Abstract
Biphasic systems can overcome the problem of low productivity in conventional media and have been exploited for biocatalysis. Solvent-tolerant microorganisms are useful in biotransformation with whole cells in biphasic reactions. A solvent-tolerant desulfurizing bacterium, Pseudomonas putida A4, was constructed by introducing the biodesulfurizing gene cluster dszABCD, which was from Rhodococcus erythropolis XP, into the solvent-tolerant strain P. putida Idaho. Biphasic reactions were performed to investigate the desulfurization of various sulfur-containing heterocyclic compounds in the presence of various organic solvents. P. putida A4 had the same substrate range as R. erythropolis XP and could degrade dibenzothiophene at a specific rate of 1.29 mM g (dry weight) of cells−1 h−1 for the first 2 h in the presence of 10% (vol/vol) p-xylene. P. putida A4 was also able to degrade dibenzothiophene in the presence of many other organic solvents at a concentration of 10% (vol/vol). This study is a significant step in the exploration of the biotechnological potential of novel biocatalysts for developing an efficient biodesulfurization process in biphasic reaction mixtures containing toxic organic solvents.
Sulfur oxides generated by the combustion of sulfur-containing fossil fuel cause severe environmental pollution. Biodesulfurization is thought to be an interesting alternative for the development of a new petroleum-refining process (2, 10, 11, 16, 18, 39). The derivatives of dibenzothiophene (DBT) and benzothiophene (BT), as well as other polycyclic aromatic sulfur heterocyclic compounds (PASHs), are the most abundant heterocyclic compounds in petroleum. Alkyl DBTs and alkyl BTs are highly recalcitrant to chemical catalysts, especially when they are alkylated at positions 4 and 6 (17, 19, 25). Many researchers have investigated biological desulfurization systems using DBT or alkyl DBTs as model compounds, and the metabolic pathway of desulfurization was proposed to be the so-called “4S” pathway, which removes sulfur while leaving the carbon backbone intact (12, 18, 20, 22, 38).
The genes responsible for the “4S” pathway of Rhodococcus erythropolis IGTS8 have been cloned and sequenced. There are three open reading frames, which are transcribed in the same orientation and are designated dszA, dszB, and dszC, as shown in Fig. 1 (28). The reaction catalyzed by the products of dszABC requires addition of reduced flavin (reduced flavin mononucleotide), which cannot be replaced by reduced pyridine nucleotide (NADH) or other flavins (flavin adenine dinucleotide or riboflavin). DszD is a flavin reductase which during NADH oxidation supplies the reducing equivalents to the desulfurizing reaction. The dszD gene encoding DszD, which is located on the chromosome, has been introduced into many strains to enhance the desulfurizing activity (8, 9, 11, 27, 37).
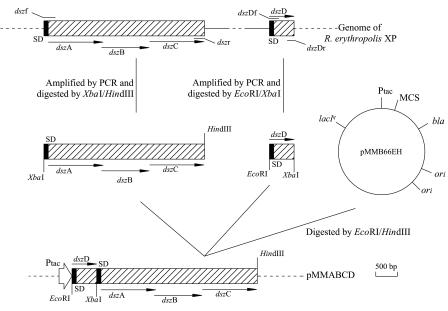
FIG. 1.
Construction of recombinant plasmid pMMABCD. The arrows indicate the direction of transcription of the genes. Only relevant restriction sites are shown (EcoRI, XbaI, HindIII). Primers dszf, dszr, dszDf, and dszDr were used for amplification of dszABCD and dszD. bla, gene encoding ampicillin resistance; lacIq, gene encoding the LacI repressor; Ptac, promoter; MCS, multiple cloning site; SD, Shine-Dalgarno sequence; ori, sequence encoding the origin of replication for duplex DNA.
A biphasic system containing water-immiscible organic solvents has been exploited for biocatalysis because it can overcome the problem of low productivity in conventional media caused by poor substrate solubility. A biphasic system can also integrate bioconversion and product recovery in a single reactor and shift the chemical equilibrium to enhance yields and selectivity (21, 34). However, many organic solvents are highly biotoxic and can kill most microorganisms even at low concentrations (0.1%, vol/vol), which has made selection of a solvent that combines adequate physicochemical properties with biocompatibility a difficult task (13, 21). Fuel oil has properties similar to those of an organic solvent and is also toxic to microorganisms. An immobilized-cell system was used previously to alleviate the harmful effects of oil in microbial desulfurization of fuels (23, 36). However, the mass transfer resistance is enhanced compared to that in free-cell systems, mainly due to internal mass transfer limitations (21). It would be preferable to have free cells that exhibit high activity in the presence of organic solvents. Bacteria isolated from an environment contaminated by organic compounds, such as toluene and xylene, were able to tolerate organic solvents due to their special structure and their characteristic physiological mechanisms (15). Some of these strains could grow even when the organic solvent concentration was more than 50% (vol/vol) (4). Microorganisms with a high tolerance to organic solvents are useful and important in many biotechnological fields, such as biodesulfurization and biocatalysis (5, 21). However, there have been no reports concerning biodesulfurization in the presence of high concentrations of toxic organic solvents. Considering the difficulty of isolating strains having both solvent tolerance and the desired catalytic activity from the environment, it may be wise to combine solvent tolerance and some unique catalytic characteristics using genetic engineering methods. The aim of this investigation was to introduce the biodesulfurizing genes into a solvent-tolerant strain in order to develop a novel biocatalyst that was effective in biphasic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmid, and growth conditions.
