Abstract
The distribution and colonization levels of the altered Schaedler flora (ASF) in their natural hosts are poorly understood. Intestinal colonization levels of the eight ASF strains in outbred Swiss Webster mice with or without Helicobacter hepaticus infection were characterized by real-time quantitative PCR. All ASF strains were detected in the cecum and colon, but some strains displayed significant variation in colonization levels with host age, gender, and H. hepaticus infection status.
The flora of the gastrointestinal tract (GIT) in mammals is highly complex and diverse. It has been estimated that there are approximately 1012 viable bacteria per g of large bowel content in humans, with the presence of at least 400 to 500 species (1, 10, 17). Dynamic changes in the composition of the normal GIT microbial species in immunocompetent hosts during aging, between genders, and after experimental infection with microbial pathogens are not well understood. This is largely due to the complexity of the microbiota and the limitation of widely used culture-based techniques. Therefore, gnotobiotic animals colonized with defined microbiota became a valuable tool for exploring microbe-host interactions (7). In the mid-1980s, a defined microbiota, namely, the altered Schaedler flora (ASF), consisting of eight murine bacterial species, was developed as a minimum set of organisms sufficient to establish normal physiological functions in the GIT (4, 11, 16). Phylogenetic comparisons based on 16S rRNA gene sequences reveal that the ASF includes two aerotolerant lactobacillus strains, ASF360 and ASF361; two Clostridium sp. strains, ASF356 and ASF502; Eubacterium sp. strain ASF492; Bacteroides sp. strain ASF519; low-G+C-content gram-positive bacterial strain ASF500; and strain ASF457, which clusters with the Flexistipes species (3). We recently developed 16S rRNA gene-based real-time quantitative PCR (QPCR) protocols for discrimination of all eight ASF strains and demonstration of their spatial distribution and stability in the GITs of gnotobiotic mice (C.B-17 SCID mice with ASF as their only flora) (15). In this study, we analyzed the natural distribution and colonization dynamics of the ASF and tabulated ASF colonization parameters in the GIT concurrently infected with the murine enteric pathogen Helicobacter hepaticus in the GITs of Swiss Webster (SW) mice which were used previously (5).
Four- to 6-week-old mice free of known murine viruses, Helicobacter spp., and parasites were obtained from Taconic Farms (Germantown, NY). Thirty male and 30 female SW mice were divided into six groups of 10 mice (either male or female) and maintained in static microisolator cages in an Association for Accreditation and Assessment of Laboratory Animal Care International-accredited facility and fed a diet (ProlabRMH3000) from PMI Nutrition International (Richmond, IN). The mice were inoculated with wild-type (WT) H. hepaticus or a cdtB-deficient H. hepaticus mutant or sham dosed with brucella broth as a control. Mice received 0.2 ml of a fresh inoculum (∼2 × 108 organisms) by gastric gavage every other day for three inoculations. This H. hepaticus mutant contains a minitransposon-mediated mutation within the cdtB gene coding for a catalytic subunit of cytolethal distending toxin; the mutant lacked cytolethal distending toxin activity (5). Five male and five female mice from each group were euthanized at 8 weeks postinfection (wpi) (15 weeks of age) and 16 wpi (23 weeks of age), respectively. Immediately after euthanasia with CO2, contents in the intestines were removed by rinsing with sterile saline. One-centimeter segments of the jejunum, ileum, cecum, and colon for RNA and DNA isolation were collected and snap-frozen in liquid nitrogen immediately after sampling and stored at −70°C prior to use. Total DNA from the harvested intestinal samples was isolated with Trizol Reagents by following the manufacturer's recommendations (Invitrogen).
