Abstract
Of 3,063 ready-to-eat food samples tested, 91 (2.97%) were positive for Listeria monocytogenes, and lineage 1 strains outnumbered lineage 2 strains 57 to 34. Seventy-one isolates (78%) exhibited multiple antibiotic resistance, and an L. monocytogenes-specific bacteriophage cocktail lysed 65 of 91 (71%) isolates. Determining phage, acid, and antibiotic susceptibility phenotypes enabled us to identify differences among strains which were otherwise indistinguishable by conventional methods.
Listeria monocytogenes is an important food-borne pathogen due to its high fatality rate. In nonpregnant adults, L. monocytogenes primarily causes septicemia, meningitis, and meningoencephalitis, and the mortality rate is 20 to 25% (22). This pathogen is particularly significant for cold-stored, ready-to-eat foods as it is frequently found in the environment and can grow at refrigerated temperatures. Our knowledge concerning the routes of food-borne transmission of L. monocytogenes has been acquired mostly through studies of epidemiological data from various prevalence studies and outbreak investigations (6, 10). The four main objectives of this study were (i) to analyze ready-to-eat food samples for the presence of L. monocytogenes; (ii) to determine the pulsed-field gel electrophoresis (PFGE) patterns and antimicrobial susceptibility profiles of the isolates; (iii) to measure the efficacy of an L. monocytogenes-specific bacteriophage cocktail for lysing the isolates; and (iv) to examine and characterize the isolates' ability to withstand acid challenge.
From January 2002 to December 2003, L. monocytogenes was cultured from ready-to-eat food samples collected in Florida using a standard randomized protocol and was characterized by using the procedure recommended by the USDA Food Safety and Inspection Service (23). Samples that tested positive for L. monocytogenes included deli-style sandwiches (n = 71), smoked turkey, beef, or ham (n = 11), salmon (n = 4), and miscellaneous (n = 5). (For further details about isolation dates, types of foods, etc. see the supplemental material.) The isolates were subjected to the PCR-based serogroup identification (2, 13) procedure using five primer sets to classify 91 L. monocytogenes strains into three serotype groups [serotype groups 1/2a(3a), 1/2b(3b), and 4b(d,e)]. The numbers of L. monocytogenes isolates belonging to lineage 1 [serotypes 1/2b(3b) and 4b(d,e)] and lineage 2 [serotype 1/2a(3a)] were 57 and 34, respectively.
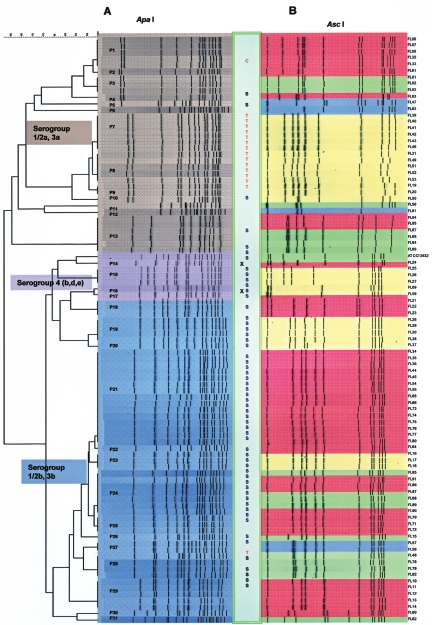
The pulsed-field gel electrophoresis patterns of genomic DNA after ApaI and AscI digestion (11) were compared in order to classify the strains into 31 PFGE groups (Fig. 1). PFGE patterns were compared using the BioNumerics software (version 3.5; Applied-Maths, Kortrijk, Belgium). Isolate relatedness was determined by the unweighted pair group method using arithmetic averages based on restriction with both enzymes. For some strains AscI was more discriminatory than ApaI, and for other strains ApaI was more discriminatory than AscI. For example, cluster analysis of the AscI digestion results grouped 29 isolates (FL16, FL17, FL18, FL34, FL35, FL36, FL44, FL45, FL54, FL55, FL64, FL65, FL66, FL70, FL71, FL72, FL73, FL74, FL75, FL76, FL77, FL80, FL85, FL86, FL87, FL88, FL89, FL90, and FL91), but ApaI digestion divided these isolates into five PFGE types, PFGE types P21 to P25. In another case, cluster analysis of the ApaI digestion results placed 13 L. monocytogenes isolates (FL19, FL20, FL31, FL39, FL40, FL41, FL42, FL43, FL46, FL49, FL51, FL52, and FL53) together in one cluster, but AscI digestion divided them into three PFGE types (PFGE types P7 to P9). PFGE type P21 was the largest PFGE type and was comprised of 15 isolates, followed by PFGE type P24 (seven strains) and PFGE types P7 and P13 (six strains each). PFGE types P1, P8, and P29 each contained five isolates while 15 PFGE types were represented by a single isolate. All of the serotype group 1/2a(3a) strains (n = 34) were clustered in 13 PFGE types, PFGE types P1 to P13 (Fig. 1); serotype group 1/2b(3b) strains (n = 51) were clustered in 14 PFGE types, PFGE types P18 to P31(Fig. 1); and serotype group 4b(d,e) strains (n = 6) were clustered in four PFGE types, PFGE types P14 to P17 (Fig. 1). For eight PFGE types (PFGE types P1, P7, P8, P9, P13, P19, P21, and P28) comprised of 47 isolates samples were collected over several months. When samples originated from the same location, the data indicated that the strain survived and proliferated for several days and persistently appeared in ready-to-eat foods. Isolates belonging to PFGE type P13 were obtained over an 11-month period (January to December 2003) from three different locations.
