Abstract
Investigations of the surface characteristics of Flavobacterium psychrophilum, an important pathogen of fish, assisted us in identifying a surface protein termed P18. In the current study, we developed a simple and efficient procedure for the purification of this protein by a two-step method. First, P18 was selectively released from flavobacteria by a heat-HEPES treatment of the cells and then subjected to anion-exchange high-performance liquid chromatography. De novo sequencing was used to generate a fragmented peptide spectrum from purified P18. Comparison of two obtained peptide sequences with a partial genome sequence of F. psychrophilum (INRA, Jouy-en-Josas, France) identified one gene encoding a 166-amino-acid OmpH-like protein that mostly likely undergoes N-terminal cleavage of the 23-residue signal peptide. The susceptibility of the OmpH-like protein to proteinase K treatment and the bacteriostatic/bactericidal activities of anti-OmpH-like protein antibodies indicated that this protein is actually exposed on the surface of F. psychrophilum. Vaccination trials showed that the OmpH-like protein can induce a high titer of anti-OmpH-like protein antibodies which are protective. Taken together, these results suggest that this surface protein produced by F. psychrophilum could be used in future vaccine development as a promising candidate antigen.
Flavobacterium psychrophilum (syn. Cytophaga psychrophila or Flexibacter psychrophilus) is a gram-negative, motile, non-spore-forming bacterium (7). This eubacterium belongs to the family Flavobacteriaceae in the Cytophaga-Flavobacterium-Bacteroides (CFB) phylum (5, 6). F. psychrophilum is the etiologic agent of “rainbow trout fry syndrome” and “bacterial cold water disease” (CWD), the two most significant systemic infections of primarily freshwater-reared salmonid fish. These diseases cause serious problems in public and private aquaculture (for a review, see reference 35). F. psychrophilum probably affects all species of salmonid fish, but coho salmon (Oncorhynchus kisutch), rainbow trout (Oncorhynchus mykiss), and ayu (Plecoglossus altivelis) (17) appear to be particularly susceptible (23). In addition, F. psychrophilum has been isolated from diseased nonsalmonid fish species (17, 26).
The molecular pathogenesis of F. psychrophilum is not well understood. However, the virulence of this flavobacterium might be related to the production of exoenzymes and/or endotoxins causing direct tissue damage in fish or enhanced invasiveness (13, 29, 43, 44). Little knowledge about the surface immunogenic factors of F. psychrophilum exists. What is known are the structures of the lipopolysaccharide O antigen (28) and some surface components, such as sialic acid-binding lectin (33) or surface blebs (34), which have been shown (or suspected) to interact with the immune system of the host. No commercial vaccine is yet available, although several studies dealing with the effects of vaccination have been done. Vaccines based on whole formalin-killed cells (19), a Sarkosyl-insoluble membrane fraction (38), or distinct molecular mass fractions (25) of F. psychrophilum were shown to confer significant protection against disease. To date, the prevention of disease outbreaks and therapeutic treatments rely on the use of approved antibiotics. However, such an approach is limited by the cost of the treatments and the potential for resistant mutants (9, 18, 42).
The identification of the major immunogenic components of an infectious agent is essential for understanding the molecular mechanism of virulence and the route of the infection, for proposing serological diagnosis of the disease, and for developing strategies for efficient immune protection. Bacterial surface components and, more particularly, the outer membrane proteins are generally very immunogenic and play a key function in virulence and the immune responses to bacterial diseases. In this context, investigations of the surface architecture of F. psychrophilum led to the identification of several dominant membrane antigens (11, 12, 31, 38) which might be used as antigenic subunits for vaccines. Because of the important role that membrane proteins play in attachment of many pathogenic bacteria to their hosts, we hypothesized that flavobacterial outer membrane proteins are likely candidates for mediating the specific attachment of these pathogens to their fish tissues. Previously, we identified two proteins produced by F. psychrophilum, termed P60 (31) and P18 (30) according to their relative molecular masses. In this study, we report the purification and characterization of the immunogenic and protective 18-kDa outer membrane-associated OmpH-like protein. This surface protein seems to be a promising candidate for the development of a future vaccine.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and isolation of surface proteins.
The different F. psychrophilum strains used in this study and their origins are listed in Table 1. Cells were cultivated in a modified Anacker and Ordal's (AOAE) liquid medium, 0.5% (wt/vol) tryptone, 0.05% (wt/vol) yeast extract, 0.02% (wt/vol) beef extract, and 0.02% (wt/vol) sodium acetate (pH 7.2) (2). Bacteria were incubated at 18°C under aerobic conditions (orbital stirring, 150 rpm). When the bacteria were harvested (optical density at 600 nm, 0.8), purity was checked by examination of Gram-stained smears, and the cells were pelleted for 10 min at 6,000 × g at 4°C and washed twice with phosphate-buffered saline (PBS; 50 mM sodium phosphate buffer and 150 mM NaCl, pH 7.4). Isolation of surface proteins from F. psychrophilum was accomplished as previously described (30). Protein content was estimated in the presence of sodium dodecyl sulfate (SDS), using bovine serum albumin as the standard (27).
TABLE 1.
F. psychrophilum strains and related type strains used in this study
| F. psychrophilum strains | Source, origin |
|---|---|
| G4 | Rainbow trout (Oncorhynchus mykiss), Poitou-Charentes, France |
| PO388 | Rainbow trout, Bretagne, France |
| JIP02.86 | Rainbow trout, Aquitaine, France |
| JIP16.00 | Rainbow trout, Finistère, France |
| JIP07.99 | Rainbow trout, France (INRA) |
| 259-93 | Rainbow trout, Idaho |
| UCD95.74 | White sturgeon (Acipenser transmontanus), California |
| NCIMB 1947T | Coho salmon (Oncorhynchus kisutch), Washington |
| FPC839 | Ayu (Plecoglossus altivelis), Tokushima, Japan |
Chromatographic techniques.
