, the KanMX marker was excised using the Cre-lox system (12). To use the pSH47 vector carrying Cre recombinase (12) in industrial strains, the positive marker ZEOR, conferring resistance to phleomycin, was introduced. The transcriptional unit carrying Tn5 ble under the control of TEF1p and CYC1t was amplified from pUT332 (CAYLA) using primers CGAGCTCGGTACCCG and CGGGTACCCTTGCAAATTAAAGCCTTCG, which introduced a KpnI site (underlined sequence). This fragment was digested by KpnI and ligated to KpnI-digested pSH47 to produce pSH47-ZEO. The KanMX marker was excised by the transformation of pSH47-ZEO and the subsequent elimination of the plasmid (12), and the additional ALD6 copy was deleted using the same strategy described above. The deletion of the additional copy was verified by PCR and restriction analysis and confirmed by Southern blot with chromosomes. The industrial strains lacking the two ALD6 alleles were transformed with pVT100U-ZEO-GPD1.
Analytical methods.
Glycerol, acetate, succinate, and ethanol concentrations were analyzed by high-pressure liquid chromatography, and acetoin and 2,3-butanediol levels were measured by gas chromatography as previously described (15). Acetaldehyde concentrations were determined enzymatically according to the method of Lundquist (13).
RESULTS
Decreasing acetate production in a model strain producing up to 30 g/liter glycerol.
A 60% reduction in acetate production has been obtained by the deletion of ALD6 from the yeast strain V5 (26). To study ALD6 inactivation in a strain in which the flux has been strongly shifted towards glycerol production, the V5 (ura3) and V5 ald6 (ura3) strains were transformed with the multicopy vector pVT100U-GPD1, producing V5 GPD1 and V5 ald6 GPD1, respectively (Table 1). As previously described (24), V5 GPD1 produced higher glycerol (28.4 g/liter) and acetate (1.5 g/liter) levels than the control strain (6.6 g/liter of glycerol and 0.6 g/liter of acetate) during wine fermentation. Strain V5 ald6 GPD1 produced an even higher glycerol concentration (33 g/liter), but the acetate accumulation (0.6 g/liter) was no higher than the wild-type level. This indicates that the ALD6 deletion effectively reduces acetate formation, even in strains exhibiting a large shift of carbon flux towards glycerol. We therefore investigated the consequences of the ALD6 deletion in combination with GPD1 overexpression in true commercial wine yeast strains during wine fermentation.
Deletion of ADL6 from three commercial wine strains overexpressing GPD1.
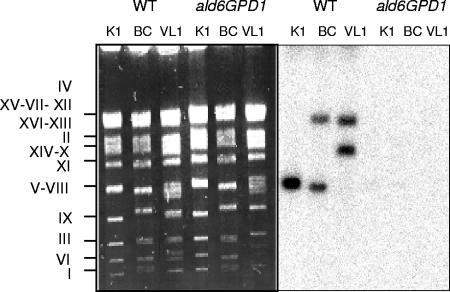
Wine yeast strains are considered to be genetically complex. Many strains are diploids, and some are aneuploids or polyploids (19, 20). In addition, they exhibit chromosome rearrangements and show a high level of chromosome size polymorphism (2, 21). To determine the number of ALD6 copies present in the three wine yeast strains K1M, VL1, and BC and their localization, we performed Southern blot analysis of chromosomes. ALD6 maps on chromosome XVI in the laboratory reference strain S288C. Chromosomal mapping analysis (Fig. 1) identified at least two copies of ALD6, each carried on a different chromosome in strains VL1 and BC. The hybridization signal for the large ALD6 copy is consistent with it mapping on chromosome XVI, whereas the other copy was detected on a smaller chromosome, corresponding to a region where chromosomes II, XIV, and X comigrate (strain VL1) or to chromosomes V or VIII (strain BC). Only one hybridization signal was observed for strain K1M, which also corresponded to chromosomes V or VIII; however, the intensity of the signal suggested that strain K1M also carried two copies of ALD6. The data obtained for VL1 and BC are consistent with those of previous studies showing that although a majority of wine yeast strains have an approximatively diploid DNA content (as VL1) (3), these strains may be aneuploids and frequently carry several homologous chromosomes of different sizes (2, 20).
FIG. 1.
Southern blot analysis of chromosomes of wild-type (WT) and ald6 GPD VL1, K1M, and BC strains. Chromosomes were separated by pulsed-field electrophoresis. Membranes were hybridized with an ALD6-specific oligonucleotide probe.
