Abstract
The rugose (also known as wrinkled or rdar) phenotype in Salmonella enterica serovar Typhimurium DT104 Rv has been associated with cell aggregation and the ability, at low temperature under low-osmolarity conditions, to form pellicles and biofilms. Two Tn5 insertion mutations in genes that are involved in lipopolysaccharide (LPS) synthesis, ddhC (A1-8) and waaG (A1-9), of Rv resulted in diminished expression of colony rugosity. Scanning electron micrographs revealed that the ddhC mutant showed reduced amounts of extracellular matrix, while there was relatively more, profuse matrix production in the waaG mutant, compared to Rv. Both mutants appeared to produce decreased levels of curli, as judged by Western blot assays probed with anti-AgfA (curli) antibodies but, surprisingly, were observed to have increased amounts of cellulose relative to Rv. Comparison with a non-curli-producing mutant suggested that the alteration in curli production may have engendered the increased presence of cellulose. While both mutants had impaired biofilm formation when grown in rich medium with low osmolarity, they constitutively formed larger amounts of biofilms when the growth medium was supplemented with either glucose or a combination of glucose and NaCl. These observations indicated that LPS alterations may have opposing effects on biofilm formation in these mutants, depending upon either the presence or the absence of these osmolytes. The phenotypes of the waaG mutant were further confirmed in a constructed, nonpolar deletion mutant of S. enterica serovar Typhimurium LT2, where restoration to the wild-type phenotypes was accomplished by complementation. These results highlight the importance of an integral LPS, at both the O-antigen and core polysaccharide levels, in the modulation of curli protein and cellulose production, as well as in biofilm formation, thereby adding another potential component to the complex regulatory system which governs multicellular behaviors in S. enterica serovar Typhimurium.
Bacteria often exist in natural environments, not only as individual (planktonic) cells but also as sessile, multicellular forms such as biofilms attached to surfaces. Multicellular behavior can also be manifested as an aggregation of cells in an extracellular matrix in colonies, as has been observed with Salmonella enterica serovar Typhimurium DT104 strain 11601 Rv cells grown on rich solid medium (1). The Salmonella colonies, which appear rugose (wrinkled), are formed only at low osmolarity and at low temperature. This morphology is also associated with cellular aggregation in liquid culture, i.e., a pellicle in standing culture and biofilms in agitated culture. Compared to its spontaneous smooth mutant, Stv, the S. enterica serovar Typhimurium DT104 Rv cells in rugose colonies differed by exhibiting resistance to oxidative and acid stresses, which concomitantly conferred survival advantages. Therefore, the multicellular forms perhaps are preferred modes of Salmonella growth in their natural environments, possibly allowing cells to survive better in the presence of environmental stresses.
It has been demonstrated that the production of a matrix composed of proteinaceous, thin, aggregative fimbriae (Tafi), also known as curli, and cellulose (25, 36, 39) is required for rugose (also called red, dry, and rough or rdar) (25) colony morphology in both S. enterica serovars Typhimurium and Enteritidis. Genes for curli production are found in the csg (formerly agf) operon, while genes for cellulose production are found in the bcs operon. In addition, White et al. (36) showed the presence of a high-molecular-weight polysaccharide that is distinct from cellulose or colanic acid that is also tightly associated with curli protein.
The machinery for synthesis of curli is encoded in two operons, csgDEFG and csgBA. CsgD is required for transcription of the csgBA operon, in which the major and minor subunits of curli are encoded by csgA and csgB, respectively. Two chaperone proteins involved in curli production are encoded by csgE and csgF, while csgG codes for a lipoprotein in the inner leaflet of the outer membrane that functions as a platform for assembly (5). Once synthesized and transported to the cell surface, the CsgA major subunits are self-assembled upon the nucleated CsgB minor subunits, a mechanism termed the nucleation-precipitation pathway. However, a thorough explanation of the entire assembly process of curli remains elusive.
Cellulose production by S. enterica serovar Typhimurium, on the other hand, is only indirectly regulated by CsgD, which is required for expression of AdrA (24). The bcs operons that code for the cellulose biosynthesis machinery are constitutively expressed; however, synthesis of cellulose occurs only when AdrA is produced, suggesting that AdrA probably regulates cellulose assembly at the posttranscriptional level (39).
For both curli and cellulose, synthesis and assembly occur in the vicinity of the cell surface. Thus, it appears that the presence of an intact cell surface is necessary for the production of these surface-associated matrix constituents. Notably, surface components, such as lipopolysaccharide (LPS) in the outer membrane of gram-negative bacteria, have been shown to play important roles in the synthesis of other surface appendages. For example, type I pilus production is defective in the absence of intact LPS, possibly because of alteration in the production of outer membrane proteins (10). LPS is also indirectly involved in the production of capsular polysaccharide and flagella, as demonstrated with the regulation by the RcsC/B two-component system, which facilitates transcription of capsular synthesis genes and repression of the transcription of flagellar genes in response to alterations in the outer membrane, including LPS (22). While intercellular complementation of curli subunits between curli-producing and non-curli-producing S. enterica serovar Enteritidis bacteria is blocked by LPS (36), the roles of LPS in the production of curli within cells have not yet been demonstrated. Also, there is no direct evidence of a role for LPS in cellulose production by salmonellae.
Herein, effects on rugose formation by transposon insertion in two genes of S. enterica serovar Typhimurium DT104 that alter LPS formation, ddhC and waaG, are described. The changes in LPS independently retarded the appearance of the rugose phenotype primarily because of alterations in the synthesis of both curli and cellulose. Conversely, both mutations resulted in increased biofilm formation under prescribed conditions. The results of the present study provide evidence that LPS plays important roles in the production of curli and cellulose, and thus in rugose colony formation, and in the development of biofilms.
(This article was presented, in part, at the ASM Conference on Salmonella: Pathogenesis, Epidemiology, and Vaccine Development, Sardinia, Italy, September 2003.)
MATERIALS AND METHODS
Strains.
All of the strains used in this study were derived from either S. enterica serovar Typhimurium DT104 Rv (1) or S. enterica serovar Typhimurium LT2 (henceforth referred to as Rv and LT2, respectively). Stv, the stable, spontaneous, smooth mutant of S. enterica serovar Typhimurium Rv, was used as a negative control for rugosity and pellicle formation (1). All of these strains and the plasmids used in this study are listed in Table 1. The high-frequency generalized transducing bacteriophage P22 mutant HT105/1 int-201 (26) was used for transduction in DT104 Rv by standard methods (17). All PCRs were performed with primers obtained from IDT DNA (Coralville, IA) (Table 2). Somatic serotyping was performed with Bacto-Salmonella O polyclonal antiserum group B factors 1, 4, 5, and 12 (Difco, Detroit, MI).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant description or genotype | Relevant phenotype | Source or reference |
|---|---|---|---|
| S. enterica serovar Typhimurium DT104 | |||
| Rv | WT | Rugose (rdar)a | 1 |
| Stv | Spontaneous smooth mutant of Rv | Smooth (saw)a | 1 |
| A1-8 | ddhC::Tn5 | Altered rugose | This study |
| A1-9 | waaG::Tn5 | Altered rugose | This study |
| S. enterica serovar Typhimurium LT2 | WT | Rugose | B. Bassler |
| YA104 | LT2(pKD46) | This study | |
| YA155 | ΔwaaG::kan | Altered rugose | This study |
| YA156 | YA155(pTOPOwaaG) | Rugose | This study |
| YA151 | ΔcsgA::kan | Semirugose (pdar)a | This study |
| YA159 | ΔbcsA::kan | Semirugose (bdar)a | This study |
| S. enterica serovar Enteritidis 4b ΔagfA | ΔagfA | Smooth (saw) | W. W. Kay (36) |
| E. coli | |||
| TOP10 | F−mcrA (mrr-hsdRMS-mcrBC) φ80 lacZ M15 lacX74 recA1 araD139 galU galK (ara-leu)7697 rpsL (Strr) endA1 | Invitrogen | |
| Plasmids | |||
| pTOPO | pCR4 cloning vector, Ampr | Invitrogen | |
| pTOPOwaaG | pTOPO with waaG insert | This study | |
| pKD46 | λ Red helper plasmid, Para, Ts ori, Ampr | B. L. Wanner (6), CGSCb | |
| pKD4 | PCR template plasmid, Flip recombinase sites flanking Kanr-encoding gene | B. L. Wanner (6), CGSC |
Designated by Romling et al. (25). rdar, red, dry, and rough; saw, smooth and white; pdar, pink, dry, and rough; bdar, brown, dry, and rough.