Anion-exchange high-performance liquid chromatography (HPLC) was performed in an Amersham Biosciences HiTrap DEAE column (diameter, 0.7 cm; length, 2.5 cm), using a Pharmacia Biosystem GradiFrac system. The sample was 1 ml of HEPES extract. The flow rate was set at 0.5 ml/min, and the eluate was collected in 0.5-ml fractions. Buffer A was used for column equilibration and for the elution of proteins (10 mM HEPES-NaOH buffer [pH 8.0]-10 mM NaCl), and buffer B was buffer A with 1 M NaCl. Elution conditions were the following: (i) 0 to 10 min with 100% buffer A and (ii) 10 to 35 min with a linear gradient to 50% buffer B. The elution profile was spectrophotometrically monitored at 280 nm, and selected eluates were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
SDS-PAGE and immunoblotting.
SDS-PAGE analyses were performed as described by Laemmli (22). Protein samples were solubilized in a reduced SDS-PAGE sample buffer and separated in SDS-polyacrylamide gels (10 by 8 by 0.075 cm; 12.5% acrylamide and 0.26% bisacrylamide; 200 V, 1 h). Protein bands were stained with silver (45) or Coomassie brilliant blue R250 or were immunolabeled. Following electrophoresis, the proteins were electroblotted onto a nitrocellulose filter (Bio-Rad) by use of a Bio-Rad TransBlot electrophoretic transfer cell as specified by the manufacturer. The blots were blocked with 5% bovine serum albumin in PBS for 1 h at room temperature and then incubated for 1 h with rabbit antiserum directed against P18 purified from F. psychrophilum G4 (dilution, 1:1,000) in 0.05% Tween 20 in PBS. The washing steps were performed three times with PBS. Immunolabeled proteins were visualized using alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (IgG) antibodies (dilution, 1:2,000; DakoCytomation, Glostrup, Denmark), followed by a color reaction with 5-bromo-4-chloro-3-indolyl phosphate and 4-nitroblue tetrazolium chloride. Primary antibodies were elicited in two rabbits (female New Zealand White rabbits) by subcutaneous inoculation of HPLC-purified P18 twice a month over a 3-month period (inoculum, 50 μg of P18 in 0.5 ml of PBS, emulsified with 0.7 ml of Freund's incomplete adjuvant [or complete adjuvant for the two first inoculations]). The two sera were pooled at the end of the immunization schedule. When trout antisera from (i) CWD-convalescent trout taken from an infected French farm or (ii) experimentally immunized trout (see below) were used as primary antibodies, an additional incubation was done with rabbit anti-trout immunoglobulins (dilution, 1:2,000).
De novo P18 sequencing.
Purified P18 was electrophoresed and stained with Coomassie brilliant blue R250, and the spots were excised from a gel. After a washing step with H2O-methanol-acetic acid (47.5:47.5:5) and acetonitrile, the gel pieces were dried in a vacuum centrifuge and rehydrated in 8 ng/μl trypsin (Sigma-Aldrich, St. Louis, MO) in 50 mM NH4HCO3. After incubation and washing steps, peptide mixtures were analyzed by online capillary chromatography (C18 PepMap column [75-μm inner diameter by 15 cm]; LC Packings, Amsterdam, The Netherlands) coupled to nanospray LCQ ion-trap mass spectrometry (MS; ThermoFinnigan, San Jose, CA). The mass spectrometer was operated in positive-ion mode at a 2-kV needle voltage and a 46-V capillary voltage. Data acquisition was performed in a data-dependent mode alternating full-scan MS over the range m/z 50 to 2,000 and a MS/MS scan of the most intense ion in the precedent MS spectrum. MS/MS data were acquired using a 2-m/z-unit ion isolation window and a 35% relative collision energy. Every MS/MS spectrum was submitted to the DeNovoX sequencing program (ThermoFinnigan, San Jose, CA). Sequences with an absolute probability and relative probability higher than 20% and 75%, respectively, and consisting of more than four amino acids were selected for further analysis. Corresponding spectra were manually checked.
In situ protease treatment.
Intact mid-log-phase F. psychrophilum cells were washed once with PBS and dispersed in PBS with 10 mM MgCl2 to a final concentration of 2 × 109 bacteria/ml. Examination of bacterial suspensions by phase-contrast light microscopy did not indicate detectable lysis of the bacteria. Cells were then incubated with 40 μg/ml soluble proteinase K for 30 min or 1 h at 25°C, after which digestion was terminated by the addition of 5 mM phenylmethylsulfonyl fluoride followed by sample boiling. As a negative control, the above procedure was repeated except that cells were incubated in buffer without proteinase K. Cells were disrupted by sonication (24 W), and about 50 μg of proteins was prepared from each sample. Bacterial lysates were subjected to SDS-PAGE, and the proteins were transferred to nitrocellulose membranes. The susceptibilities of individual proteins to proteinase K digestion were assessed by immunoblotting with the appropriate rabbit polyclonal antibodies. To exclude the possibility of outer membrane damage, the lysates were also immunoblotted with antibodies directed against GldJ, a lipoprotein involved in the gliding mobility of Flavobacterium johnsoniae which is not exposed at the cell surface and is thus protected against protease digestion in intact bacteria (8). To assess the protease susceptibilities of the proteins when not in situ, an additional control was added. Cells were treated with proteinase K as described above in the presence of Triton X-100 to a final concentration of 0.05%, which disrupts F. psychrophilum membranes.
Growth inhibition by anti-P18 antibodies.
F. psychrophilum G4 was grown to mid-exponential phase, and a 10-μl aliquot (about 50 × 106 bacteria/ml) was placed in each well of a 96-well tissue culture dish (Techno Plastic Products, Trasadingen, Switzerland). Growth inhibition tests were performed by inoculating 200 μl of twofold serial dilutions of decomplemented (56°C, 30 min) anti-P18 polyclonal rabbit antiserum in AOAE medium. Growth was determined spectrophotometrically. The growth inhibition titer was the reciprocal of the highest dilution at which no absorbance variation was observed after 4 days of incubation at 18°C. Culture aliquots were also examined at that time by phase-contrast light microscopy for the formation of immobile bacterial aggregates. Alternatively, fetal calf serum was added (5% [vol/vol]) to visualize the cell lysis due to complement activation. As negative controls, cells were also cultivated as described above in medium supplemented with PBS (5% [vol/vol]) or with preimmune rabbit antiserum. The viability of the cells was evaluated by using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, Oreg.) according to the manufacturer's instructions. The stained cells were observed by fluorescence microscopy (Olympus BX40; magnification, ×1,000). The living and dead cells were counted in 10 areas, and the proportion of viable cells was calculated as a percentage of all the cells. Experiments were performed in triplicate, and values are expressed as means ± standard deviations. Statistical analysis was performed by one-way analysis of variance (ANOVA) on square root-transformed data, and pairwise comparisons were made using Bonferroni's test. Differences were considered significant at P values of <0.05.