To delete ALD6 from the three strains, we used a modification of the short flanking homology PCR-based gene disruption method: the integrative fragments were flanked by large homologous regions to increase the recombination efficiency. Extensions of 154 bp and 239 bp were used, allowing the selection of several transformants with correct integration for each strain. Analysis of the transformants obtained after one round of transformation was consistent with the presence of two ALD6 genes in each strain, with only one being deleted (data not shown). After excision of the KanMX module, a second transformation step was used to generate the double deletion mutants. The absence of any ALD6 open reading frame in the mutants was confirmed by Southern blotting with chromosomes (Fig. 1).
Growth and fermentation of the engineered industrial strains.
The ald6 GPD1 strains were obtained by transformation of the ald6 null mutants with pVT100U-ZEO-GPD1. Growth and fermentation kinetics of the three series of four strains (each series being wild-type, GPD1, ald6, and ald6 GPD1 strains) (Table 1) were monitored during standard wine fermentation on MS medium.
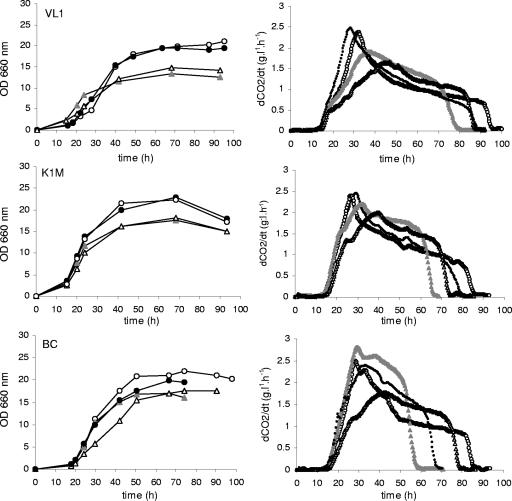
During wine fermentation, growth typically becomes rapidly limited by the low nitrogen concentration in grape must. Therefore, wine yeast strains exhibit a short exponential growth phase followed by stationary phase, during which most of the sugars are consumed (Fig. 2). Overexpression of GPD1 may alter the cell number, depending on the level of acetaldehyde accumulated during the growth phase, with this effect being directly proportional to the level of glycerol formed (24). Indeed, the numbers of VL1 GPD1 and K1M GPD1 cells were lower than control strain numbers. Deletion of ALD6 did not affect growth, confirming previous data for the wine yeast-derived strain V5 (26). In contrast, there were substantial differences in fermentation rates between ald6 GPD1 and GPD1 strains. Strains overexpressing GPD1 had higher fermentation rates during stationary phase, resulting in a shorter fermentation. In contrast, the duration of fermentation was longer than that of the wild type for ald6 strains. Interestingly, the combination of GPD1 overexpression and ALD6 deletion resulted in an intermediate profile, with a duration of fermentation similar to that of the wild-type strains, despite a lower maximal fermentation rate.
FIG. 2.
Growth and fermentation rate of the wine yeast strains VL1, K1M, and BC (•) and the corresponding engineered ald6 (○), GPD1 (▴), and ald6 GPD1 (▵) strains. Fermentation experiments were performed on MS medium at a 1.1-liter scale under conditions simulating wine fermentation. One representative of three independent fermentation experiments is shown. OD, optical density.
Impact on the main fermentation by-products.
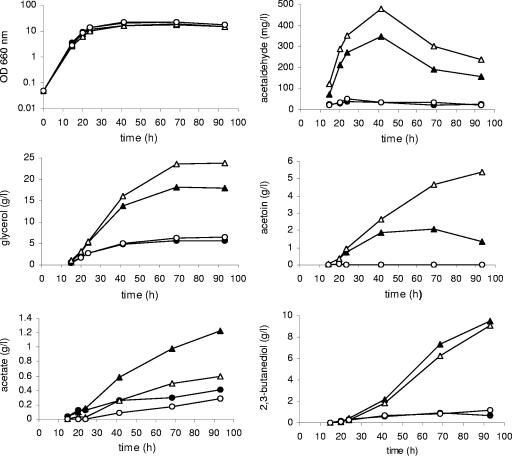
The concentration of the main fermentation by-products was measured after complete sugar exhaustion (Table 2). The biosynthesis of acetate by the three ald6 GPD1 strains was low, similar to that of wild-type strains. The production of succinate by ald6 GPD1 strains was slightly higher than that by GPD1 strains, indicating that the flux towards the tricarboxylic acid cycle was not greatly increased in these strains. Surprisingly, the ald6 GPD1 strains produced significantly more glycerol than did the GPD1 strains. Ethanol production by ald6 GPD1 strains was much lower than that in GPD1 strains, consistent with the greater glycerol production. The production of acetaldehyde and derived metabolites (Table 2) was also monitored throughout fermentation for the set of strains constructed in the K1M background (Fig. 3). Acetaldehyde production was markedly higher in the GPD1 strains than in the control strains and was even higher in the ald6 GPD1 strains, consistent with the limitation of ACDH. This was accompanied by higher levels of acetoin and 2,3-butanediol production. The ald6 GPD1 strains accumulated higher levels of acetoin than the GPD1 strains but were unable to convert this surplus into 2,3-butanediol.