In order to enumerate the numbers of bacteria of eight ASF strains in the intestines of SW mice by QPCR, genomic DNAs from cultured ASF bacteria were used to generate standard curves of six 10-fold dilutions, ranging from 106 to 10 pg. QPCR was performed with a Prism Sequence Detection Systems 7700 (Applied Biosystems, Foster City, CA) as described previously (15). Subsequently, the quantities of the bacterial DNA were converted into copy numbers of the respective ASF genomes on the basis of their 16S rRNA gene copies. The copy numbers of the ASF genomes were then normalized to micrograms of mouse chromosomal DNA whose quantities in the samples were measured by QPCR with the 18S rRNA gene-based primers and probe mixture (Applied Biosystems). Data on the levels of H. hepaticus were analyzed among multiple groups by the Kruskal-Wallis test and between two groups by a two-tailed t test for normally distributed data or a Mann-Whitney U test for abnormally distributed data. The normality of the data sets was analyzed with the Kolmogorov-Smirnov test. Values of P < 0.05 were considered significant.
The numbers of 16S rRNA gene copies of six ASF strains (ASF356, ASF360, ASF361, ASF457, ASF492, and ASF519) were estimated previously by Southern blotting or QPCR (15). For determination of the numbers of 16S rRNA gene copies within ASF500 and ASF502, approximately 1 μg of genomic DNA from ASF strain 500 or 502 was digested with BamHI, EcoRI, or HindIII overnight, separated on a 1% agarose gel, and then transferred to a nylon membrane. For preparing a hybridization probe, the 262-bp or 405-bp PCR fragment was amplified from the 16S rRNA gene of strain ASF500 or ASF502, respectively, with the specific primers as described previously (15). The probe was labeled with horseradish peroxidase with a direct nucleic acid labeling and detection system, and hybridization was performed by following the manufacturer's recommendations (Amersham Biosciences, Piscataway, NJ). Southern blotting indicated that strains ASF500 and ASF502 contained one and two copies of the 16S rRNA gene, respectively (data not shown).
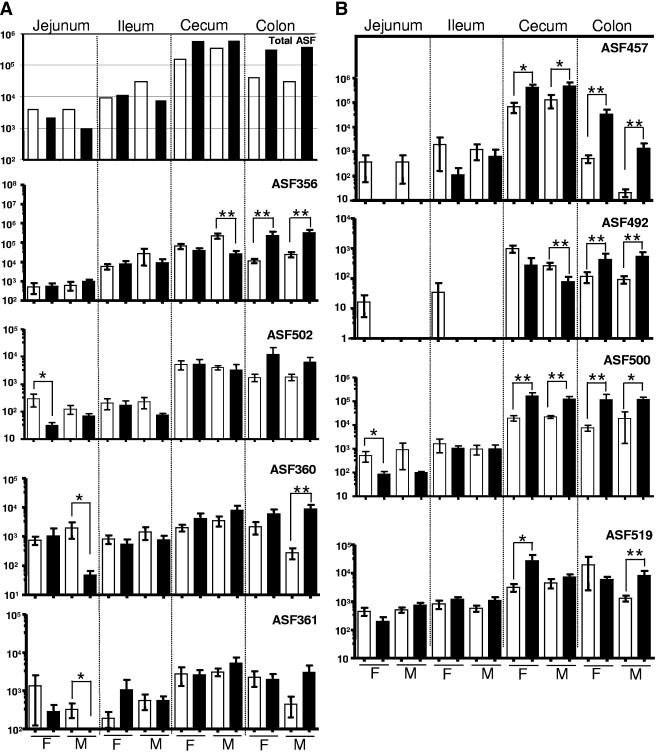
Our data demonstrated that all eight ASF strains persistently colonized the ceca and colons of SW mice; six ASF strains (except for strains ASF457 and ASF492) were also present in their jejuna and ilea. The total number of mucosa-associated ASF strains in SW mice increased from the small intestine (103 to 104) to the large bowel (104 to 106) at both 15 and 23 weeks of age (Fig. 1). Between 15 and 23 weeks of age, the total numbers of ASF bacteria increased in the ceca (∼5-fold) and colons (∼10-fold) whereas the level of ASF bacteria slightly decreased in the jejuna and ilea; these results were due to the increase in several ASF strains, such as ASF457, ASF492, and ASF356, in the large intestine during aging or the significant reduction or elimination of some ASF strains, such as ASF457 and ASF492, in the small intestine. Given that ∼1 μg of mouse DNA was isolated from 1 mg of wet intestinal tissue with Trizol reagent (unpublished data), the total number of ASF bacteria per gram of wet intestinal tissue was approximately 106 in the jejunum, 107 in the ileum, >109 in the cecum, and 108 in the colon at 23 weeks of age. This trend is consistent with our previous observation in germ-free C.B-17 SCID mice colonized with ASF strains (5 × 106 bacteria/g in the small intestine, ∼1010 bacteria/g in the cecum, and >108 bacteria/g in the colon) (15). These data suggest that ASF bacteria may be used as indicator organisms for monitoring dynamic changes in the overall microflora in the GITs of experimental mice.