Vaccination, challenge trials, and trout antibody quantification.
Immunization trials were conducted on rainbow trout groups (20 fish per group; mean weight, 2.8 g). Fish were anesthetized with 100 μg/ml tricaine methane sulfonate (MS-222; Argent, Redmond, WA) and immunized intraperitoneally with 50 μl of P18-enriched fraction (P18-EF), with or without Freund's complete adjuvant (FCA) (approximately 7 μg and 14 μg of protein, respectively). The remaining treatments included fish injected with 50 μl of buffer (PBS) with and without FCA. At 9 and 14 weeks postimmunization, rainbow trout were challenged with live F. psychrophilum 259-93 cells as described previously (24). A single 20-fish group for each treatment was mock infected and served as a negative control group. The fish were monitored for 28 days after challenge for mortality. A minimum of 20% of the fish that died each day were cultured on tryptic yeast extract agar in an attempt to reisolate F. psychrophilum and confirm the cause of death. The cumulative percent mortality (CPM) was determined after 28 days, and the relative percent survival (RPS) was calculated using the following equation: RPS = [1 − (CPM of immunized trout)/(CPM of PBS-injected trout)] × 100.
Sera from the mock-infected control rainbow trout were collected at the end of the challenge, and the F. psychrophilum-specific antibody titers were determined by enzyme-linked immunosorbent assay (ELISA) as previously described (23). An analysis of differences in serum ELISA antibody titers between treatment groups was performed by one-way ANOVA on log10-transformed titer data, and pairwise comparisons were made using Dunnett's multiple comparison test. The mean cumulative percent mortality following bacterial challenge was analyzed by ANOVA using Tukey's test. Differences were considered significant at P values of <0.05.
DNA manipulation and nucleic acid sequencing.
Standard procedures were used to isolate genomic DNA and to clone and analyze DNA fragments (39). Restriction enzymes, T4 DNA ligase, extensor Hi-Fidelity PCR enzyme, and deoxynucleotides were obtained from Promega or ABgene and used according to the instructions of the suppliers. Nucleic acid sequencing was performed by a standard protocol (Genome Express, Meylan, France). All reported DNA sequence data were confirmed by sequencing both DNA strands from at least two independent cloned PCR products. Comparisons to database sequences were made by using the BLAST (1) and FASTA (37) algorithms. Multiple sequence alignment was performed by the ClustalW program. A prediction of membrane-spanning regions and their orientations was performed using the TMpred program (16). The method of Kyte and Doolittle was used to analyze hydrophobicity (21). Signal peptide prediction was done with SignalP 3.0 software (4, 36). Protein homology searches were carried out with the SWISS-PROT database with the EMBL BLAST and EMBL FASTA servers.
Nucleotide sequence accession numbers.
The sequences reported in this paper have been deposited in the EMBL database under accession no. AM161038, AM161039, AM161040, AM161041, and AM161042.
RESULTS
Purification of P18.
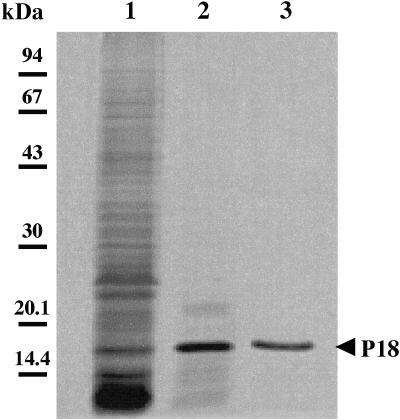
P18 was first selectively released from a cell suspension by treatment with 10 mM HEPES-NaOH buffer (pH 7.5) for 10 min at 65°C. The supernatant collected after centrifugation contained mainly P18 (>73%), which was weakly contaminated by several low-molecular-weight components and P22 as the major contaminants (Fig. 1, lane 2). Silver-stained SDS-PAGE did not reveal any lipopolysaccharide in this preparation. Thereafter, the P18-enriched fraction was subjected to anion-exchange HPLC. P18 was eluted from the column with 0.25 mM NaCl, as revealed by SDS-PAGE analysis (Fig. 1, lane 3). The protein was recovered with a yield of about 80%. The whole purification procedure is summarized in Table 2.
FIG. 1.
SDS-PAGE analysis of F. psychrophilum G4 proteins. Samples were whole cell proteins (10 μg) solubilized with 1% SDS (lane 1), proteins released after treatment (10 min, 65°C) of the cells with 10 mM HEPES-NaOH (pH 7.5) (lane 2), and HPLC-purified P18 (lane 3). Protein bands were silver stained.
TABLE 2.
Purification of P18 from intact cells of F. psychrophilum G4
| Purification step | Total protein (mg) | Amt of P18 (mg) | Purification yield (%) | Purification factor (fold) |
|---|---|---|---|---|
| Cellular extract | 522.08 | 10.96 | 100.0 | 1.0 |
| P18-enriched fraction | 13.05 | 9.51 | 86.8 | 34.7 |
| Anion-exchange HPLC eluate | 8.74 | 8.70 | 79.4 | 47.4 |
In situ protease sensitivity of P18.
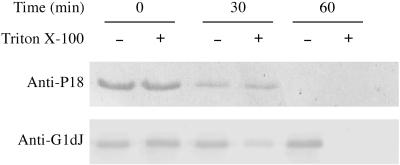
Protein surface exposure was studied by incubation of intact F. psychrophilum G4 with proteinase K on the premise that surface-exposed proteins would be degraded while subsurface proteins would be protected against proteolysis. First, we verified that proteinase K did not penetrate or destroy F. psychrophilum cells, as the A280-to-A260 ratio of the supernatant obtained was not different before or after the protease treatment. Western blot analysis of proteinase K-treated and untreated whole cells (Fig. 2) revealed that P18 disappeared completely after proteinase K treatment, indicating that it was located on the outside surfaces of the cells. Analysis of the signal intensities revealed that >80% of the P18 signal disappeared after 30 min of such treatment. The bacterial outer membrane remained intact during the in situ proteolysis treatments since there was no detectable degradation of a 65-kDa component recognized with antibodies directed against F. johnsoniae GldJ. As a periplasmic exposed outer membrane lipoprotein, GldJ could only be digested with proteinase K when the cells were solubilized with Triton X-100 prior to protease addition.