TABLE 2.
Main fermentation by-products formed by engineered wine yeast strains during fermentation on MS medium
| Strain | Concn of:
|
||||||
|---|---|---|---|---|---|---|---|
| Glycerol (g liter−1) | Ethanol (g liter−1) | Acetate (g liter−1) | Succinate (g liter−1) | Acetaldehyde (mg liter−1) | Acetoin (g liter−1)a | Butanediol (g liter−1) | |
| VL1 | 6.5 | 97.2 | 0.57 | 0.31 | 16 | ND | 1.14 |
| ald6 | 7.4 | 96.6 | 0.19 | 0.29 | 8 | ND | 1.30 |
| GPD1 | 24.4 | 80.2 | 2.64 | 0.48 | 105 | 5.4 | 4.01 |
| GPD1 ald6 | 26.8 | 74.7 | 0.62 | 0.62 | 182 | 6.2 | 6.05 |
| K1M | 5.8 | 93.3 | 0.38 | 0.31 | 39 | ND | 0.61 |
| ald6 | 6.8 | 93.6 | 0.10 | 0.43 | 18 | ND | 0.52 |
| GPD1 | 18.2 | 82.6 | 0.98 | 0.78 | 183 | 4.2 | 4.85 |
| GPD1 ald6 | 21.9 | 78.8 | 0.43 | 0.88 | 228 | 5.8 | 3.93 |
| BC | 7.2 | 92.4 | 0.38 | 0.37 | 9 | ND | 1.45 |
| ald6 | 7.1 | 90.4 | 0.05 | 0.64 | 10 | ND | 0.99 |
| GPD1 | 16.5 | 82.7 | 1.36 | 0.58 | 95 | 2.9 | 3.9 |
| GPD1 ald6 | 26.9 | 75.5 | 0.50 | 0.74 | 320 | 9.5 | 5.09 |
ND, not detected.
FIG. 3.
By-product concentrations in fermentations with the strains K1M (•), K1M GPD1 (▴), K1M ald6 (○), and K1M ald6 GPD1 (▵) at various time points. Fermentation conditions are described in the legend of Fig. 2. OD, optical density.
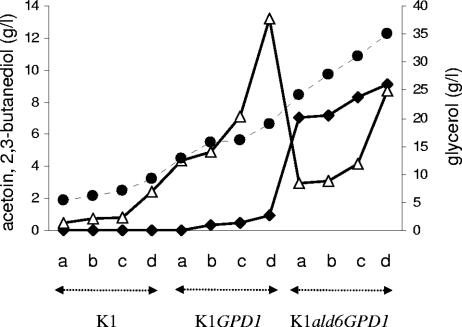
To further understand the relationships between the production of glycerol, acetoin, and 2,3-butanediol, a large range of glycerol production levels was generated by cultivating the strains K1M, K1M GPD1, and K1M ald6 GPD1 on MS medium with initial glucose contents of 140, 170, 200, and 250 g/liter (Fig. 4). V5 ald6 GPD1 clearly produced higher levels of acetoin and lower levels of 2,3-butanediol than did V5 GPD1. Interestingly, the production of these two compounds was related to the level of glycerol produced. The production of acetoin increased progressively as a function of glycerol concentration and then sharply increased for glycerol production to higher than 16 g/liter, while the 2,3-butanediol concentration markedly decreased. Although factors other than the level of glycerol might contribute to this effect, since different strains were used in this experiment, these data strongly suggest that the 2,3-butanediol dehydrogenase reaction becomes suddenly limited above a certain level of production of glycerol.
FIG. 4.
Glycerol (•), acetoin (⧫), and 2,3-butanediol (▵) production by the strains K1M, K1M GPD1, and K1M ald6 GPD1 grown on MS medium containing 140 (a), 170 (b), 200 (c), and 250 (d) g/liter glucose.