FIG. 1.
Cluster analysis of L. monocytogenes isolates along with phenotypic characteristics. PFGE was performed after genomic DNA was digested with ApaI (A) or AscI (B) using the CDC PulseNet standardized procedure (11). Digestion patterns for both restriction enzymes were used to determine PFGE types (PFGE types P1 to P31 in panel A). PCR-based serotype groups are indicated by brown [serotype group 1/2a(3a)], violet [serotype group 4b(d,e)], and blue [serotype group 1/2b(3b)]. The box between the two panels indicates antibiotic resistance (T, tetracycline; C, carbenicillin; S, sulfomethoxazole) and an inability to induce glutamate-dependent acid resistance under aerobic conditions (X). Susceptibility to the listeriophage cocktail was expressed as the number of PFU generated per ml by individual isolates and is indicated as follows: yellow, 0 to 101 PFU/ml; blue, 102 to 104 PFU/ml; green, 105 to 106 PFU/ml; and red, >107 PFU/ml.
Epidemiological studies of outbreaks of human disease have demonstrated that L. monocytogenes can cause gastrointestinal disorders with no or low associated mortality (1, 5). The ability of L. monocytogenes strains to cause listeriosis depends on their survival in the gastrointestinal tract. In addition to σB-dependent- and -independent pH homeostasis (7, 8, 24), the utilization of exogenous glutamate and glutamate decarboxylase of the pathogen plays a significant role (4, 17). The glutamate-dependent acid resistance system utilizes exogenous glutamate and was tested in EG medium containing 1.5 mM glutamate at pH 2.5. Most of the isolates (89/91 isolates) successfully induced the glutamate-dependent acid resistance system when they were grown to the stationary phase under aerobic conditions, and the addition of 1.5 mM glutamate during acid challenge increased the cell survival 100-fold or more (Table 1). Two serotype group 4b(d,e) isolates, FL24 and FL59, did not induce this system.
TABLE 1.
Listeriophage susceptibility and glutamate-dependent acid resistance phenotypes of serotype group 4b(d,e) isolatesa
| Strain (isolation date) | Pulse type | Phage susceptibility (PFU/ml) | Glutamate-dependent acid resistance (% survival) |
|---|---|---|---|
| ATCC 13932 | ND | 3.1 × 107 ± 0.8 × 107 | 7.6 ± 0.91 |
| ATCC 19115 | ND | 4.4 × 107 ± 0.8 × 107 | 4.4 ± 0.84 |
| FL9 (6 March 2003) | P16 | R | 7.75 ± 2.6 |
| FL24 (12 June 2003) | P14 | 5.2 × 107 ± 0.6 × 107 | 0.145 ± 0.02 |
| FL25 (18 June 2003) | P15 | R | 2.5 ± 0.26 |
| FL26 (18 June 2003) | P15 | R | 2.7 ± 0.33 |
| FL27 (18 June 2003) | P15 | R | 2.45 ± 1.2 |
| FL59 (14 October 2003) | P17 | R | 0.33 ± 0.11 |
ND, not determined. Tests to determine the susceptibility to the listeriophage cocktail and acid shock assays were performed three to five times, and the results are expressed as means ± standard deviations. R, resistant to the listeriophage cocktail (101 to 0 PFU/ml). For the glutamate-dependent acid resistance system the percent survival was determined after acid shock in EG medium containing 1.5 mM glutamate (pH 2.5) for 1 h.