FIG. 1.
Eight ASF strains persistently colonize the intestines of sham-dosed SW mice as quantified by QPCR. Data are presented as means ± standard errors. Values represent the number of copies of each ASF genome per microgram of mouse DNA. Open (□) and closed (▪) bars represent mice euthanized at 15 and 23 weeks of age, respectively. F, female SW mice; M, male SW mice; *, P < 0.05; **, P < 0.008.
Previous studies reported only part of the ASF flora present in the mouse GIT (2, 14); however, these were based on sequencing of denaturing gradient gel electrophoresis (DGGE) fragments and PCR-generated clones, and these techniques have a low probability of detection of low-frequency species. For example, only strains ASF360 (L. murinus) and ASF519 (Bacteroides sp.) were present in the distal colons of inbred 129 Sv/Ev mice when being characterized by sequencing of the 16S rRNA gene fragments derived by PCR-DGGE (2). Nonetheless, Salzman et al. identified four members of the ASF in the cloned microbial 16S rRNA gene sequences generated from the GITs of FVB mice, including strains ASF360 and ASF519 in the large intestine, strain ASF361 in the small intestine, and strain ASF502 in both the cecum and the large intestine (14). The detection of all ASF species by QPCR thus confirms that these species are part of the natural murine GIT flora, albeit some at relatively low proportions of the total flora (6, 8, 15). In addition, the use of mouse strains with various genetic backgrounds and different vendor sources and types of diets could also contribute to differences in the colonization status of ASF in the murine GIT (4).
Ages of mice influence colonization dynamics or levels of certain ASF strains (Fig. 1). Between 15 and 23 weeks of age, the levels of strains ASF356 and ASF492 in the colon and of strains ASF457 and ASF500 in both the colon and the cecum significantly increased (P < 0.05 or 0.008). In contrast, the levels of strain ASF457 in the jejunum and strain ASF492 in the small intestine significantly decreased between these two time points, whereas strain ASF492 was not detected in the small intestine at 23 weeks of age. Also, alteration of the colonization levels of some ASF strains during aging was found to be gender dependent. The cecal levels of strains ASF356 and ASF492 and the jejunal levels of strains ASF360 and ASF361 in males and the jejunal levels of strains ASF500 and ASF502 in females were significantly higher at 15 weeks of age than at 23 weeks of age (P < 0.05). In addition, Bacteroides sp. strain ASF519 in the ceca of female mice (P < 0.016) and the colons of male mice (P < 0.008) significantly increased between 15 and 23 weeks of age. These results suggest that mouse age and gender play an import role in the colonization levels of some of ASF strains in the intestinal tracts of SW mice.