CGSC, E. coli Genetic Stock Center.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Source or referencec |
|---|---|---|
| pwaaG11 | CCC AAG CTT CGA CGA CAT CAT TCA GTT Ta | 5848-5830d |
| pwaaG12 | CGG GGT ACC TAC CTT TAT GCC ACT TCA Ga | 4401-4419d |
| pwaaGD11 | GAA AAA ATG CTG CCG CAT GAG GCA CGC ACC ATA GAT TTG GAC AGC CTG CTG TGT AGG CTG GAG CTG CTT Cb | 5681-5730d |
| pwaaGD12 | TAG TGT GGT TAA CGG CGC TTT CAG CTC AAC CAT CTA AAT CAC CTG TAA TAC ATA TGA ATA TCC TCC TTA Gb | 4531-4580d |
| pwaaGIN11 | GAG GGC ATC AGG TTC GTG T | 5547-5566d |
| pwaaGIN12 | AGG CGT TGC GTA AGG AAG G | 4643-4662d |
| pwaaPIN11 | TTC CTG AAG TGG CAT AAA G | 4387-4405d |
| pwaaPIN12 | AAA TAC TCA CGC ATA AAC C | 3872-3890d |
| pcsgAD11 | ATG GCT ATT CGC GTC ACC CAA CGC TAA TAC CGT TAC GAC TTT TAA ATC AAG TGT AGG CTG GAG CTG CTT Cb | 17701-17750e |
| pcsgAD12 | TTT GAA AGT GCG GCA AGG AGC AAT AAA GTA TGC ATA ATT TCC TCC CGA AAC ATA TGA ATA TCC TCC TTA Gb | 18271-18320e |
| pcsgA11 | TCA CCC AAC GCT AAT ACC G | 17713-17732e |
| pcsgA12 | CCT TGC TGA GTC GTG GTA A | 18332-18350e |
| pbcsAD11 | ACG TCC GCC GGG AGC CTG CGA TGA GCG CCC TTT CCC GGT GGC TGC TTA TCG TGT AGG CTG GAG CTG CTT Cb | 11930-11881f |
| pbcsAD12 | TAT CAT CAT TGT TGA GCC TGA GCC ATA ACC CGA TCC GAC GGC TGT ATC GCC ATA TGA ATA TCC TCC TTA Gb | 9281-9330f |
| pbcsA11 | ATG ATG CGG GCG ACA AAA C | 4937-4955f |
| pbcsA12 | CCT ATT ACC GCC GCA CAC A | 14221-14239f |
| p16s rRNA11 | GAA GAG TTT GAT CAT GGC TC | 21441-21460g |
| p16s rRNA12 | TAC GGT TAC CTT GTT ACG AC | 19953-19972g |
| pKanKD3 | CAG TCA TAG CCG AAT AGC CT | Datsenko and Wanner (6) |
| inv2 | GAA CTT TTG CTG AGT TGA AGG ATC A | Ducey and Dyer (9) |
HindIII and KpnI sites are underlined.
Sequence homologous to template plasmid pKD4 underlined.
Sequence numbering from S. enterica serovar Typhimurium LT2.
Numbering from sequence with accession no. AF026386.
Numbering from sequence with accession no. AE008749.
Numbering from sequence with accession no. AJ315148.
Numbering from sequence with accession no. AE008820.
Media and growth conditions.
All strains were stored at −80°C in LB medium containing 25% glycerol. The strains were grown either in LB Miller (LB) base broth or agar (Difco), which contains only 0.5 g NaCl/liter, or in M63 minimal medium supplemented with 0.5% vitamin-free Casamino Acids (CAA; Difco) and 0.2% glucose. To generate growth curves for Rv, Stv, LT2, the ddhC and waaG mutants, and the waaG-complemented strains, overnight cultures grown at 37°C in LB with or without antibiotics were used to inoculate fresh medium in tubes at a dilution of 1:100. The tubes were incubated with shaking at 210 rpm at either 28°C or 37°C, and the optical density at 600 nm (OD600) was measured periodically through 24 h. Antibiotics were used at the following final concentrations: sodium ampicillin, 100 μg/ml; kanamycin sulfate, 50 μg/ml. To obtain isolated colonies and to observe progressive rugose colony formation, 100 μl of 10−6 to 10−8 dilutions of overnight broth cultures of salmonellae were spread plated onto LB plates and incubated at 22 to 25°C for 6 days. Salmonellae were also streaked for individual colony isolation on LB agar, where on the denser area of the streak some colonies of the WT strain turned rugose in as few as 2 to 3 days while some of the mutant colonies required 3 to 4 days to reveal rugosity. When streaked in straight lines on LB plates with a denser inoculum, the rugosity of the WT formed at 2 days while it took 3 days for the mutants streaked in the same fashion to reach their relatively maximum rugosity. LB agar supplemented with Congo red (40 μg/ml) and Coomassie brilliant blue (20 μg/ml) was also used to determine the Congo red-binding property of the colonies. Calcofluor (fluorescent brightener 28; Sigma), used to detect cellulose, was dissolved in water at a concentration of 20 mg/ml and then added to agar medium before autoclaving at a final concentration of 40 μg/ml.
Random mutagenesis and screening for transposon mutants with altered rugose phenotypes.
Random mutagenesis was performed by transforming the wild-type (WT) strain S. enterica serovar Typhimurium DT104 Rv with Tn5 in the transposon-transposase (transposome) complex (EZ:TN Kan-2; EPICENTRE, Madison, WI) by electroporation (12). The transposase was activated by endogenous magnesium, resulting in transposition of Tn5 into the chromosome. Transformants were selected on LB-kanamycin plates. After observation at 25°C for 4 to 7 days, colonies that did not exhibit full rugosity compared to WT Rv were selected. The stability of the altered rugose phenotype was confirmed by three passages on LB plates at 25°C for 4 days.
Analysis of transposon mutants.
Random amplification of transposon ends was employed to rapidly analyze EZ:TN transposon mutants (9). Employing a three-step PCR, the method used a single primer (inv2) which is homologous to one end of the transposon but has minimum homology to the whole genome of S. enterica serovar Typhimurium DT104. The reactions usually produced several bands because of some homology with some Salmonella sequences adjacent to the transposon end where the inv2 primer binds; however, only a few of the PCR fragments had a transposon end. The PCR products were sequenced in the DNA sequencing facility at the Center for Biosystems Research at the University of Maryland with the transposon-specific sequencing primer provided in the EZ:TN kit.
Deletion of waaG, csgA, and bcsA and complementation of waaG.