FIG. 2.
Proteinase K sensitivities of P18 and GldJ in 2 × 109 intact F. psychrophilum G4 cells in the absence (−) or presence (+) of 0.05% Triton X-100. After incubation with proteinase K for 0, 30, or 60 min at 25°C, cells were lysed in 1% SDS. Protein samples were then analyzed by SDS-PAGE, and immunoblots were developed with anti-P18 or anti-F. johnsoniae GldJ rabbit serum.
Immunoreactivity of P18.
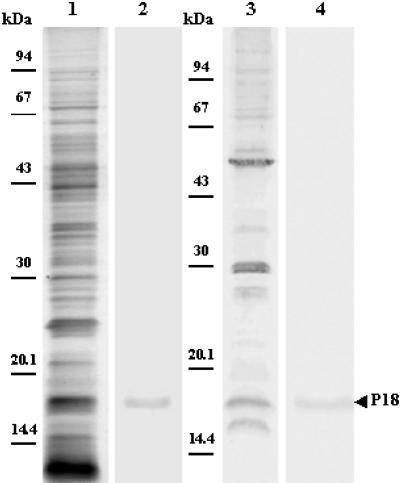
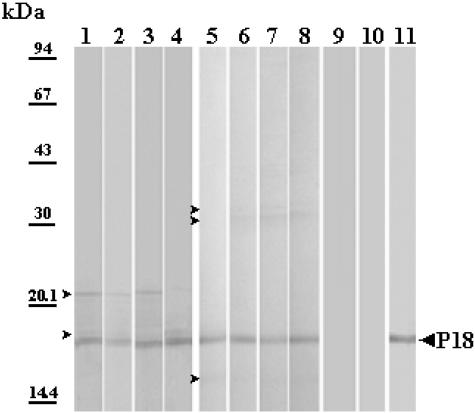
The specificity of rabbit anti-P18 serum was checked by Western blotting of the whole membrane protein extract of F. psychrophilum G4. The results clearly indicated that the antibodies prepared against P18 were specific because a single band was detected (Fig. 3, lane 2). The immunologic reactions of whole-cell extracts of the F. psychrophilum strains listed in Table 1 were analyzed with antiserum against the purified P18 protein of F. psychrophilum G4. An immune reaction was observed with proteins of similar sizes (i.e., apparent molecular mass of 18 kDa) and in the same range (Fig. 4). This supports the high conservation of P18 among F. psychrophilum strains. Additionally, a serum collected from a naturally infected trout (i.e., a CWD-convalescent fish) was also used to identify major immunogens. P18 was one of the major antigens recognized by the trout antibodies (Fig. 3, lanes 3 and 4). The reactivity of P18 was also assayed with sera derived from rainbow trout immunized with P18-EF. As expected, immunoblots exhibited a strong reaction of fish anti-P18 antibodies with the protein (Fig. 5). However, sera from fish immunized without FCA supplementation also showed weak immune reactions, with two minor components with apparent molecular masses of 22 and 19 kDa (Fig. 5, lanes 1 to 4 [four representative samples]). When FCA was added to the antigenic preparation, fish sera contained higher antibody titers (Table 3), which were mainly directed against P18 (dilution of 1:2,000 versus 1:200 for sera from fish immunized with P18-EF alone). Additionally, sera from this group (Fig. 5, lanes 5 to 8 [four representative samples]) showed weak reactions, with three different contaminants with apparent molecular masses of 37, 33, and 15 kDa. Antibodies did not react with an aliquot of the whole flavobacterial cells which had been incubated with proteinase K prior to loading in the SDS-PAGE gel (Fig. 5, lane 9), while they strongly reacted with HPLC-purified P18 (Fig. 5, lane 11). Sera from fish which had been inoculated with PBS supplemented or not with FCA were used as negative controls (one representative sample is shown in Fig. 5, lane 10). They did not contain any antibodies capable of recognition of F. psychrophilum antigens.
FIG. 3.
Identification of antigen P18 of F. psychrophilum G4 by Western blotting. Samples were whole cell proteins (20 μg; lanes 1, 2, and 3) and HPLC-purified P18 (5 μg; lane 4) solubilized with 1% SDS. Protein bands were silver stained (lane 1) or probed with rabbit serum directed against HPLC-purified P18 (lane 2) or with a serum collected from a CWD-convalescent trout (lanes 3 and 4).
FIG. 4.
Immunoblot of membrane proteins of eight different strains of F. psychrophilum. Samples were 20 μg of membrane proteins from strains G4 (lane 1), PO388 (lane 2), JIP02.86 (lane 3), JIP16.00 (lane 4), JIP07.99 (lane 5), UCD95.74 (lane 6), NCIMB 1947T (lane 7), and FPC839 (lane 8), revealed with a hyperimmune rabbit antiserum directed against P18.
FIG. 5.
Western blot analysis of a selection of representative serum samples collected from rainbow trout immunized with a P18-enriched fraction obtained through heat/HEPES treatment of F. psychrophilum G4, either not supplemented (lanes 1 to 4; dilution, 1:200) or supplemented with FCA (lanes 5 to 9 and 11; dilution, 1:2,000), or injected with PBS+FCA as a negative control (lane 10; dilution, 1:50). Sample proteins were F. psychrophilum G4 crude SDS extract (lanes 1 to 10; 10 μg of proteins/well) or HPLC-purified P18 (lane 11; 5 μg). The protein sample was digested with proteinase K for 1 h at 37°C prior to solubilization and SDS-PAGE (lane 9).
TABLE 3.