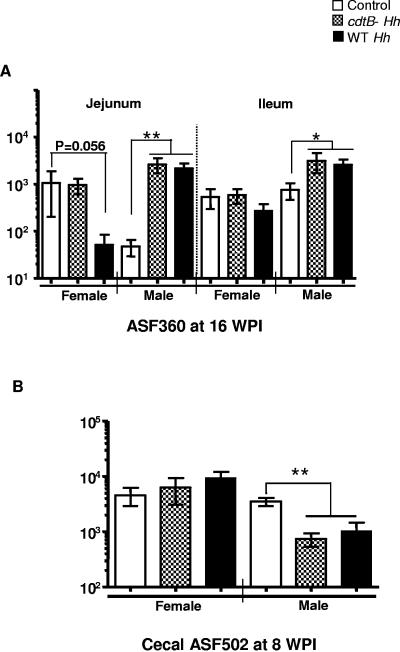
We previously demonstrated that WT H. hepaticus colonized the jejunum, ileum, cecum, and colon at similar levels at 8 wpi between male and female SW mice; however, the level of H. hepaticus in the ceca and colons of females mice was less by ∼1,000-fold than in the male mice at 16 wpi (5). In addition, the cdtB-deficient H. hepaticus mutant was able to transiently colonize the intestines of male SW mice through 8 wpi but was cleared at 16 wpi; in contrast, this mutant did not colonize the intestines of female SW mice (5). Infection with WT H. hepaticus or the cdtB-deficient H. hepaticus mutant led to an approximately 10-fold increase in strain ASF360 over the sham-dosed controls in the jejuna and ilea of 23-week-old male mice at 16 wpi (Fig. 2A). In contrast, cecal levels of strain ASF502 were decreased by infection with WT H. hepaticus or cdtB-deficient H. hepaticus mutant in male SW mice (15 weeks old) (P < 0.008 for the mutant and <0.016 for WT H. hepaticus 3B1) by 8 wpi (Fig. 2B). H. hepaticus infection had a minimal effect on the colonization level or distribution of strains ASF356, ASF361, ASF457, ASF492, ASF500, and ASF519 along the intestinal tract (data not shown).
FIG. 2.
Influence of H. hepaticus infection on colonization by ASF strains ASF360 and ASF502. Data are presented as means ± standard errors. Control, sham-dosed mice; cdtB- Hh, a cdtB-deficient H. hepaticus mutant; WT Hh, WT H. hepaticus strain 3B1; *, P < 0.05; **, P < 0.008.
Interestingly, Lactobacillus sp. strain ASF360, but not Lactobacillus murinus ASF361, increased in response to H. hepaticus infection. Recently, it has been reported that the Lactobacillus Acidophilus group, which is phylogenetically related to strain ASF360 (3), was missing from the DGGE profiles of germfree, interleukin-10 (IL-10)-deficient mice which developed severe colitis 3 and 6 weeks after microbial exposure; in contrast, L. murinus ASF361 was identified in both the IL-10-deficient and wild-type mice (on an inbred 129 Sv/Ev background) (2). In addition, approximately 10% of murine Lactobacillus isolates had an inhibitory effect on tumor necrosis factor alpha production by lipopolysaccharide-stimulated mouse macrophages (12). Experimental inoculation of selected probiotic lactobacilli attenuated pathogenic or commensal intestinal microbe-induced typhlocolitis in several IL-10-deficient mouse strains (9, 13). Whether the increased levels of strain ASF360 in H. hepaticus-infected SW mice, which have no demonstrable intestinal pathology, plays a probiotic role requires further investigation.
Overall, H. hepaticus infection had a limited impact on colonization levels of ASF strains. This is consistent with the observation that H. hepaticus infection in SW mice did not cause enteritis. One possible explanation for this result is that immunocompetent, outbred SW mice have the ability to immunologically respond to H. hepaticus infection and simultaneously to sustain homeostasis of commensal intestinal microbiota. This explanation is supported by our recent finding that the levels of strains ASF361, ASF500, and ASF502 were significantly increased in the colons of T-cell receptor β-deficient mice with H. hepaticus-induced typhlocolitis (unpublished observations). However, this study did not rule out the possibility that H. hepaticus infection affected the distribution and colonization of other non-ASF intestinal microbes. Recently, Kuehl et al. demonstrated that H. hepaticus colonization was associated with a decrease in the overall diversity of the microbial community in the ceca of C57BL/6 mice (8a).