DISCUSSION
We show that the deletion of ALD6 efficiently reduces acetate production both by wild-type industrial wine yeasts and by engineered strains in which the carbon flux has been strongly shifted towards glycerol. Interestingly, the production of glycerol was increased as a result of the ALD6 deletion, and this effect was particularly clear when ALD6 was deleted from GPD1 strains. The high glycerol production, which in turn results in the production of NAD+, is surprising, since Ald6p preferably uses NADPH (11, 36); the deletion of ALD6 was therefore expected to result in an NADPH shortage. We show here that the redistribution of carbon in the GPD1 ald6 strains involves mainly acetaldehyde accumulation and increased flux through the acetoin-butanediol pathway (Fig. 5), consistent with the limitation of both alcohol dehydrogenase and ACDH reactions. Since the synthesis of acetaldehyde and acetoin from glucose results in a net production of 2 mol NADH and the synthesis of 2,3-butanediol generates 1 mol NADH, glycerol production might be increased to balance this NADH surplus. Indeed, the amount of NADH generated by the surplus of acetaldehyde, acetoin, and 2,3-butanediol in GPD1 ald6 compared to GPD1 strains is similar to the amount of NAD+ generated by the excess of glycerol (data not shown).
FIG. 5.
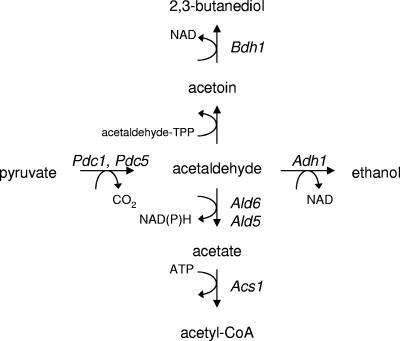
Pathways for acetaldehyde metabolism in Saccharomyces cerevisiae. The main isoforms active during glucose fermentation are indicated. Pdc1 and Pdc5, pyruvate decarboxylase; Adh1, alcohol dehydrogenase; Ald6, Ald5, acetaldehyde dehydrogenase; Acs1, acetyl coenzyme A (CoA) synthase; Bdh1, butanediol dehydrogenase; TPP, thiamine pyrophosphate.
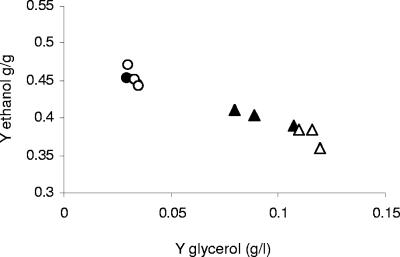
An inverse correlation was clearly shown between the ethanol and glycerol yields of the strains (Fig. 6). The ethanol yield of the ald6 GPD1 strains was the lowest, 15 to 20% lower than that of the wild type.
FIG. 6.
Ethanol yield as a function of glycerol yield. Strains K1M (•), K1M GPD1 (▴), K1M ald6 (○), and K1M ald6GPD1 (▵) were cultivated under fermentation conditions described in the legend of Fig. 2.
While GPD1 overexpression per se largely affects central metabolism, in particular at the acetaldehyde node (15, 16, 17, 24), additional changes are triggered by the ALD6 deletion. The production of acetaldehyde of ald6 GPD1 strains was higher than that of GPD1 strains (up to 300 mg/liter instead of 50 to 150 mg/liter) but remained in the upper range of the concentrations found in wine (28). The concentration of 2,3-butanediol can reach 1.3 g/liter in some wines and is not expected to appreciably affect the sensory qualities of wine. In contrast, acetoin levels higher than the detection threshold (150 mg/liter) are undesirable in table wines (29). The reduction of acetoin to 2,3-butanediol is catalyzed by the butanediol dehydrogenase Bdh1p, which uses NADH as a preferred cofactor (9, 10). We show in this study that this reaction was less efficient in ald6 GPD1 strains than in GPD1 strains, resulting in the accumulation of considerable amounts of acetoin. Furthermore, the efficiency of this reaction seems to be directly correlated to the amount of glycerol produced. Since the production of 2,3-butanediol from acetoin consumes 1 mol of NADH, acetoin accumulation may therefore be due to the low availability of NADH in ald6 GPD1 strains, as they produce more glycerol than GPD1 strains. Alternatively, it is possible that butanediol dehydrogenase becomes rate limiting in ald6 GPD1 strains.