Protection of vaccinated rainbow trout following challenge with F. psychrophilum 259-93a
| Antigen | ELISA titer (mean ± SEM)b,c | % Protection efficacy
|
|
|---|---|---|---|
| CPM (mean ± SEM)c | RPSd | ||
| PBS | 360a ± 83.3 | 21.8a ± 2.6 | 0.0 |
| PBS+FCA | 2,060b ± 578.2 | 28.4a ± 3.2 | −30.5 |
| P18-EF alone | 1,060c ± 279.8 | 10.0a ± 1.9 | 54.1 |
| P18-EF+FCA | 9,600d ± 1,141.6 | 2.5b ± 1.0 | 88.5 |
Rainbow trout (mean weight, 16 g) were challenged at 14 weeks postvaccination with 6.25 × 106 CFU/fish.
F. psychrophilum-specific antibody titers were measured after vaccination in mock-infected rainbow trout sera.
Mean CPM and titer values with different superscripts indicate significant differences, with P values of <0.05.
RPS (n = 2 per treatment) was determined relative to PBS treatment.
Bacteriostatic and bactericidal activities of anti-P18 antibodies.
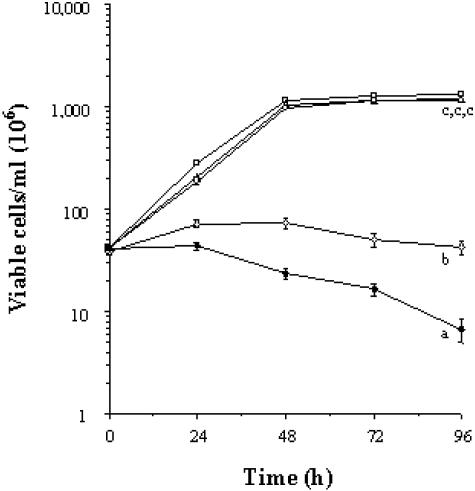
As a component located on the surface of F. psychrophilum G4, P18 should lead to the production of specific antibodies which could be deposited on the bacterial cell surface and trigger a set of biological phenomena related to this deposition (i.e., activation of complement, bacteriostasis, and bacteriolysis). Growth inhibition tests were performed with anti-P18 antibodies to confirm the position of P18 with respect to the lipid bilayer of the flavobacterial outer membrane. The anti-P18 serum exhibited a threshold inhibiting dilution of 1:200, while the preimmune serum did not inhibit the growth of F. psychrophilum G4. This observation confirmed that P18 is exposed at the surface of the flavobacterial cell. To assess the bacteriostatic and bactericidal activities of anti-P18 antibodies, F. psychrophilum cells were thereafter grown in the presence of heat-treated anti-P18 serum (dilution, 1:200) supplemented or not with fetal bovine serum containing complement. Preliminary experiments showed that the concentration of fetal calf serum that was used was not bactericidal for F. psychrophilum in the absence of flavobacterium-specific antibodies (Fig. 6). In the absence of complement, no significant F. psychrophilum growth was observed at 4 days when a 1:200 dilution of anti-P18 antibodies was added compared to that of cells treated with PBS, fetal bovine serum, or a similar dilution of preimmunization serum. The growth inhibition that was observed was bacteriostatic, since the number and viability of bacterial cells were almost constant over this time period (Fig. 6). In the presence of fetal calf serum (which contained bovine complement), anti-P18 antibodies were bacteriostatic on the first day, since the cell number did not increase and the cell viability was constant. On the other hand, a drastic decrease in cell viability was observed between the second and fourth day compared to that in control samples containing only anti-P18 antibodies and no complement. Viable flavobacteria were estimated to comprise <16% of initially inoculated cells. Moreover, bacterial agglutination was seen in the presence of anti-P18 but not with fetal calf serum (data not shown).
FIG. 6.
Bacteriostatic and bactericidal activities of anti-P18 antibodies toward F. psychrophilum G4 during in vitro growth. The growth medium was supplemented with the following: □, PBS (5% [vol/vol]); ▵, anti-P18 preimmune serum (1:200); ○, fetal bovine serum containing complement (5% [vol/vol]); ⋄, anti-P18 decomplemented serum (1:200); ⧫, anti-P18 decomplemented serum (1:200) and fetal bovine serum containing complement (5% [vol/vol]). Values are expressed as means ± standard deviations (n = 3 per treatment). Different superscripts indicate significant differences, with P values of <0.05.
Protection studies.
Duplicate groups of 20 rainbow trout were used in the study to evaluate protection, including groups receiving the P18-enriched fraction supplemented (P18-EF+FCA) or not (P18-EF) with FCA. Additionally, PBS- and PBS+FCA-injected groups of fish were included in the evaluation. No significant protection was observed at 9 weeks postvaccination. The results of protection studies performed at 14 weeks postvaccination are summarized in Table 3. With FCA, P18-EF gave an RPS value of 88.5, which was significantly (P < 0.05) larger than that of the control group (PBS), while P18-EF alone elicited a decrease in mortality which was not statistically significant (P > 0.05). F. psychrophilum was reisolated from 74% of the fish that died in the study and were checked for the presence of the bacterium by culture. The highest F. psychrophilum-specific antibody titers were observed in sera from P18-EF+FCA-vaccinated fish, followed by sera from PBS+FCA-injected trout. Fish injected with P18-EF only generated a weaker antibody response than P18-EF+FCA-injected fish (about 10-fold less) but a significantly different response (3-fold rise) than PBS-injected controls. Together, these data indicate that P18 is capable of inducing antibody synthesis by fish and that such antibodies are protective.
P18 sequencing and identification of the ompH-like gene sequence.