Direct knockout of waaG, bcsA, and csgA in S. enterica serovar Typhimurium LT2 was performed according to Datsenko and Wanner (6). For deletion of waaG, primers pwaaGD11 and pwaaGD12 were used in a PCR with plasmid pKD4 as the template to amplify a Kanr-encoding gene flanked by DNA regions flanking waaG. The PCR products were treated with DpnI and electroporated into LT2 carrying λ Red helper plasmid pKD46 (Ampr, strainYA104). Transformants were selected on LB plates containing kanamycin. Primers pwaaG11 and pwaaG12, corresponding to the sequence just outside of the deleted region, were used in PCRs to verify the deletion of waaG and insertion of the Kanr-encoding gene in strain YA155. In addition, primers pKanKD3 and pwaaG11 were used to confirm the presence of the junction region between the sequence just outside of the deleted region and the Kanr-encoding gene. The same procedure was followed for csgA and bcsA deletions with primer pairs pcsgAD11-pcsgAD12 and pbcsAD11-pbcsAD12, creating strains YA151 and YA159, respectively, and with primer pairs pcsgA11-pcsgA12 and pbcsA11-pbcsA12 for PCR verification. For gene complementation, waaG was amplified from the LT2 strain with primers pwaaG11 and pwaaG12. The PCR product was cloned into pCR4 (TOPO Cloning; Invitrogen, Carlsbad, CA), and the ligation mixture was transformed into Escherichia coli TOP10. Plasmid pTOPO-waaG was isolated from the Ampr transformants, and the presence of an insert was verified by restriction analysis. Plasmid pTOPO-waaG was transformed into YA155 to create YA156.
RT-PCR.
Total RNA was isolated from 4-day-old colonies of LT2 and YA155 with the RNeasy kit (QIAGEN, Valencia, CA). Approximately 1 μg of isolated RNA was treated with 1 U of DNase (Invitrogen, Carlsbad, CA) for 30 min at 25°C. After inactivation of DNase by heating at 65°C for 10 min, the RNA was used as a template for reverse transcription (RT) with internal primers pwaaGIN11 and pwaaGIN12 or pwaaPIN11 and pwaaPIN12 and Superscript III reverse transcriptase (Invitrogen). The cDNA was amplified in separate PCRs with the same internal primers. The amount of total RNA was equalized by assessing the RT-PCR products of the two strains with primers p16srRNA11 and p16srRNA12 for 16S rRNA. No amplification product was obtained when the total RNA was used as the template in PCRs with these primers.
LPS profiling.
A 1:100 dilution of an overnight culture of Salmonella strains (Rv, Stv, LT2, the ddhC and waaG mutants, and the waaG-complemented strain) in LB either with or without antibiotics was grown at 37°C to an OD600 of 0.6. Preparation of the cell lysate was done according to Hitchcock and Brown (11). LPS was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) in Tris-Tricine gel (Bio-Rad, Hercules, CA), and the bands were visualized by silver staining.
Electron microscopy.
Electron microscopy was performed on 4-day-old colonies (Rv, Stv, LT2, the ddhC and waaG mutants, and the waaG-complemented strain) as described previously (1). The colonies were obtained after streaking the cells on LB plates and selecting those that had produced maximum rugosity. Briefly, colonies, including a thin layer of agar underneath, were excised and fixed in 2% glutaraldehyde in phosphate-buffered saline, pH 7.4, at room temperature for 1 h and at 4°C overnight. They were then postfixed with osmium tetroxide, followed by 2% aqueous uranyl acetate. The samples were then dehydrated with increasing concentrations of ethanol (75%, 90%, 95%, and 100%). They were critical point dried, coated with gold palladium alloy, and observed with a Hitachi S-4700 scanning electron microscope (Hitachi Scientific Instruments, Gaithersburg, MD).
Isolation of curli protein.
Diluted (10−6 to 10−7) overnight broth cultures of each strain (Rv, Stv, LT2, the ddhC and waaG mutants, and the waaG-complemented strain) were plated on LB and grown at 25°C to yield individual colonies. After either 2, 4, or 6 days, colonies were isolated and resuspended by vigorous vortexing in 10 mM Tris buffer, pH 6.8, in the presence of 2.0-mm-diameter sterile glass beads. One milliliter of suspension (OD600 adjusted to 3.0) was centrifuged, and the cell pellet was resuspended in 100 μl of SDS sample buffer (62.5 mM Tris Cl [pH 6.8], 10% glycerol, 2% SDS) and boiled for 10 min. The cell lysate was centrifuged, and the pellet was washed once with sterile water, dissolved in 100 μl of 97% formic acid (Sigma), frozen at −80°C, and lyophilized (25, 36).
SDS-PAGE and Western blotting.
The lyophilized samples (formic acid-resistant proteins) were resuspended in 100 μl of SDS sample buffer and sonicated for 5 s (Vibra Cell; Sonics & Materials, Danbury, CT) before being loaded onto a 16% polyacrylamide gel. Prestained Kaleidoscope standard proteins (Bio-Rad, Hercules, CA) were used as molecular weight markers. Equal loading was assessed visually by comparing the overall profile of formic acid-resistant proteins on the Coomassie blue-stained gel from each strain. Separated proteins were transferred onto polyvinylidene fluoride Immobilon-P membranes (Millipore, Bedford, MA). The membranes were then incubated overnight in 3% (wt/vol) skim milk in TTBS buffer (Tris-buffered saline containing 0.1% Tween 20) at room temperature and probed with mouse monoclonal anti-SEF17 antibody (a kind gift from William Kay) (36) diluted 1:1,000 in TTBS buffer, followed by horseradish peroxidase-conjugated goat anti-mouse antibodies. The antibody-bound proteins were detected with ECL detection reagents (Amersham Biosciences, Piscataway, NJ) and exposed to Hyperfilm (Amersham Biosciences). The density of several bands on the film, visually similar in intensity, was measured with the Quantity One program. Curli isolation was repeated and tested on three occasions with essentially the same pattern of proteins being recovered by Western blotting.
Cellulose assay.
Cells of each strain (Rv, Stv, LT2, the ddhC and waaG mutants, and the waaG-complemented strain) were streaked with dense inocula in 5-cm-long straight lines spaced approximately 3 cm apart on 15 large (150 by 15 mm) LB plates (with antibiotics added for the mutants). At 4 days, all of the strains, including the mutants, had developed their relative maximum rugosity when streaked in this fashion. Cells were harvested by scraping the growth from the agar surface with loops. To prevent dehydration of the cell masses during collection, the cell masses obtained from each plate were left accumulated on the agar on one side of the plate, keeping the plate lid on, and were then removed just immediately before weighing. Approximately 2 g (wet weight) of cells were harvested. The cell masses were placed in 50-ml polystyrene conical tubes, which were subsequently covered with Parafilm and lyophilized. From 2 g of wet weight, the yield was approximately 400 mg of dry weight in the WT strain and between 310 and 350 mg in the LPS mutants. The lyophilized cells (200 mg) were mixed with 3 ml of acetic-nitric reagent (8:2:1 acetic acid-nitric acid-distilled H2O [dH2O]) in 25-ml polystyrene centrifuge bottles and boiled for 30 min. The mixture was centrifuged, and the pellet was transferred to 25-ml Corex centrifuge bottles and washed once with 3 ml dH2O and once with 3 ml acetone. After drying with slow shaking at room temperature overnight, each pellet was dissolved in 125 μl of concentrated H2SO4 (Fisher) with slow shaking for 1 h at room temperature. Five microliters of each sample was diluted by mixing with 400 μl filtered dH2O in a boil-proof 1.7-ml Eppendorf tube (Eppendorf, Brinkmann Instruments, Inc.). The amount of cellulose isolated was detected by adding 750 μl of chilled anthrone (Sigma) reagent (0.2 g in 100 ml H2SO4) to each tube, boiling the tubes for 5 min, and reading the absorbance at 620 nm. The amount of cellulose in the samples was extrapolated from a linear standard curve derived from A620 readings of increasing volumes (5 to 40 μl) of 1:100 dilutions of Avicel cellulose (10 mg; Fluka Biochemika, Buchs, Switzerland; dissolved in 125 ml of H2SO4 for 2 min at 40°C), diluted in 400 ml dH2O, and reacted with 750 μl of anthrone reagent (35). Results are based on data from duplicate experiments, except for YA151, which was analyzed in replicate.