While our understanding of the ecology and virulence of L. monocytogenes has clearly improved over the past decade, there is still limited information concerning the antibiotic resistance patterns of L. monocytogenes strains isolated during routine surveys of foods that have not been implicated in illness (9, 19). The isolates' antibiotic susceptibility patterns were determined using the broth microdilution method of the National Committee for Clinical Laboratory Standards (16). The resistance breakpoint concentrations used were 512 μg/ml for sulfomethoxazole, 4 μg/ml for ciprofloxacin, and 16 μg/ml for tetracycline. As expected, all 91 isolates were resistant to nalidixic acid. One isolate (1.1%) from smoked ham was found to be resistant to ciprofloxacin. Fifteen isolates (16%) exhibited tetracycline resistance, and 14 of these isolates belonged to lineage 2 [serotype group 1/2a(3a)] and to four PFGE types (PFGE types P7 to P10); the exception was strain FL11, which was a serotype group 1/2b(3b) strain belonging to PFGE type P29. Sulfomethoxazole resistance was detected for 55 isolates (60%), 45 of which were members of the pool of 51 serotype group 1/2b(3b) isolates. All isolates belonging to serotype group 4b(d,e) were resistant to sulfomethoxazole, while 30 of the 34 isolates belonging to serotype group 1/2a(3a) were sensitive to sulfomethoxazole. No resistance to ampicillin, gentamicin, penicillin G, or trimethoprim was observed for any L. monocytogenes isolate.
An additional approach that we used to differentiate the strains was to determine and compare their sensitivities to a bacteriophage mixture (LMP-102) containing six distinct lytic phages specific for L. monocytogenes serotypes 1/2a, 1/2b, 1/2c, 3a, 3b, 4b, and 4d (14). The phages were selected based on their ability to lyse L. monocytogenes isolates during a screen involving more than 200 isolates belonging to different serotypes (A. Sulakvelidze, personal communication). Most isolates (64/91 isolates) were lysed by the phage cocktail and generated a phage titer of >105 PFU/ml in the culture supernatants after they were infected with the phage cocktail. Although the majority of the serotype group 1/2b(3b) isolates (40/51 isolates [80%]) were susceptible to the listeriophage cocktail, several serotype group 4b(d,e) isolates were resistant. It is interesting that L. monocytogenes strains from the ATCC belonging to serogroup 4b (ATCC 13932 and ATCC 19115) (Table 1), as well as food-borne outbreak strain LCDC 81-861 (3, 20) (data not shown), were effectively lysed by the listeriophage cocktail. In order for this control strategy to be effective, phages that lyse several different L. monocytogenes strains, especially strains belonging to serotypes 4b and 1/2b, must be found. The data also provided a preliminary insight into the efficacy with which the L. monocytogenes isolates from ready-to-eat foods in Florida may be reduced or eliminated by treatment with L. monocytogenes-specific bacteriophages, an approach that has been gaining increased attention lately (12, 15, 21).
The combination of phage and antibiotic susceptibility phenotypes enabled us to identify differences among some of the PFGE type P13 isolates which were otherwise indistinguishable by PFGE and PCR-based serotype analyses (Table 2). Isolates FL68, FL69, and FL84 exhibited moderate resistance to the phage cocktail (they generated 100-fold fewer PFU than FL4 and FL5 generated), and isolate FL84 also exhibited resistance to sulfomethoxazole (>512 μg/ml). It was difficult to determine precisely if this strain generated genetic variants during the time that it was present in the implicated food service facilities. However, the indistinguishable PFGE patterns after individual digestion with two restriction enzymes suggest that the isolates may have had a common ancestor and undergone minor genetic modifications, resulting in reduced susceptibility to listeriophages and resistance to sulfomethoxazole.
TABLE 2.
Differentiation of food-borne isolates belonging to pulse type P13 based on antibiotic and listeriophage susceptibility phenotypesa
| Strain | Date of isolation | Antibiotic resistance | Phage susceptibility |
|---|---|---|---|
| FL4 | 13 February 2003 | 2.9 × 108 ± 2.8 × 108 | |
| FL5 | 21 February 2003 | 3.1 × 108 ± 2.5 × 108 | |
| FL67 | 6 November 2003 | 1.3 × 107 ± 1.0 × 107 | |
| FL68 | 6 November 2003 | 1.7 × 106 ± 0.3 × 106 | |
| FL69 | 6 November 2003 | 3.6 × 106 ± 1.0 × 106 | |
| FL84 | 11 December 2003 | Sulfomethoxazole | 2.4 × 106 ± 0.6 × 106 |
Tests to determine susceptibility to the listeriophage cocktail were performed three to five times, and the results are expressed as means ± standard deviations.