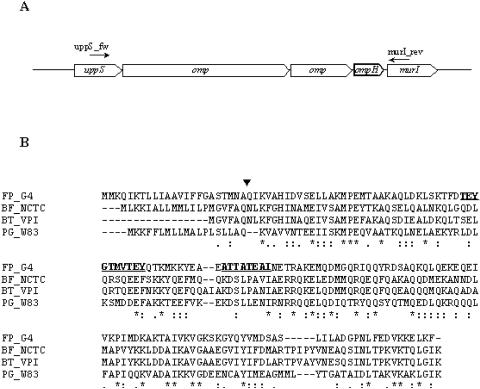
For further characterization of P18, the protein was subjected to de novo sequencing. Several peptides were identified, among which the following two sequences were found: (i) TEYGTMVTEY and (ii) ATTATEAI. These peptides were searched against an ongoing genome sequence of F. psychrophilum strain JIP02.86 (INRA, Jouy-en-Josas, France). Both peptides matched (100% identity) an OmpH-like protein encoded by one gene belonging to a cluster of six genes, including two genes for additional probable outer membrane proteins (Fig. 7A). Protein identity was ascertained by submitting MS/MS data obtained with P18 to a search against a database harboring the OmpH-like protein sequence. Two highly conserved genes within the CFB phylum (uppS [di-trans,poly-cis-decaprenylcistransferase] and murI [glutamate racemase]) belonging to the locus/operon were used to design primers for amplification of the whole locus. They were (i) uppS_fw (CATTATTATGGACGGGAATGG) and (ii) murI_rev (GCCGCCTATTCCTGAATCGAA). Using PCR, the ompH gene and surrounding regions were amplified (5 kb) and sequenced. Analysis of the deduced amino acid sequence of the F. psychrophilum G4 OmpH-like protein revealed that it is a 166-amino-acid protein (Fig. 7B) that is almost totally conserved (except for a single Ile25/Thr25 substitution) among the F. psychrophilum strains used in this study (Table 1). Sequences producing significant alignments were (i) the cationic outer membrane OmpH-like protein from Porphyromonas gingivalis and (ii) the putative outer membrane proteins from Bacteroides fragilis, Bacteroides thetaiotaomicron, and Cytophaga hutchinsonii, all of which belong to the CFB phylum (Fig. 7B). However, the best hit found was with P. gingivalis OmpH (163 amino acids long) and was only 32%. A search of the NCBI conserved domain database revealed significant alignments with almost the whole sequence of the pfam03938 domain of OmpH, an outer membrane protein (OmpH-like) (E value = 3 × 10−5), and with the COG2825 domain of the histone-like protein (HlpA), another outer membrane protein (E value = 1 × 10−7), confirming the similarity to the OmpH analogs. The program SignalP does in fact predict that the OmpH-like protein is an outer membrane protein with an N-terminal signal peptide that may be cleaved between Ala23 and Gln24 (signal peptide probability, 0.999). The leader sequence shows a typical transmembrane helix structure with a significant outside-to-inside helix formation score of 1,422 on the transmembrane prediction scale, suggesting strongly that OmpH is a surface-located protein. The mature protein (143 amino acids) gives rise to a predicted 16,248.6-Da protein, which is in very close agreement with the observed mass (18 kDa).
FIG. 7.
(A) ompH locus organization in F. psychrophilum JIP02.86. (B) Multiple sequence alignment of F. psychrophilum G4 P18 with OmpH-like homologues from the following strains belonging to the CFB phylum: Bacteroides fragilis NCTC 9343 (BF_NCTC), Bacteroides thetaiotaomicron VPI-5482 (BT_VPI), and Porphyromonas gingivalis W83 (PG_W83). Identical amino acids, conservative substitutions, and semiconservative substitutions are indicated by stars, stacked double dots, and single dots, respectively. Dashes are gaps introduced to maximize alignment. The predicted cleavage site of the F. psychrophilum OmpH signal peptide is indicated (▾). Peptides sequenced de novo are underlined and shown in bold.
DISCUSSION
This study was undertaken to characterize P18 and to assess the potential relevance of such a protein in eliciting a protective immune response in fish. First, we developed a purification method allowing quantitative recovery of the native protein by applying the following simple two-step purification protocol: heat-HEPES extraction of P18 from the cells followed by anion-exchange HPLC. SDS-PAGE analysis of the purified P18 revealed a high degree of purity (Fig. 1; Table 2). This is the first report of the purification of an F. psychrophilum surface protein allowing its detailed biochemical characterization. We have shown that this protein of 18 kDa is encoded by a gene homologous to the ompH genes of some bacteria belonging to the CFB phylum and some other gram-negative and -positive bacteria. The DNA sequence of the cloned F. psychrophilum ompH gene was determined, and the deduced amino acid sequence revealed significant identity with the OmpH-like protein of Porphyromonas gingivalis. The amino acid sequence shows a typical tripartite “positive-hydrophobic-polar” sequence for a precursor signal sequence, with the presence of a cleavage site for signal peptidase I, used to remove the signal peptide for exported proteins (46). The final location of the OmpH-like protein at the cell surface had been suspected since the protein is released from F. psychrophilum by heat-HEPES treatment (30). The surface exposure of the OmpH-like protein was confirmed here by the following two approaches: first, the protein was totally digested in intact cells by proteinase K, while the outer membrane lipoprotein GldJ, as expected from its periplasmic exposition, was not digested (Fig. 2); second, anti-OmpH-like protein antibodies were capable of bacteriostatic and bactericidal activities toward F. psychrophilum in the absence and presence of complement, respectively (Fig. 6). The processed OmpH-like protein in F. psychrophilum has an apparent molecular mass of 18 kDa (30; this study) (Fig. 1), while the molecular mass for the amino acid sequence of the protein after cleavage of the probable signal peptide was calculated to be 16.2 kDa. This might be due to the experimental conditions used for molecular mass determination. However, we cannot rule out the possible presence of an alternative signal cleavage site in F. psychrophilum.
We previously hypothesized that P18 might be an S-layer protein (30) since it is actually recovered from flavobacterial cells by treatments known to release such components. However, the protein appears to be a true member of the OmpH family (also known as Skp or HlpA) (15, 20, 32). The OmpH-like protein produced by F. psychrophilum is surface located and immunoaccessible and induces neutralizing antibodies, as recently reported for Chlamydia pneumoniae (14). In Escherichia coli, OmpH has been described as a molecular chaperone required for efficient release of translocated proteins from the plasma membrane. It interacts with unfolded proteins emerging from the sec translocation machinery and contributes to the correct folding of some outer membrane proteins and their insertion into the outer membrane (e.g., OmpA [10, 40]) as well as to preventing the aggregation of soluble proteins such as lysozyme (47). Such a key function in the periplasmic space for OmpH (41) is not consistent with the surface exposition observed in F. psychrophilum (this study) as well as in Chlamydia trachomatis (3) and C. pneumoniae (14). The flavobacterial OmpH protein might have different functions, including the chaperone periplasmic function generally assigned to the OmpH family and an interaction function with the environment, which might be an adaptation to some peculiar aspect of flavobacterial physiology. Further studies are needed to determine whether surface-exposed P18 has chaperone-like activities similar to those of periplasmic OmpH proteins from other bacteria.