Escherichia coli DH5α was used for general cloning. The genes responsible for DBT degradation were obtained from R. erythropolis XP, a DBT-desulfurizing bacterium that uses the “4S” pathway. R. erythropolis XP was cultivated as previously described (38). A solvent-tolerant strain, Pseudomonas putida Idaho, was used as the host strain (4). The broad-host-range expression vector pMMB66EH was also used in this study (7).
Pseudomonas cells were initially grown in modified Pseudomonas medium 187 (M187) containing (per liter of distilled water) 10 g of yeast extract, 10 g of Bacto Tryptone (Difco), 5 g of K2HPO4, 10 ml of glycerol, and 5 ml of a metal salts solution. The metal salts solution contained (per liter of distilled water) 0.4 g FeSO4, 0.2 g NaCl, 0.4 g MgSO4 · 7H2O, and 0.2 g MnSO4 · 4H2O, and H2SO4 was added until the pH was less than 3.0. The medium without the salts solution was autoclaved for 20 min; the salts solution was sterilized by passage through a 0.22-μm membrane filter. The cells were cultivated at 30°C on a rotary shaker at 180 rpm. For cultivation of P. putida A4, 1 mM isopropyl-β-d-thiogalactoside (IPTG) was added to induce expression of dszABCD. For the two-phase reactions, Pseudomonas cells were resuspended and cultivated in M9 minimal medium (24). Growth media were supplemented with ampicillin (100 mg liter−1 for E. coli and 1 g liter−1 for Pseudomonas) if necessary.
General cloning procedures.
Restriction digestion, agarose gel electrophoresis, isolation of plasmids, and other DNA manipulations were carried out by using standard protocols (26, 31). Primers were designed based on the sequences of dszD and dszABC of R. erythropolis IGTS8. The dszD gene of R. erythropolis XP was amplified with primers dszDf and dszDr using R. erythropolis XP genomic DNA as the template. Similarly, the dszABC gene cluster was amplified with dszf and dszr. The sequences of the four primers were as follows: dszDf, 5′-GAGGAATTCATGTCTGACAAGCCGAATGCC-3′ (EcoRI restriction site underlined); dszDr, 5′-CACTCTAGACTATTGACCTAACGGAGTCGG-3′ (XbaI restriction site underlined); dszf, 5′-CACTCTAGAAGGACGCATACGCGATGACTC-3′ (XbaI restriction site underlined); and dszr, 5′-GATCAAAGCTTCAGATCCTCAGGAGGTGAA-3′ (HindIII restriction site underlined). The 0.6-kb dszD PCR product was digested with EcoRI and XbaI, and the 3.7-kb dszABC PCR product was digested with XbaI and HindIII. Then the two fragments were ligated into EcoRI-HindIII-digested pMMB66EH. The resulting plasmid was designated pMMABCD (Fig. 1).
A recombinant Pseudomonas strain harboring pMMABCD was constructed by the triparental mating method (35) with helper plasmid pRK2013 (a gift from David H. Figurski, Department of Microbiology, Columbia University, New York, NY). The cell mass was plated on M9 minimal medium plates supplemented with citrate and 1 g liter−1 ampicillin. Colonies were transferred onto M187 agar plates supplemented with 1 g liter−1 ampicillin and flooded with pure p-xylene. Then the plates were sealed and incubated at 30°C for 72 h. The solvent-tolerant transformants that appeared were tested for DBT degradation.
Bioavailability analysis.
Seed cultures of P. putida A4, P. putida Idaho, and R. erythropolis XP were diluted 25-fold using basal salts medium (BSM) (38) supplemented with 0.5 mM DBT or Na2SO4 as a sulfur source, with or without 10% (vol/vol) p-xylene. Then incubation was performed in seal-capped 300-ml flasks at 30°C and 180 rpm for 24 h on a rotary shaker; 1 g liter−1 ampicillin and 1 mM IPTG were added for cultivation of P. putida A4. Growth was determined by measuring the absorbance at 600 nm using a UV-Vis spectrophotometer.