In summary, these results highlight the great potential of yeast strains overexpressing GPD1 and producing ALD6 for producing low-alcohol wines. An alcohol content at least 2% (vol/vol) lower might be expected in wines produced using a yeast strain overproducing around 20 g/liter glycerol. However, accumulation of acetoin limits the extent to which carbon flux can be diverted to glycerol. Further improvement of these strains requires new efforts to minimize the formation of undesirable compounds, in particular at the acetaldehyde branch point.
Acknowledgments
We thank Marc Perez for help with gas chromatography analyses.
REFERENCES
- 1.Bely, M., J. M. Sablayrolles, and P. Barre. 1990. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in enological conditions. J. Ferm. Bioeng. 70:246-252. [Google Scholar]
- 2.Bidenne, C., B. Blondin, S. Dequin, and F. Vezinhet. 1992. Analysis of the chromosomal DNA polymorphism of wine strains of Saccharomyces cerevisiae. Curr. Genet. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Bradbury, J. E., K. D. Richards, H. A. Niederer, S. A. Lee, P. Dunbar, and R. C. Gardner. 2005. A homozygous diploid subset of commercial wine yeast strains. Antonie Leeuwenhoek 3:1-12. [DOI] [PubMed] [Google Scholar]
- 4.Church, G., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Barros Lopes, M., A. Rehman, H. Gockowiak, A. J. Heinrich, P. Langridge, and P. A. Henschke. 2000. Fermentation properties of a wine yeast overexpressing the Saccharomyces glycerol 3-phosphate dehydrogenase (GPD2). Aust. J. Grape Wine Res. 6:208-215. [Google Scholar]
- 6.Dequin, S. 2001. The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl. Microbiol. Biotechnol. 56:577-588. [DOI] [PubMed] [Google Scholar]
- 7.Eglinton, J. M., A. J. Heinrich, A. P. Pollnitz, P. Langridge, P. A. Henschke, and M. de Barros Lopes. 2002. Decreasing acetic acid accumulation by a glycerol overproducing strain of Saccharomyces cerevisiae by deleting the ALD6 aldehyde dehydrogenase gene. Yeast 19:295-301. [DOI] [PubMed] [Google Scholar]
- 8.Eustace, R., and R. J. Thornton. 1986. Selective hybridization of wine yeast for higher yields of glycerol. Can. J. Microbiol. 33:112-117. [Google Scholar]
- 9.Gonzalez, E., M. R. Fernandez, C. Larroy, L. Sola, M. A. Pericas, X. Pares, and J. A. Biosca. 2000. Characterization of a (2R,3R)-2,3-butanediol dehydrogenase as the Saccharomyces cerevisiae YAL060W gene product. Disruption and induction of the gene. J. Biol. Chem. 275:35876-35885. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez, E., M. R. Fernandez, C. Larroy, X. Pares, and J. A. Biosca. 2001. Characterization and functional role of Saccharomyces cerevisiae 2,3-butanediol dehydrogenase. Chem. Biol. Interact. 130-132:425-434. [DOI] [PubMed] [Google Scholar]
- 11.Grabowska, D., and A. Chelstowska. 2003. The ALD6 gene product is indispensable for providing NADPH in yeast cells lacking glucose-6-phosphate dehydrogenase activity. J. Biol. Chem. 278:13984-13988. [DOI] [PubMed] [Google Scholar]
- 12.Güldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundquist, F. 1974. Acetaldehyd: Bestimmung mit Aldehyd Dehydrogenase, p. 1509-1513. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis. Academic Press, Inc., New York, N.Y.