Proteins belonging to the OmpH family have been described for a wide variety of bacterial species. Some studies have provided results indicating that members of the OmpH family may be immunogenic antigens of some bacteria (e.g., Chlamydia trachomatis [3]). This was also observed with F. psychrophilum. Indeed, sera from convalescent fish that were naturally infected were used successfully in this study to identify the OmpH-like protein in a set of other antigenic flavobacterial proteins recognized predominantly in the context of infection (Fig. 3). Consequently, the native OmpH-like protein was involved in serological and immunological experiments. High-titer anti-OmpH-like protein antibodies were raised in rabbits. The monospecific antiserum reacted with the OmpH-like protein only since one single band of 18 kDa was detected on immunoblots of F. psychrophilum lysates resolved by SDS-PAGE (Fig. 3 and 4), and the OmpH-like protein was shown to be highly conserved. Moreover, such antibodies binding the OmpH-like protein were capable of inducing protein and/or membrane distortions, resulting in the lysis of F. psychrophilum in the presence of complement. However, it is well known that the correlation is limited between protective immunity and the level of antibodies in fish after vaccination and that fish elicit a nonspecific immune response upon injection of non-antigen-containing agents (e.g., Freund's adjuvant). Here we observed good protective immunity against F. psychrophilum, suggesting that specific immunity was obtained. Indeed, vaccination trials with a fraction highly enriched with OmpH-like protein induced significant protective immunity in fish, with this protection very probably being a result of the relatively high titers of antibodies produced against the flavobacterial OmpH-like protein (Fig. 5; Table 3). The bacteriostatic/bactericidal actions of the monospecific anti-OmpH-like protein rabbit serum observed in the present study also suggest that a sufficiently strong immune response against this protein could be related to protection.
Taking these results as a whole, we concluded that the OmpH-like protein in F. psychrophilum presents the capability to induce high antibody titers in rainbow trout and results in significant protective immunity. While additional studies are needed to assess the possible involvement of the OmpH-like protein in the pathogenesis of this bacterium, this work should be a prelude to the development of recombinant vaccines against F. psychrophilum, using the antigen characterized here as a possible immunity target.
Acknowledgments
This work was partly supported by the Région Aquitaine and by a grant to F.D. from the Ecole Doctorale Université Bordeaux 2, Ministère de la Recherche.
We thank M. J. McBride, C. Quentel, and C. Michel for the generous gifts of sera directed against F. johnsoniae GldJ and against trout antibodies and for some F. psychrophilum strains, respectively. We also thank S. Papillon and A.-M. Richard (from Enitab) and B. Shewmaker, M. Higgins, and A. Weighall (from Clear Springs Foods) for their excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anacker, R. L., and E. J. Ordal. 1955. Study of a bacteriophage infecting the myxobacterium Chondrococcus columnaris. J. Bacteriol. 70:738-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., and D. D. Rockey. 1999. Use of primate model system to identify Chlamydia trachomatis protein antigens recognized uniquely in the context of infection. Microbiology 145:2077-2085. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. vonHeijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bernardet, J.-F., P. Segers, M. Vancanneyt, F. Berthe, K. Kersters, and P. Vandamme. 1996. Cutting a gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978). Int. J. Syst. Bacteriol. 46:128-148. [Google Scholar]
- 6.Bernardet, J.-F., Y. Nakagawa, and B. Holmes. 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52:1049-1070. [DOI] [PubMed] [Google Scholar]
- 7.Borg, A. F. 1960. Studies on myxobacteria associated with diseases in salmonid fishes. Wildlife disease, no. 8. American Association for the Advancement of Science, Washington, D.C.
- 8.Braun, T. F., and M. J. McBride. 2005. Flavobacterium johnsoniae GldJ is a lipoprotein that is required for gliding motility. J. Bacteriol. 187:2628-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruun, M. S., A. S. Schimdt, L. Madsen, and I. Dalsgaard. 2000. Antimicrobial resistance patterns in Danish isolates of Flavobacterium psychrophilum. Aquaculture 189:201-212. [Google Scholar]
- 10.Bulieris, P. V., S. Behrens, O. Holst, and J. H. Kleinschmidt. 2003. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J. Biol. Chem. 278:9092-9099. [DOI] [PubMed] [Google Scholar]
- 11.Crump, E. M., M. B. Perry, S. C. Clouthier, and W. W. Kay. 2001. Antigenic characterization of the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:750-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump, E. M., J. Burian, P. D. Allen, and W. W. Kay. 2005. Identification and expression of a host-recognized antigen, FspA, from Flavobacterium psychrophilum. Microbiology 151:3127-3135. [DOI] [PubMed] [Google Scholar]
- 13.Dalsgaard, I., and L. Madsen. 2000. Bacterial pathogens in rainbow trout, Oncorhynchus mykiss (Walbaum), reared at Danish freshwater farms. J. Fish Dis. 23:199-209. [Google Scholar]
- 14.Finco, O., A. Bonci, M. Agnusdei, M. Scarselli, R. Petracca, N. Norais, G. Ferrari, I. Garaguso, M. Donati, V. Sambri, R. Cevenini, G. Ratti, and G. Grandi. 2005. Identification of new potential vaccine candidates against Chlamydia pneumoniae by multiple screenings. Vaccine 23:1178-1188. [DOI] [PubMed] [Google Scholar]
- 15.Hirvas, L., J. Coleman, P. Koski, and M. Vaara. 1990. Bacterial ‘histone-like protein I’ (HLP-I) is an outer membrane constituent? FEBS Lett. 262:123-126. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 374:166. [Google Scholar]