PFGE, either alone or in combination with serotyping, is currently the method of choice for investigating food-borne outbreaks of listeriosis. This strategy is also used for tracing the outbreak-causing strain to the source of contamination, information which has significant epidemiological and public health ramifications and has been used in recent surveys (10, 18, 19). Our data indicate that a number of L. monocytogenes isolates that were indistinguishable by PFGE may not necessarily be identical isolates. We were able to differentiate food-borne isolates based on antibiotic resistance and acid tolerance phenotypes in combination with phenotypic analysis. The information obtained should be useful for epidemiological and public health studies of L. monocytogenes.
Supplementary Material
Acknowledgments
We thank Frances Trouth, Michelle Orton, and Amy Blodgett for their excellent technical assistance. We also thank Carl Schroeder for comments and suggestions during preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aureli, P., G. C. Fiorucci, D. Caroli, G. Marchiaro, O. Novara, L. Leone, and S. Salmaso. 2000. An outbreak of febrile gastroenteritis associated with corn contaminated by Listeria monocytogenes. N. Engl. J. Med. 342:1236-1241. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, M. K., and D. R. Call. 2003. Listeria monocytogenes serotype identification by PCR. J. Clin. Microbiol. 41:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conner, D. E., R. E. Brackett, and L. R. Beuchat. 1986. Effect of temperature, sodium chloride, and pH on growth of Listeria monocytogenes in cabbage juice. Appl. Environ. Microbiol. 52:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 5.Dalton, C. B., C. C. Austin, J. Sobel, P. S. Hayes, W. F. Bibb, L. M. Graves, B. Swaminathan, M. E. Proctor, and P. M. Griffin. 1997. An outbreak of gastroenteritis and fever due to Listeria monocytogenes in milk. N. Engl. J. Med. 336:100-106. [DOI] [PubMed] [Google Scholar]
- 6.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of sigma-B in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes sigma-B in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 10.Graves, L. M., S. B. Hunter, A. R. Ong, D. Schoonmaker-Bopp, K. Hise, L. Kornstein, W. E. DeWitt, P. S. Hayes, E. Dunne, P. Mead, and B. Swaminathan. 2005. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 43:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves, L. M., and B. Swaminathan. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulse-field gel electrophoresis. Int. J. Food Microbiol. 65:55-62. [DOI] [PubMed] [Google Scholar]
- 12.Hudson, J. A., C. Billington, G. Carey-Smith, and G. Greening. 2005. Bacteriophages as biocontrol agents in food. J. Food Prot. 68:426-437. [DOI] [PubMed] [Google Scholar]
- 13.Jinneman, K. C., and C. Hill. 2001. Listeria monocytogenes lineage group classification by MAMA-PCR of the listeriolysin gene. Curr. Microbiol. 43:129-133. [DOI] [PubMed] [Google Scholar]
- 14.Leverentz, B., W. S. Conway, M. J. Camp, W. J. Janisiewicz, T. Abuladze, M. Yang, R. A. Saftner, and A. Sulakvelidze. 2003. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl. Environ. Microbiol. 69:4519-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leverentz, B., W. S. Conway, W. Janisiewicz, and M. J. Camp. 2004. Optimizing concentration and timing of a phage spray application to reduce Listeria monocytogenes on honeydew melon tissue. J. Food Prot. 67:1682-1686. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2003. National Committee for Clinical Laboratory Standards methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed., vol. M100, p. A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 17.Oliver, M., S. Rousseaux, P. Piveteau, J. P. Lemaitre, A. Rousset, and J. Guzzo. 2004. Screening of glutamate decarboxylase activity and bile salt resistance of human asymptomatic carriage, clinical, food, and environmental isolates of Listeria monocytogenes. Int. J. Food Microbiol. 93:87-99. [DOI] [PubMed] [Google Scholar]
- 18.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders, B. D., K. Mangione, C. Vincent, J. Schermerhorn, C. Farchione, N. Dumas, C. A. Bopp, L. Kornstein, E. D. Fortes, K. Windham, and M. Wiedmann. 2004. Distribution of Listeria monocytogenes molecular subtypes among human and food isolates from New York state shows persistence of human disease-associated Listeria monocytogenes strains in retail environments. J. Food Prot. 67:1417-1428. [DOI] [PubMed] [Google Scholar]
- 20.Schlech, W. F., P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 21.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swaminathan, B. 2001. Listeria monocytogenes, p. 383-410. In M. P. Doyle, L. R. Beuchat, and T. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 23.USDA Food Safety and Inspection Service. 13 September 2005, posting date. Isolation and identification of Listeria monocytogenes from red meat, poultry, egg and environmental samples. [Online.] United States Department of Agriculture Food Safety and Inspection Service Office of Public Health Science, Washington, D.C. http://www.fsis.usda.gov/Ophs/Microlab/Mlg_8_04.pdf.
- 24.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. P. L. A. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.