Biofilm assays and microscopic observations.
To visualize the production of biofilms in all Salmonella strains in glass test tubes, cells were inoculated into 5 ml of either M63 medium supplemented with 0.5% CAA and 0.2% glucose (M63) or LB Miller base broth (either alone [L] or supplemented with 0.2% glucose [LG] or with 0.2% glucose and 0.4% NaCl [LGS]) (3) in loosely capped borosilicate glass culture tubes (16 by 125 mm). The tubes were incubated with shaking at 210 rpm at 37°C for 20 h. To measure early adherence of cells to a polyvinyl chloride (PVC) surface, 100-μl volumes of overnight cultures (OD600 adjusted to 1.5) grown in M63 medium supplemented with 0.5% CAA and 0.2% glucose with (for the mutants) or without antibiotics were added to the wells of a Falcon PVC microtiter plate (35-3912 and 35-3913) and incubated without shaking at 37°C for 8 h. To determine the amount of crystal violet binding to the attached cells, 100 μl of M63 medium and 50 μl of 0.5% crystal violet solution were added to each well in the plate to stain the attached cells. After 15 min at room temperature, the plate was rinsed gently by immersion in water several times until the rinse water ran clear. The plate was air dried for 30 min, and 200 μl of 80:20 acetone-ethanol was added to each well for 10 min to release the crystal violet from the cells, after which the absorbance at 595 nm was read in a microtiter reader (Multiskan Ascent; Labsystems, Thermo Electron, Waltham, MA) (21). Values from replicate assays in 8 to 15 wells were averaged.
RESULTS
Random mutagenesis.
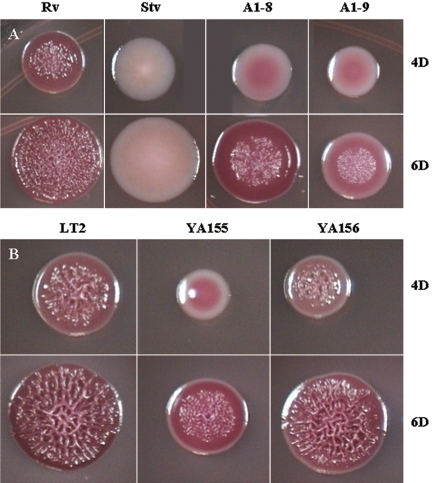
In order to identify mutants with an altered rugose phenotype, transposon mutants were screened at 4 days on LB plates with low osmolarity at 25°C, the conditions required for the WT strain S. enterica serovar Typhimurium DT104 Rv to develop the full rugose phenotype. Two mutants, A1-8 and A1-9, which showed a reduced rugose morphology, were selected for further study. The development of the mutant colonies could be observed more closely in LB plates containing approximately 10 colonies, which were obtained by spreading serially diluted overnight cultures of the mutants. Thus, with an inoculation from a highly diluted suspension, growth was more likely to begin from a single cell, with much slower formation of rugose characteristics (in 6 days) than if growth had originated from a heavy inoculum. Compared to the WT rugose Rv colonies that exhibited the rdar phenotype on the Congo red plate at 4 days, the A1-8 and A1-9 mutant colonies were pink and smooth, with white peripheries (Fig. 1A). A1-8 colonies had irregular edges, while A1-9 colonies were slightly smaller with entire edges. At 6 days, both mutants had developed some corrugation and A1-8 showed more-pronounced Congo red binding, similar to that of WT Rv; however, both mutants still had not developed full rugosity compared to the WT. Only the center of the colonies was corrugated and, interestingly, the corrugation lacked the complex pattern exhibited by the WT strain. A1-9 developed more WT-like rugose colonies than did A1-8 when streaked with a high-density inoculum on the plate. The colonies of A1-9 were very elastic when pulled with a loop, and in some streak areas they became very adhesive and stuck so strongly to the agar medium that they could not be removed with a loop.
FIG. 1.
Growth of S. enterica serovar Typhimurium DT104 Rv and LT2 and their derivatives on LB Congo red plates at 4 days (4D) and 6 days (6D) at 25°C. (A) Stv is a spontaneous mutant of Rv; A1-8 and A1-9 are ddhC::Tn5 and waaG::Tn5 mutants, respectively. (B) YA155 is a knockout mutant of waaG (ΔwaaG::kan), and YA156 is a complemented strain derived from YA155. Cells from overnight cultures grown at 37°C were serially diluted (10−6 to 10−8), spread on LB plates to obtain individual colonies, and grown at 25°C for 4 days (top) or 6 days (bottom).
We considered the possibility that reduced rugosity may have resulted from the slower growth of the LPS mutants on plates whose colonies appeared to be smaller than those of the WT. However, this did not appear to be the case, since the growth rates of A1-8 and A1-9 in LB broth at 28°C were comparable to that of WT Rv (data not shown). Only a very slight decrease in the rate of A1-9 growth was observed early in the exponential phase, and all three strains reached approximately the same OD600 value at 11 h. Assuming that the cells have reached stationary phase after 3 days of growth on the plates at 25°C and that the growth rate in broth would be relatively comparable to that of a colony on a plate, this observation rules out the possibility that a difference in growth rate affected the changes seen in the colonies of both mutants compared to the WT.
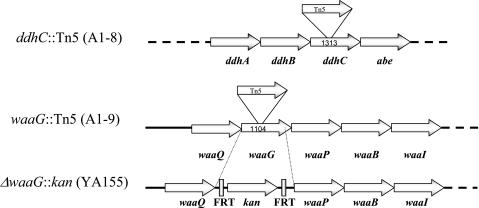
Analysis of the sites of transposon insertion in A1-8 and A1-9 revealed that the transposon had been inserted into two genes that function in the production of LPS, ddhC (formerly rfbH) and waaG (formerly rfaG), after nucleotides (nt) 553 and 612, respectively, with the Kanr-encoding gene of the transposon transcribed in the same direction as the two genes (Fig. 2).
FIG. 2.
Sites of transposon insertion in strains A1-8 (ddhC::Tn5) and A1-9 (waaG::Tn5) and deletion in strain YA155 (ΔwaaG::kan). Values indicate the sizes of the respective genes in base pairs. Tn5 was inserted at nt 553 in the ddhC gene and at nt 612 in waaG. In strain YA155, the entire waaG gene, except for the last 25 bp, was deleted and replaced with a kanR gene flanked by Flip recombinase sites (FRT) that were created in the gene knockout procedure. Dotted lines indicate the presence of additional genes in the operon.
Phenotypic characterization of waaG and ddhC mutants.
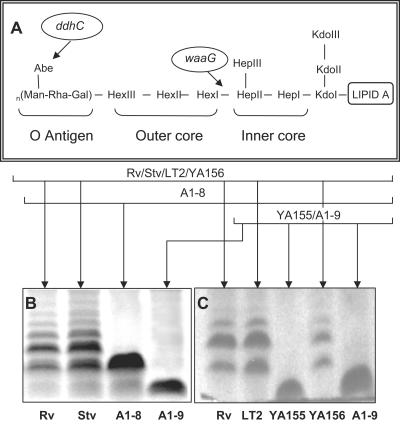
Because ddhC and waaG both code for proteins involved in LPS synthesis, the LPS profiles of the WT and mutant strains were analyzed to verify that alteration of LPS had occurred in the mutant strains. ddhC codes for CDP-4-keto-6-deoxy-d-glucose-3-dehydrase, which is involved in the synthesis of abequose, the last sugar component of the O antigen in LPS, and waaG codes for the enzyme UDP-glucose-LPS α1,3-glucosyltransferase, which functions in the addition of a glucose molecule to heptose II of the core polysaccharide of LPS (Fig. 3A), making the latter mutant a “deep rough” LPS mutant.