- 17.Iida, Y., and A. Mizokami. 1996. Outbreaks of coldwater disease in wild ayu and pale chub. Fish Pathol. 31:157-164. [Google Scholar]
- 18.Izumi, S., and F. Aranishi. 2004. Relationship between gyrA mutations and quinolone resistance in Flavobacterium psychrophilum isolates. Appl. Environ. Microbiol. 70:3968-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, M., K. Kawai, M. Okabe, N. Nakano, and S. Oshima. 2003. Efficacy of oral vaccine against bacterial coldwater disease in ayu Plecoglossus altivelis. Dis. Aquat. Organ. 55:261-264. [DOI] [PubMed] [Google Scholar]
- 20.Koski, P., L. Hirvas, and M. Vaara. 1990. Complete sequence of the ompH gene encoding the 16-kDa cationic outer membrane protein of Salmonella typhimurium. Gene 88:117-120. [DOI] [PubMed] [Google Scholar]
- 21.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.LaFrentz, B. R., S. E. LaPatra, G. R. Jones, J. L. Congleton, B. Sun, and K. D. Cain. 2002. Characterization of serum and mucosal antibody responses and relative per cent survival in rainbow trout, Oncorhynchus mykiss (Walbaum), following immunization and challenge with Flavobacterium psychrophilum. J. Fish Dis. 25:703-713. [Google Scholar]
- 24.LaFrentz, B. R., S. E. LaPatra, G. R. Jones, and K. D. Cain. 2003. Passive immunization of rainbow trout, Oncorhynchus mykiss (Walbaum), against Flavobacterium psychrophilum, the causative agent of bacterial coldwater disease and rainbow trout fry syndrome. J. Fish Dis. 26:371-384. [DOI] [PubMed] [Google Scholar]
- 25.LaFrentz, B. R., S. E. LaPatra, G. R. Jones, and K. D. Cain. 2004. Protective immunity in rainbow trout Oncorhynchus mykiss following immunization with distinct molecular mass fractions isolated from Flavobacterium psychrophilum. Dis. Aquat. Organ. 59:17-26. [DOI] [PubMed] [Google Scholar]
- 26.Lehman, J., D. Mock, F. J. Stürenberg, and J.-F. Bernardet. 1991. First isolation of Cytophaga psychrophila from a systemic disease in eel and cyprinids. Dis. Aquat. Organ. 10:217-220. [Google Scholar]
- 27.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. L. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 28.MacLean, L. L., E. Vinogradov, E. M. Crump, M. B. Perry, and W. W. Kay. 2001. The structure of the lipopolysaccharide O-antigen produced by Flavobacterium psychrophilum (259-93). Eur. J. Biochem. 268:2710-2716. [DOI] [PubMed] [Google Scholar]
- 29.Madsen, L., and I. Dalsgaard. 1998. Characterization of Flavobacterium psychrophilum: a comparison of proteolytic activity and virulence of strains isolated from rainbow trout (Oncorhynchus mykiss), p. 45-52. In A. C. Barnes, G. A. Davidson, M. P. Hiney, and D. McIntosh (ed.), Methodology in fish diseases research. Fisheries Research Services, Aberdeen, United Kingdom.
- 30.Massias, B., F. Dumetz, M.-C. Urdaci, and M. Le Hénaff. 2004. Identification of P18, a surface protein produced by the fish pathogen Flavobacterium psychrophilum. J. Appl. Microbiol. 97:574-580. [DOI] [PubMed] [Google Scholar]
- 31.Merle, C., D. Faure, M.-C. Urdaci, and M. Le Hénaff. 2003. Purification and characterization of a membrane glycoprotein from the fish pathogen Flavobacterium psychrophilum. J. Appl. Microbiol. 94:1120-1127. [DOI] [PubMed] [Google Scholar]
- 32.Missiakas, D., J. M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 33.Moller, J. D., J. L. Larsen, L. Madsen, and I. Dalsgaard. 2003. Involvement of a sialic acid-binding lectin with hemagglutination and hydrophobicity of Flavobacterium psychrophilum. Appl. Environ. Microbiol. 69:5275-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moller, J. D., A. C. Barnes, I. Dalsgaard, and A. E. Ellis. 2005. Characterisation of surface blebbing and membrane vesicles produced by Flavobacterium psychrophilum. Dis. Aquat. Organ. 64:201-209. [DOI] [PubMed] [Google Scholar]
- 35.Nematollahi, A., A. Decostere, F. Pasmans, and F. Haesebrouck. 2003. Flavobacterium psychrophilum infections in salmonid fish. J. Fish Dis. 26:563-574. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of procaryotic and eucaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 38.Rahman, H., A. Kuroda, J. M. Dijkstra, I. Kiryu, T. Nakanishi, and M. Ototake. 2002. The outer membrane fraction of Flavobacterium psychrophilum induces protective immunity in rainbow trout and ayu. Fish Shellfish Immunol. 12:169-179. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Schäfer, U., K. Beck, and M. Muller. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274:24567-24574. [DOI] [PubMed] [Google Scholar]
- 41.Schlapschy, M., M. K. Dommel, K. Hadian, M. Fogarasi, I. P. Korndorfer, and A. Skerra. 2004. The periplasmic E. coli chaperone Skp is a trimer in solution: biophysical and preliminary crystallographic characterization. Biol. Chem. 385:137-143. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, A. S., M. S. Bruun, I. Dalsgaard, K. Pedersen, and J. L. Larsen. 2000. Occurrence of antimicrobial resistance in fish-pathogenic and environmental bacteria associated with four Danish rainbow trout farms. Appl. Environ. Microbiol. 66:4908-4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Secades, P., B. Alvarez, and J. A. Guijarro. 2001. Purification and characterization of a psychrophilic, calcium-induced, growth-phase-dependent metalloprotease from the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 67:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Secades, P., B. Alvarez, and J. A. Guijarro. 2003. Purification and properties of a new psychrophilic metalloprotease (Fpp2) in the fish pathogen Flavobacterium psychrophilum. FEMS Microbiol. Lett. 226:273-279. [DOI] [PubMed] [Google Scholar]
- 45.Tunón, P., and K.-E. Johansson. 1984. Yet another improved staining method for the detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 9:171-179. [DOI] [PubMed] [Google Scholar]
- 46.Tuteja, R. 2005. Type I signal peptidase: an overview. Arch. Biochem. Biophys. 441:107-111. [DOI] [PubMed] [Google Scholar]
- 47.Walton, T. A., and M. C. Sousa. 2004. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol. Cell 15:367-374. [DOI] [PubMed] [Google Scholar]