Southern hybridization analysis.
Southern hybridization experiments were performed using a DIG DNA labeling and detection kit (Roche). Probes were prepared by random primer labeling with digoxigenin according to the manufacturer's instructions. Hybridization was performed overnight at 52°C to detect the dsz gene cluster. Filters (positively charged nylon transfer membranes) were washed under high-stringency conditions twice for 5 min at room temperature in 2× SSC-0.1% sodium dodecyl sulfate and then twice for 15 min at 68°C in 0.1× SSC-0.1% sodium dodecyl sulfate (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Biphasic reaction.
All the reactions involving organic solvents (except the growth of P. putida A4 in the presence of p-xylene) were performed using cell suspensions, and organic solvents were added to a final concentration of 10%. Pseudomonas cells for this type of reaction were grown in M187 supplemented with 1 mM IPTG and 0.1% p-xylene at 30°C for 12 h (1 g liter−1 ampicillin was added when P. putida A4 was grown), centrifuged, and resuspended in M9 minimal medium to obtain a concentration of 7.31 g liter−1. These experiments were carried out in 250-ml seal-capped flasks which were incubated at 180 rpm for 12 h. At each time, an entire reaction flask was analyzed, which minimized the error due to sampling of an aqueous-organic emulsion. The controls were reaction mixtures without bacteria or with heat-inactivated bacteria as described above.
Analytical methods.
Substrate consumption and product formation were analyzed by gas chromatography with flame ionization detection (CP3380; Varian Associates) using an SPB-5 column (inside diameter, 0.32 mm; length, 30 m; Supelco) after the reaction solution was extracted with 0.5 volume ethyl acetate at a pH less than 2.0. Chromatography was performed with nitrogen gas as the carrier gas, using an oven temperature program in which the temperature started at 50°C and then increased to 300°C at a rate of 20°C min−1 and was kept at 300°C for 5 min. The injector and detector temperatures were 275°C and 280°C, respectively. The volume injected was 1 μl.
The molecular structures of the metabolites were analyzed using gas chromatography-mass spectrometry (GCD 1800C; Hewlett-Packard) with a 50-m DB-5 mass spectrometry column (J&W Scientific, Folsom, CA), and the metabolites were identified using the WILEY 275 mass spectral database. Typically, 50 ml of an induced P. putida A4 cell suspension in M9 minimal medium (7.31 g liter−1) was incubated with 0.5 mM PASHs, such as DBT, at 30°C and 180 rpm for 24 h on a rotary shaker. Then the broth was extracted with 0.5 volume of ethyl acetate at a pH less than 2.0. The ethyl acetate extract was then concentrated under nitrogen gas before injection. Chromatography was performed with nitrogen as the carrier gas, using an oven temperature program in which the temperature started at 60°C, increased to 150°C at a rate of 10°C min−1, and then increased to 280°C at a rate of 15°C min−1. The volume injected was 1 μl.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in the GenBank database under accession numbers AY278323 (dszABC) and AY569038 (dszD).
RESULTS
Construction of P. putida A4.
Recombinant plasmid pMMABCD was introduced into the solvent-tolerant strain P. putida Idaho by the triparental mating method. Many transformants were obtained, and some of them were selected for further study. One, designated P. putida A4, was selected from the solvent-tolerant transformants because of its desulfurization ability. To confirm that P. putida A4 was the desired transformant, Southern hybridization analysis was performed, and a 4.3-kb restriction fragment was detected (see Fig. S1 in the supplemental material). The partial 16S rRNA gene sequence in the diagnostic region (nucleotides 1 to 500) of P. putida A4 was 100% homologous to sequences of P. putida strains that have been deposited in the GenBank database (nucleotide sequence accession numbers AY772474, AY574282, AY647158, DQ192174, DQ192173, and AE016778). In addition, bioavailability experiments were performed with DBT or Na2SO4 as the sole sulfur source. P. putida A4 was able to grow in BSM supplemented with 0.5 mM DBT, 1 mM IPTG, and 10%(vol/vol) p-xylene, while no growth of P. putida Idaho or R. erythropolis XP was detected under the same conditions (Table 1). All these results indicated that the dszABCD gene cluster was successfully expressed in P. putida A4, which enabled P. putida A4 to grow with DBT as the sole sulfur source.
TABLE 1.
Growth of R. erythropolis XP, P. putida Idaho, and P. putida A4 in BSM with or without DBT or Na2SO4
| Strain | Growtha
|
|||||
|---|---|---|---|---|---|---|
| BSM (no sulfur)
|
BSM + Na2SO4
|
BSM + DBT
|
||||
| No solvent | 10% p-Xylene | No solvent | 10% p-Xylene | No solvent | 10% p-Xylene | |
| R. erythropolis XP | − | − | + | − | + | − |
| P. putida Idaho | − | − | + | + | − | − |
| P. putida A4 | − | − | + | + | + | + |
+, growth; −, no growth.