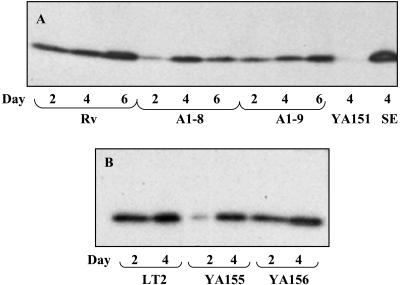
FIG. 3.
General structure of LPS in S. enterica serovar Typhimurium and roles of both ddhC and waaG in LPS synthesis. (A) Structure of Salmonella LPS with the sites affected by ddhC and waaG mutations identified. (B) LPS electrophoretic profiles of the A1-8 (ddhC::Tn5) and A1-9 (waaG::Tn5) mutants of S. enterica serovar Typhimurium DT104 showing the production of low-molecular-weight LPS compared with the ladder pattern of WT strain Rv. (C) A profile of LPS alteration similar to that of A1-9 was shown in the waaG knockout strain of the WT LT2 strain, YA155, which was complemented to the WT phenotype in strain YA156 by plasmid-borne waaG. The production of low-molecular-weight LPS in mutants further confirmed the roles of both ddhC and waaG in determining the structure of LPS. Proteinase K-treated whole-cell lysates were prepared from cells growing in early log phase at 37°C in LB, run on an SDS-PAGE gel, and silver stained. Abe, abequose; Kdo, 2-keto-3-deoxyoctulosonic acid.
WT S. enterica serovar Typhimurium DT104 Rv produced the typical ladder pattern on the SDS-PAGE gel, with different-molecular-weight bands separating LPS molecules containing lipid A and core polysaccharide with various lengths of the O-antigen polysaccharide chain (Fig. 3B). In comparison, the ddhC mutant expressed mostly a low-molecular-weight LPS corresponding to a band that was located slightly higher than the core polysaccharide with one repeat of O antigen, possibly indicating a lack of complete O antigen in the absence of abequose. The waaG mutant primarily produced an incomplete core polysaccharide of LPS. The alteration of the LPS O antigen in both the ddhC and waaG mutants was further verified by somatic O-antigen analysis. While Rv cells agglutinated in the presence of group B Salmonella antiserum, no agglutination was observed when A1-8 and A1-9 cells were suspended in the antiserum. The absence of O antigen was also confirmed by the inability of phage P22, which requires complete O antigen for attachment, to infect these two mutants (data not shown).
Matrix production.
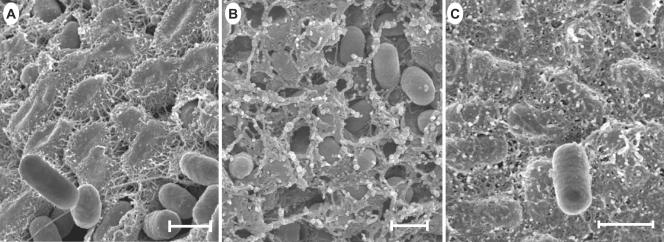
Because the rugose morphotype is associated with the production of extracellular matrix (1, 25), scanning electron micrographs of the mutant and WT cells of colonies streaked on plates at 4 days of growth, when some colonies appeared to produce relatively maximum rugosity, were compared. WT Rv produced a fibrous matrix that was seen at high magnification as multiple fibers projecting from the perimeter of the cells, interconnecting them with adjacent cells, and forming tightly configured fibrillar mats (Fig. 4A). Only a few loose cells were found occasionally throughout the colonies. The ddhC mutant A1-8, however, produced reduced amounts of extracellular matrix with differential characteristics compared with that of the WT. The matrix was fibrous and nodular, and the cells were more loosely associated with the matrix, showing an “eggs in the nest” profile (Fig. 4B). Similar pictures were obtained from different areas (the shallow grooves and hills) of the colony. It was also noted that the cells were shorter than those in the WT. In contrast, mutant A1-9 produced a profuse and thicker matrix, which completely covered the cells such that individual cells were difficult to discern (Fig. 4C). The matrix appeared to be more fibrous and had an adhesive-like characteristic. Therefore, differences in the rugose phenotype were partly due to changes in matrix production.
FIG. 4.
Scanning electron micrographs of cells from intact colonies of S. enterica serovar Typhimurium DT104 Rv and mutants A1-8 and A1-9 grown on LB agar for 4 days at 25°C. Cells were streaked on LB agar for isolated colonies, and those reaching relatively full rugosity for the corresponding strain after 4 days were prepared for scanning electron microscopic observation. A thick fibrous matrix covered most of the cells in Rv colonies, as shown in panel A, and was contrasted with some smooth cells occasionally seen lying on the cellular matrix. The ddhC::Tn5 mutant (A1-8) produced a thinner fibrous matrix (B). The waaG::Tn5 mutant (A1-9), however, produced a much thicker, adhesive-like matrix (C) than that of the WT. Bars, 1 μm.
Curli production.
Differences in the extracellular matrix which affect the rugose phenotype and Congo red binding of colonies can, at least in part, be due to changes in curli production (36). Therefore, curli protein was isolated from WT, A1-8, and A1-9 cells and Western blotting was performed to quantify CsgA, the major subunit of curli protein. Curli protein was produced by WT Rv starting at day 2, when the cells still appeared to be smooth, with slightly increased production when the cells were obviously rugose at days 4 and 6 (Fig. 5A). Comparatively, a reduced amount of curli was produced by A1-8 and A1-9 at days 2, 4, and 6 relative to that produced by WT Rv. The amounts of CsgA produced by Rv and A1-8 at day 4 appeared to be similar, but further quantification of band density showed that A1-8 actually produced only 84% of that produced by Rv.
FIG. 5.
Curli production as judged from the major subunit CsgA production of WT Rv and LT2 and their derivatives. Curli production is shown for WT Rv and its ddhC::Tn5 (A1-8) and waaG::Tn5 (A1-9) mutants after 2, 4, and 6 days of growth (A) and for WT LT2 with its ΔwaaG::kan (YA155) derivative and the waaG-complemented strain of YA155 (YA156) (B) after 2 and 4 days of growth on LB at 25°C. Strains S. enterica serovar Typhimurium LT2 YA151 (ΔcsgA) and S. enterica serovar Enteritidis 4b (SE) were included as negative and positive controls, respectively, in panel A. Curli protein was isolated by formic acid treatment of whole-cell lysates, separated by SDS-PAGE, and probed with anti-S. enterica serovar Enteritidis 4b AgfA monoclonal antibodies.
Direct knockout of waaG resulted in phenotypes similar to those of the transposon insertion mutants.
To exclude the possibility of multiple transposon insertions, spontaneous mutations, and polar effects, and because of an inability to transduce the LPS mutations into a WT Rv background, a single-gene knockout was performed. The waaG mutant was selected for deletion, since this mutant showed a more profound change in matrix production compared to WT Rv. Because of the multiantibiotic resistance phenotype of S. enterica serovar Typhimurium DT104, transformation of WT strain Rv with the λ Red helper plasmid (Ampr) was not possible (6). Therefore, the deletion was performed on non-antibiotic-resistant strain S. enterica serovar Typhimurium LT2. The LT2 strain has either an mviA defect (2) or a rare UUG start codon in the rpoS gene (15) resulting in altered expression of RpoS. Nevertheless, this strain exhibited a normal rugose phenotype comparable to that of strain DT104 Rv (Fig. 1A and B).