In summary, we have characterized the colonization dynamics of eight ASF strains in the intestinal tracts of outbred SW mice by QPCR. This molecular approach overcomes culture-dependent limitations and provides a powerful tool for quantitatively dissecting bacterium-host interactions with high resolution. In addition, the information obtained in this study regarding the influence of age, gender, and H. hepaticus infection on the distribution and colonization levels of the respective ASF strains will be useful in designing in vivo experiments for elucidating the roles of ASF strains in mice challenged with enteric pathogens or subjected to other environmental stimuli.
Acknowledgments
We thank Kathleen Cormier, Erinn Stefanich, Jeff Bajko, Vivian, Kristen Clapp, and Amy Lee for technical assistance.
This study was supported in part by NIH grants R01 CA67529 (J.G.F.), P01 CA26731 (J.G.F. and D.B.S), P30ES02109, and AI50952 (J.G.F., D.B.S., and M.F.P.).
REFERENCES
- 1.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 2.Bibiloni, R., M. A. Simon, C. Albright, B. Sartor, and G. W. Tannock. 2005. Analysis of the large bowel microbiota of colitic mice using PCR/DGGE. Lett. Appl. Microbiol. 41:45-51. [DOI] [PubMed] [Google Scholar]
- 3.Dewhirst, F. E., C.-C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubos, R., R. W. Schaedler, R. Costello, and P. Hoet. 1965. Indigenous, normal, and autochthonous flora of the gastrointestinal tract. J. Exp. Med. 122:67-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge, Z., Y. Feng, M. T. Whary, P. R. Nambiar, S. Xu, V. Ng, N. S. Taylor, and J. G. Fox. 2005. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect. Immun. 73:3559-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge, Z., D. A. White, M. T. Whary, and J. G. Fox. 2001. Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus. J. Clin. Microbiol. 39:2598-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon, H. A., and L. Pesti. 1971. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol. Rev. 35:390-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huijsdens, X. W., R. K. Linskens, M. Mak, S. G. Meuwissen, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2002. Quantification of bacteria adherent to gastrointestinal mucosa by real-time PCR. J. Clin. Microbiol. 40:4423-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Kuehl, C. J., H. D. Wood, T. L. Marsh, T. M. Schmidt, and V. B. Young. 2005. Colonization of the cecal mucosa by Helicobacter hepaticus impacts the diversity of the indigenous microbiota. Infect. Immun. 73:6952-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy, J., L. O'Mahony, L. O'Callaghan, B. Sheil, E. E. Vaughan, N. Fitzsimons, J. Fitzgibbon, G. C. O'Sullivan, B. Kiely, J. K. Collins, and F. Shanahan. 2003. Double blind, placebo controlled trial of two probiotic strains in interleukin 10 knockout mice and mechanistic link with cytokine balance. Gut 52:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orcutt, R. P., F. J. Gianni, and R. J. Judge. 1987. Development of an “altered Schaedler flora” for NCI gnotobiotic rodents. Microecol. Ther. 17:59. [Google Scholar]
- 12.Pena, J. A., S. Y. Li, P. H. Wilson, S. A. Thibodeau, A. J. Szary, and J. Versalovic. 2004. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl. Environ. Microbiol. 70:558-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pena, J. A., A. B. Rogers, Z. Ge, V. Ng, S. Y. Li, J. G. Fox, and J. Versalovic. 2005. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect. Immun. 73:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman, N. H., H. de Jong, Y. Paterson, H. J. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 15.Sarma-Rupavtarm, R. B., Z. Ge, D. B. Schauer, J. G. Fox, and M. F. Polz. 2004. Spatial distribution and stability of the eight microbial species of the altered Schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 70:2791-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaedler, R. W., R. Dubos, and R. Costello. 1965. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122:77-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon, G. L., and S. L. Gorbach. 1984. Intestinal flora in health and disease. Gastroenterology 86:174-193. [PubMed] [Google Scholar]