- 14.Meaden, P. G., F. M. Dickinson, A. Mifsud, W. Tessier, J. Westwater, H. Bussey, and M. Midgley. 1997. The ALD6 gene of Saccharomyces cerevisiae encodes a cytosolic, Mg2+-activated acetaldehyde dehydrogenase. Yeast 13:1319-1327. [DOI] [PubMed] [Google Scholar]
- 15.Michnick, S., J. L. Roustan, F. Remize, P. Barre, and S. Dequin. 1997. Modulation of glycerol and ethanol yields during alcoholic fermentation in Saccharomyces cerevisiae strains overexpressed or disrupted for GPD1 encoding glycerol-3-phosphate dehydrogenase. Yeast 13:783-793. [DOI] [PubMed] [Google Scholar]
- 16.Nevoigt, E., and U. Stahl. 1996. Reduced pyruvate decarboxylase and increased glycerol-3-phosphate dehydrogenase [NAD+] levels enhance glycerol production in Saccharomyces cerevisiae. Yeast 12:1331-1337. [DOI] [PubMed] [Google Scholar]
- 17.Nevoigt, E., R. Pilger, E. Mast-Gerlach, U. Schmidt, S. Freihammer, M. Eschenbrenner, L. Garbe, and U. Stahl. 2002. Genetic engineering of brewing yeast to reduce the content of ethanol in beer. FEMS Yeast Res. 22:225-232. [DOI] [PubMed] [Google Scholar]
- 18.Noble, A. C., and G. F. Bursick. 1984. The contribution of glycerol to perceived viscosity and sweetness in white wine. Am. J. Enol. Vitic. 35:110-112. [Google Scholar]
- 19.Pretorius, I. S. 2000. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675-729. [DOI] [PubMed] [Google Scholar]
- 20.Puig, S., A. Querol, E. Barrio, and J. E. Perez-Ortin. 2000. Mitotic recombination and genetic changes in Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 66:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachidi, N., P. Barre, and B. Blondin. 2000. Examination of the transcriptional specificity of an enological yeast. A pilot experiment on the chromosome-III right arm. Curr. Genet. 37:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Rankine, B. C., and D. A. Bridson. 1971. Glycerol in Australian wines and factors influencing its formation. Am. J. Enol. Vitic. 22:2-12. [Google Scholar]
- 23.Remize, F. 1999. Ph.D. thesis. University of Montpellier II, Montpellier, France.
- 24.Remize, F., J. L. Roustan, J. M. Sablayrolles, P. Barre, and S. Dequin. 1999. Glycerol overproduction by engineered Saccharomyces cerevisiae wine yeast strains leads to substantial changes in by-product formation and to a stimulation of fermentation rate in stationary phase. Appl. Environ. Microbiol. 65:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remize, F., J. M. Sablayrolles, and S. Dequin. 2000. Re-assessment of the influence of yeast strain and environmental factors on glycerol production in wine. J. Appl. Microbiol. 88:371-378. [DOI] [PubMed] [Google Scholar]
- 26.Remize, F., E. Andrieu, and S. Dequin. 2000. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl. Environ. Microbiol. 66:3151-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remize, F., L. Barnavon, and S. Dequin. 2001. Glycerol export and glycerol 3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting steps for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 3:301-312. [DOI] [PubMed] [Google Scholar]
- 28.Romano, P., G. Suzzi, L. Turbanti, and M. Polsinelli. 1994. Acetaldehyde production in Saccharomyces cerevisiae wine yeasts. FEMS Microbiol. Lett. 118:213-218. [DOI] [PubMed] [Google Scholar]
- 29.Romano, P., G. Suzzi, V. Brandolini, E. Menziani, and P. Domizio. 1996. Determination of 2,3-butanediol in high and low acetoin producers of Saccharomyces cerevisiae wine yeasts by automated multiple development (AMD). Lett. Appl. Microbiol. 22:299-302. [DOI] [PubMed] [Google Scholar]
- 30.Sablayrolles, J. M., P. Barre, and P. Grenier. 1987. Design of laboratory automatic system for studying alcoholic fermentations in anisothermal enological conditions. Biotechnol. Technol. 1:181-184. [Google Scholar]
- 31.Saint-Prix, F., L. Bönquist, and S. Dequin. 2004. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p play a major role in acetate formation. Microbiology 150:2209-2220. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact cells using single stranded nucleic acid as carrier. Curr. Genet. 16:339-446. [DOI] [PubMed] [Google Scholar]
- 34.Taherzadeh, M. J., L. Adler, and G. Liden. 2002. Strategies for enhancing fermentative production of glycerol—a review. Enz. Microb. Technol. 31:53-66. [Google Scholar]
- 35.Vezinhet, F., J. N. Hallet, and B. Blondin. 1990. Chromosomal DNA patterns and mitochondrial polymorphisms as tools for identification of enological strains of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 32:568-571. [Google Scholar]
- 36.Wang, X., C. J. Mann, Y. Bai, L. Ni, and H. Weiner. 1998. Molecular cloning, characterization, and potential roles of cytosolic and mitochondrial aldehyde dehydrogenases in ethanol metabolism in Saccharomyces cerevisiae. J. Bacteriol. 180:822-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Z. X., J. Zhuge, H. Fang, and B. Prior. 2001. Glycerol production by microbial fermentation: a review. Biotechnol. Adv. 19:201-223. [DOI] [PubMed] [Google Scholar]