The waaG gene is part of the waaQGPBIJ operon, which encodes the enzymes for synthesis and modification of the core polysaccharide of LPS (27). The selected procedure deletes only a single gene without affecting the adjacent gene in an operon because of the presence of a ribosome binding site at the 3′end of the PCR product (6). Except for the last 25 bp, the mutation deleted the entire waaG gene, which was replaced with the Kanr-encoding gene, later shown to be transcribed in the same direction as waaG (Fig. 2). RT-PCR analysis confirmed that the mutation was not polar since waaP, the gene downstream of waaG in the operon, was still transcribed in knockout strain YA155 (data not shown).
As shown in Fig. 1B, the waaG mutant of LT2, strain YA155, was smaller in colony size than WT LT2. The growth rate of YA155 at 28°C was somewhat reduced compared to that of the WT in the log phase (data not shown); however, just before reaching the stationary phase, there was a crossover at about 30 h in the growth curves and the waaG mutant reached an OD even higher than that of the WT. A similar pattern of growth was observed at 37°C, and the OD readings were confirmed by viable counts at periodic points during the growth curve (data not shown). Thus, at stationary phase, there were slightly more mutant cells, although they were visibly much smaller (data not shown), than WT cells. When one colony each of WT strain LT2 and mutant strain YA155 grown for the same time (26 h at 37°C), with colony sizes of 2.5 mm and 2 mm, respectively, were each serially diluted and viable cells were counted, approximately 50% more cells were present in the YA155 colony (6.4 × 108 CFU) than in the WT colony (4.4 × 108 CFU). These observations suggested that, in this case, the colony size did not correspond to the number of cells and the rate of growth. This further suggested that changes in the waaG mutants were probably not due to slower growth and consequently fewer numbers of cells but were more likely the result of physiological changes exerted by the LPS core alteration.
Strain YA155 was similar phenotypically to transposon mutant A1-9 in colony morphology and Congo red binding (Fig. 1B). The original phenotype could be restored when the gene was complemented with a plasmid in strain YA156 (Fig. 1B). The similarity of the LPS profiles between the transposon and deletion mutants was also further confirmed (Fig. 3B and C). The complemented strain produced an LPS chain comparable to that of the WT, indicating that a single gene was likely responsible for the phenotypic changes in the waaG mutant (Fig. 3C). Impaired curli production was also displayed by knockout mutant YA155 at day 2 (Fig. 5B). Although the amount of curli protein produced appeared to be similar to that of the WT visually at day 4, further quantification of the band densities showed that the production of YA155 was approximately 70% of that of LT2. Complementation of YA155 with a plasmid increased curli production to a level similar to that exhibited by the WT strain.
Cellulose production.
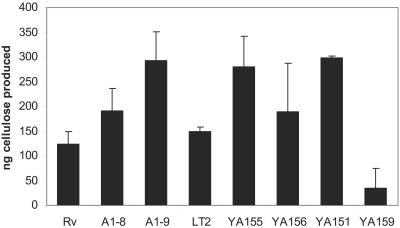
Preliminary studies had shown that, visualized under UV light, calcofluor-bound A1-8 colonies seemed to have a fluorescence intensity similar to that of WT Rv, while A1-9 had relatively brighter fluorescence than the parent strain (data not shown), perhaps indicating that the waaG mutant produced more polysaccharide, probably cellulose, than the WT strain. Hyperproduction of cellulose was further confirmed by quantitative analysis. Both LPS mutants produced more cellulose than the WT strain from the same amount of dry cell pellet, while waaG mutants A1-9 (paired, one-tailed Student t test, P = 0.025) and YA155 had the highest yield, almost double the amount of cellulose produced by the WT strain (P = 0.03), and complementation of strain YA156 with waaG reversed its cellulose production, making it similar to that of the WT strain (P = 0.29) (Fig. 6). Therefore, the increase in calcofluor binding seemed to reflect the increase in cellulose production. Additionally, non-curli-producing csgA mutant YA151 showed an overall greater amount of cellulose than the WT (P = 0.03), approximating the amount of cellulose produced by the waaG mutant. This observation raised the possibility that production of curli may actually impede cellulose production. As expected, a very small amount of cellulose was produced by bcsA mutant YA159 (P = 0.03).
FIG. 6.
Cellulose production by WT Rv and LT2 and their derivatives. Greater amounts of cellulose were produced by both the ddhC::Tn5 (A1-8) and waaG::Tn5 (A1-9) mutants than by WT Rv. Greater cellulose production was also obtained from the waaG deletion mutant YA155 than from WT LT2 and waaG-complemented strain YA156. Cellulose was isolated from 200 mg of lyophilized cell mass obtained from colonies of each strain streaked with a high-density inoculum on LB plates, followed by a 4-day incubation at 28°C. Control strains YA151 (ΔcsgA::kan) and YA159 (ΔbcsA::kan) were included as positive and negative controls, respectively. Error bars represent the standard deviations.
Both mutants are biofilm formers.
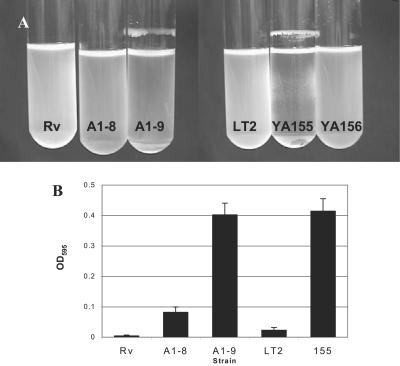
Since the appearance of a rugose phenotype and extracellular matrix production had been associated with biofilm formation, the ability of the mutants to form biofilms was investigated under different growth conditions. Previous studies have shown that the ability of salmonellae to form biofilms may depend on the type of medium in which they are grown (29). When grown in rich medium (LB) at 37°C with shaking, neither the WT nor the mutants were able to form biofilms (Table 3). This response was expected, since both curli and cellulose, components of the extracellular matrix required for biofilm formation, are not produced at high temperatures (39). However, when grown in M63 minimal medium supplemented with glucose and incubated with shaking at 37°C, ddhC mutant A1-8 and waaG mutants A1-9 and YA155 formed thick biofilms appearing as a ring at the liquid-air interface. When 0.2% glucose was added to the LB medium either alone (LG) or in combination with 0.4% NaCl (LGS), both mutants again formed biofilms that were similar to those seen in glucose-supplemented minimal medium (Table 3), although that produced by A1-8 was more fragile (Fig. 7A). The waaG mutant YA155 also appeared as spots stuck to the walls of the tubes at the liquid-solid interface (Fig. 7A). Thus, it seems that it was the presence of glucose and salt in the emended medium, and not the minimal medium per se, that promoted biofilm formation in the waaG mutant under these conditions. In contrast, WT strains Rv and LT2 formed no visible biofilms in any of the three different media at 37°C (Table 3).
TABLE 3.
Biofilm formation by S. enterica serovar Typhimurium
| Strain | Biofilma formation in indicated mediumb at:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 37°C
|
28°C
|
|||||||
| L | LG | LGS | M63 | L | LG | LGS | M63 | |
| Rv | − | − | − | − | +++ | − | − | − |
| A1-8 | − | + | + | ++ | − | + | + | ++ |
| A1-9 | − | ++ | +++ | +++ | + | ++ | ++ | +++ |
| LT2 | − | − | − | − | ++ | − | − | − |
| YA155 | − | +++ | +++ | ++++ | − | + | ++ | +++ |
| YA151 | − | − | − | − | − | − | − | − |
| YA159 | − | − | − | − | − | − | − | − |
Symbols: +, barely visible ring; ++, distinctly visible ring; +++, presence of a thick ring on the tube wall at the liquid-air interface similar to that produced by WT Rv in L medium at 28°C; ++++, presence of an additional spotty adherence on the tube wall below the surface of the broth; −, absence of a ring.
The media used were LB only (L), LB supplemented with 0.2% glucose (LG), LB supplemented with 0.2% glucose and 0.4% NaCl (LGS), and M63 minimal medium supplemented with 0.5% CAA and 0.2% glucose (M63).
FIG. 7.
Production of biofilms by ddhC::Tn5 (A1-8) and waaG::Tn5 (A1-9) mutants. (A) Biofilm production by waaG mutants A1-9 (waaG::Tn5) and YA155 (ΔwaaG::kan) was observed as rings on the tube walls at the liquid-air interface. Strain A1-8 produced fragile biofilms which detached easily and cannot be seen here. Note the less dense medium in both A1-8 and A1-9 caused by the formation of cell clumps, which caused the cells to settle to the bottom of the tube. Bacteria were grown in LGS at 37°C with shaking at 250 rpm for 20 h. Complementation of waaG in strain YA156 eliminated the ability of cells to form biofilms under this condition. (B) The ddhC::Tn5 (A1-8) and waaG::Tn5 (A1-9) mutants were shown to exhibit higher levels of adherence to PVC surfaces compared to WT Rv. The higher level of adherence of A1-8 than of WT LT2 was similar to that of ΔwaaG::kan mutant strain YA155. Cells were grown in M63 medium supplemented with 0.2% glucose and 0.5% CAA in microtiter wells for 8 h. Attached cells were quantified by staining the biofilms with crystal violet and determining the absorbance of the solubilized dye at 595 nm.
At a lower temperature, 28°C, the WT strains formed biofilms only in L medium, as expected for curli and cellulose induction, and which was further supported by the results showing that neither csgA mutant YA151 nor bcsA mutant YA159 formed any biofilms in any of the media under these growth conditions (Table 3). However, the ddhC showed a thick ring of growth in all of the media except L, while the waaG mutant produced biofilms in all four media (L, LG, LGS, and M63). Various conditions thus induced biofilm formation differently in these strains.
Formation of biofilms was also assessed quantitatively in the initial adherence stage on the PVC surface of microtiter wells of cells grown in M63 medium with glucose supplementation at 37°C. Mutants A1-9 and YA155 displayed approximately 10 times as much crystal violet binding as parent strains Rv and LT2, respectively, after incubation for 8 h (Fig. 7B). The ddhC mutant also showed increased adhesion, although not equal to that observed for the waaG mutant. Additional experiments with LB broth produced inconclusive results because the medium components, without any cells, adhered to the PVC surface (data not shown). LPS alterations resulting from mutations in tol in Pseudomonas fluorescens SBW25 was shown to reduce the strength of an air-liquid biofilm (pellicles) (30). However, we did not observe a significant difference in pellicle strength between the WT and both ddhC and waaG mutants in the present study (data not shown).
DISCUSSION
LPS alteration and its effect on the rugose phenotype.
During these investigations of the rugosity phenomenon in salmonellae, the possibility of involvement of cellular factors in rugosity expression was considered. Two semirugose mutants, A1-8 and A1-9, derived from random mutagenesis were selected as candidates for further study because of aberrant rugosity expression. Each had a transposon insertion in a gene involved in LPS production, i.e., ddhC or waaG. In S. enterica serovar Typhimurium, the LPS is composed of lipid A, 2-keto-3-deoxyoctulosonic acid, the inner core composed of three phosphorylated heptose groups (HepI, HepII, and HepIII), an outer core composed of three hexose groups, and repeating units of O antigen. ddhC is a member of the rfb operon encoding the locus for the chromosomal O-antigen biosynthesis of LPS. The LPS profile of this mutant showed that the absence of ddhC prevented the cells from repeating the O-antigen chain. On the other hand, disruption of waaG resulted in the production of a more severely truncated LPS that lacked the outer core of LPS (Rd1 chemotype) (16). The fact that these two different levels of LPS mutations both retarded rugose morphology implied a positive role for LPS in rugose appearance. A similar phenomenon of a rugose phenotype in Vibrio cholerae was also shown to be affected by mutations in two genes involved in LPS synthesis, rfdB and rfbE, which blocked the switching of V. cholerae from smooth to rugose (23). Although rugose morphology in V. cholerae is attributed to the presence of the exopolysaccharide (EPS) VPS and its regulator VpsR (38), LPS was still shown to be involved, thus indicating that the synthesis and/or assembly of some of the components of the rugose matrix may depend on an intact cell surface in V. cholerae, as well.
Effects of ddhC and waaG mutations on curli production.
The ddhC mutation, which affects the O antigen of LPS, appeared to have a minimal effect on Congo red binding. This corresponds to the finding of White et al. (36) that introduction of a galE mutation into S. enterica serovar Enteritidis affected O-antigen production, thus resulting in reduced aggregative morphology but little alteration in Congo red binding. However, the present work showed that there was some retardation of curli production, which may have been sufficient to alter the rugose matrix. The waaG mutant also showed a decrease in curli production compared to the WT, which was restored after the mutant YA155 was complemented by plasmid-borne waaG. Lowering of curli production may have been due to the effect of the waaG mutation on the integrity of the outer membrane, as suggested by the major increase in susceptibility to the hydrophobic compound novobiocin (a MIC of <1.6 μg/ml compared to 50 μg/ml) in the waaG mutant compared to the WT (data not shown). In E. coli, a waaG mutation had been shown to reduce the level of phosphorylation in the core polysaccharide, which may diminish membrane stability by affecting the level of cross-linking between LPS molecules (37). Therefore, the waaG mutation may have impaired normal curli production by affecting the transport and/or assembly of curli subunits. However, this explanation may not be relevant because the ddhC mutant, for which the novobiocin MIC is even greater than that for the WT (100 μg/ml compared to 50 μg/ml), also had impaired curli production.
Effect of LPS alteration on cellulose production.
Although curli production was depressed in both A1-8 and A1-9, it was found that cellulose was overproduced in each of them. The increase in the ddhC mutant was not at the level of the waaG mutant, and cellulose overproduction was also shown by the curli mutant YA151 (csgA). These results seemed to signify that normal curli production actually hinders cellulose production. Therefore, it is possible that it was the decrease in curli production in the waaG mutant that was in large part responsible for the increase in the amount of measured cellulose. White et al. (36) have suggested that both curli and cellulose are under similar regulation and are components of the same matrix; however, they may be produced at different times during the development of rugose morphology. Therefore, the synchrony of production may be important in establishing the balance between the two matrix components, since disruption in the timing of curli production may be a signal for increased cellulose synthesis. LPS may be significantly involved in maintaining this balance, thus serving as yet another possible mechanism or mediator of regulation of cellulose synthesis.
Although colanic acid may possibly play a role, the absence of mucoidy in the waaG mutant in this study, while present in a previous study (22, 34), prompted an alternative explanation. Pathways for EPS and LPS synthesis, in some instances, are shared. For example, in salmonellae, the same precursor molecule, UDP-glucose, is used for LPS O antigen and capsular polysaccharide (32), as well as for cellulose. Thus, the waaG mutation, which disrupts the transfer of UDP-glucose to LPS, may redirect the precursor to another polysaccharide pathway in which cellulose overproduction could occur. This switching has been shown in algC in P. aeruginosa, between the production of alginate and O antigen (7). Interestingly, V. cholerae and Lactobacillus lactis galU and galE mutants coding for UDP-glucose phosphorylase and UDP-galactose epimerase for LPS synthesis showed a lack of EPS production (4, 20).
Effect of LPS mutations on biofilm production.
Biofilm formation in L medium with low osmolarity by the WT strains at 28°C seemed to be dependent on curli and cellulose, as shown by control strains YA151 (csgA) and YA159 (bcsA), both of which failed to form biofilms under these conditions. However, in the presence of glucose or of glucose and NaCl at 37°C or 28°C, the amount of biofilm produced was reversed, with both mutants actually forming more biofilm than the WT. Thus, these observations were similar to those made with a ddhC mutant of Salmonella in a previous study (19), where there was an increase in adherence compared to the WT when bacteria were grown in LB medium with glucose at 30°C. However, our findings were in contrast to those on a waaG mutant of E. coli K-12 (10) in which the mutant exhibited reduced adherence to polystyrene surfaces compared to the WT strain grown in LB medium at 37°C. Our results showed that none of the strains produced biofilms in LB at 37°C. The difference between the adherence properties of the waaG mutants in our study and those in the E. coli study may possibly be due to the difference in their genetic backgrounds, as revealed by the differences in colanic acid production caused by mutations in this gene. Alternatively, the difference in the NaCl concentration in the growth medium, albeit small, may affect adherence of the waaG mutant quite significantly. However, the inverse relationship between LPS and attachment has been shown previously (14, 33, 34), and the present study further confirmed that observation.
Overexpression of cellulose in Agrobacterium tumefaciens may lead to more abundant biofilms (18). However, biofilm production by salmonellae has been shown to be dependent on osmolarity and temperature. Thus, the observation that adherence of the LPS mutants increased at both 28°C and 37°C in the presence of either glucose or a combination of NaCl and glucose, in which synthesis of both curli and cellulose is probably repressed in the WT strain, raises the distinct possibility that these mutants may form biofilms under these conditions by activating another, unique, biofilm-forming pathway. The use of an alternative pathway has been shown in several bacteria, whereby different appendages or cellular structures are produced by the bacteria forming biofilms under different growth conditions (13, 21, 28), thus revealing the ability of these bacteria to adapt to alteration in cell surfaces, as well as changing growth environments, to form biofilms. Whether novel Salmonella EPS (8) or Salmonella colanic acid (31) is involved remains an important question to address. The results of the present study indicate that salmonellae having LPS alteration may be able to induce alternative pathways in which the extracellular matrix is produced constitutively in medium with high osmolarity. Our results, in part, show that careful consideration of growth or assay conditions must be made when interpreting the effect of a single mutation on adherence, since the same gene disruption may appear to either reduce or increase the biofilm-forming ability and multicellular aggregation of cells, depending on the assay conditions. Further identification of the matrix components of biofilms formed in the presence of glucose and NaCl would help to reach more definitive answers to the complexity of biofilm formation in these mutants.
In summary, this study has shown that changes in the cell surface caused by LPS alteration in the ddhC and waaG mutants of S. enterica serovar Typhimurium result in significant changes in the production of both curli and cellulose, as well as biofilm formation. Balance in the production of the two matrix components curli and cellulose appears to depend, at least in part, on LPS. Curiously, the same effects seen in the ddhC and waaG mutants may be reproduced in the WT strains having mutations in other genes involved in LPS synthesis. Cellulose production in enteric bacteria has been only recently discovered (29, 30, 39), and overproduction of cellulose in Salmonella LPS mutants, as shown in this study, represents a novel phenomenon that suggests that cellulose production could possibly be regulated in a manner independent of that for curli. Additionally, further studies to elucidate the alternative pathways of biofilm formation by salmonellae are essential and may prove beneficial to the design of a method for the eradication of cellulose-containing biofilms, thus promoting more effective interventions against biofilm-forming pathogens.
Acknowledgments
We thank Tim Maugel for expert advice on electron microscopy methodology. We also thank Nathan Head for suggestions on the adherence assay, Barbara Triplett for suggestions on the cellulose assay, William Kay for valuable advice, and Richard Stewart and Clay Fuqua for suggestions and critical reading of the manuscript.
This research was supported in part by a grant from the Joint Institute for Food Safety and Applied Nutrition (JIFSAN), University of Maryland, College Park.
Footnotes
The electron microscopy work described herein is contribution 102 of the Laboratory of Biological Ultrastructure at the University of Maryland, College Park.
REFERENCES
- 1.Anriany, Y. A., R. M. Weiner, J. A. Johnson, C. E. DeRezende, and S. W. Joseph. 2001. Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl. Environ. Microbiol. 67:4048-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearson, S., J. Benjamin, W. Swords, and J. Foster. 1996. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 178:2572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies, D. 2000. Physiological events in biofilm formation, p. 37-52. In P. G. D. G. Allison, H. M. Lapin-Scott, and M. Wilson (ed.), Community structures and cooperation in biofilms. Cambridge University Press, New York, N.Y.
- 8.de Rezende, C. E., Y. Anriany, L. E. Carr, S. W. Joseph, and R. M. Weiner. 2005. Capsular polysaccharide surrounds smooth and rugose types of Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 71:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ducey, T., and D. Dyer. 2002. Rapid identification of EZ:TN transposon insertion sites in the genome of Neisseria gonorrheae. Epicentre Forum 9:6-7. [Google Scholar]
- 10.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 in the rfaG, rfaP, and gaIU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman, L., and J. Jendrisak. 1999. Use of EZ:TN transposomes for genetic analysis and direct sequencing of bacterial genomic DNA. Epicentre Forum 6:1-7. [Google Scholar]
- 13.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landini, P., and A. J. Zehnder. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating the expression of lipopolysaccharide and flagellar genes. J. Bacteriol. 184:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, I. S., J. Lin, H. K. Hall, B. Bearson, and J. W. Foster. 1995. The stationary phase sigma factor σS (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol. Microbiol. 17:155-167. [DOI] [PubMed] [Google Scholar]
- 16.MacLachlan, P. R., and K. E. Sanderson. 1985. Transformation of Salmonella typhimurium with plasmid DNA: differences between rough and smooth strains. J. Bacteriol. 161:442-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Matthysse, A. G., M. Marry, L. Krall, M. Kaye, B. E. Ramey, C. Fuqua, and A. R. White. 2005. The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol. Plant-Microbe Interact. 18:1002-1010. [DOI] [PubMed] [Google Scholar]
- 19.Mireles, J. R., II, A. Toguchi, and R. M. Harshey. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent, signaling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 22.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashid, M. H., C. Rajanna, A. Ali, and D. K. Karaolis. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113-119. [DOI] [PubMed] [Google Scholar]
- 24.Romling, U., M. Gomelsky, and M. Galperin. 2005. C-di-GMP: the dawning of a novel bacterial signaling system. Mol. Microbiol. 57:629-639. [DOI] [PubMed] [Google Scholar]
- 25.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 26.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transductional abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 27.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikh, J., S. Hicks, M. Dall'Agnol, A. D. Phillips, and J. P. Nataro. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41:983-997. [DOI] [PubMed] [Google Scholar]
- 29.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 30.Spiers, A. J., and P. B. Rainey. 2005. The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology 151:2829-2839. [DOI] [PubMed] [Google Scholar]
- 31.Stevenson, G., S. J. Lee, L. K. Romana, and P. R. Reeves. 1991. The cps gene cluster of Salmonella strain LT2 includes a second mannose pathway: sequence of two genes and relationship to genes in the rfb gene cluster. Mol. Gen. Genet. 227:173-180. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland, I. 1990. Biotechnology of microbial exopolysaccharides. Cambridge University Press, New York, N.Y.
- 33.Thomsen, L. E., M. S. Chadfield, J. Bispham, T. S. Wallis, J. E. Olsen, and H. Ingmer. 2003. Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 228:225-231. [DOI] [PubMed] [Google Scholar]
- 34.Toguchi, A., M. Siano, M. Burkart, and R. M. Hersey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Updegraff, D. M. 1969. Semimicro determination of cellulose in biological materials. Anal. Biochem. 32:420-424. [DOI] [PubMed] [Google Scholar]
- 36.White, A. P., D. L. Gibson, S. K. Collinson, P. A. Banser, and W. W. Kay. 2003. Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar Enteritidis. J. Bacteriol. 185:5398-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]