Growth of P. putida A4 with 10% (vol/vol) p-xylene.
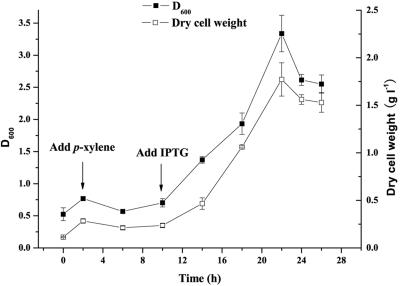
P. putida A4 was able to grow in M187 supplemented with p-xylene. When 10% p-xylene and a 4% inoculum were added, the absorbance at 600 nm of the broth reached 3.3 (dry cell weight, 1.77 g liter−1) after 22 h of shaking at 30°C. The growth curve in Fig. 2 shows that there was a transient decrease in biomass and a lag phase after 10% p-xylene was added.
FIG. 2.
Growth of P. putida A4 in M187 supplemented with 10% p-xylene and 1 g liter−1 ampicillin. IPTG was added to induce expression of the desulfurizing genes, which was driven by the tac promoter. ▪, absorbance at 600 nm (D600) of P. putida A4 culture; □, dry cell weight of P. putida A4 culture. The values are means of at least three replicates, and the error bars indicate standard deviations.
Desulfurization of heterocyclic sulfur compounds.
To investigate the effect of the host strain on the desulfurizing enzyme system, induced cells of P. putida A4 were incubated in M9 minimal medium (without p-xylene) supplemented with some PASHs, such as DBT, methyl DBTs, and methyl BTs, at 30°C and 180 rpm for 24 h. Then the broth was extracted and analyzed by gas chromatography-mass spectrometry to detect the metabolites of PASH degradation by the recombinant strain P. putida A4. 2-Hydroxybiphenyl and 2-hydroxy-3,3′-dimethyl-biphenyl were detected as metabolites of DBT and 4,6-dimethyldibenzothiophene (4,6-DM-DBT), respectively; 2-hydroxy-3′-methyl-biphenyl and 2-hydroxyl-3-methyl-biphenyl were metabolites of 4-methyldibenzothiophene (4-M-DBT). Moreover, 2-isopropenylphenol was the metabolite of 3-methylbenzothiophene (3-M-BT), as shown in Fig. S2 in the supplemental material.
Desulfurization in organic solvent medium.
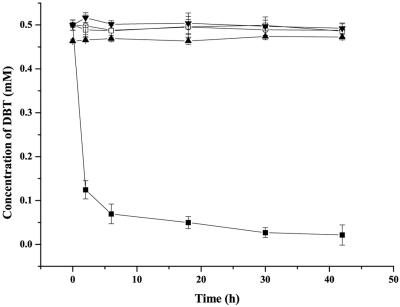
Free cells of P. putida A4, P. putida Idaho, and R. erythropolis XP were resuspended in M9 minimal medium and shaken in 300-ml seal-capped flasks (20 ml of fluid in each flask) at 30°C and 180 rpm with 10% (vol/vol) p-xylene and 0.5 mM (91.13 mg liter−1) DBT in order to investigate the degradation with an organic solvent present. After a 40-h reaction, 97% of the DBT was degraded by P. putida A4, and the majority (86%) was degraded in the initial 6 h; no decrease in the amount of DBT was observed in the reactions performed with R. erythropolis XP, P. putida Idaho, or the controls (Fig. 3). The specific rate of degradation in the first 2 h was 1.29 mM DBT g (dry weight) of cells−1 h−1.
FIG. 3.
Degradation of DBT. Experiments were performed in M9 minimal medium containing 10% (vol/vol) p-xylene. ▪, P. putida A4; □, P. putida Idaho; ○, R. erythropolis XP; ▾, M9 minimal medium (no cells); ▴, heat-inactivated P. putida A4. The values are means of at least three replicates, and the error bars indicate standard deviations.
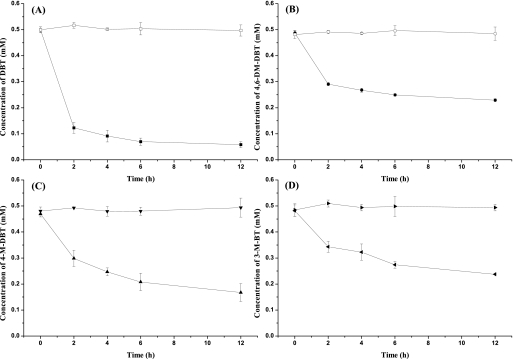
The PASHs 3-M-BT, DBT, 4-M-DBT, and 4,6-DM-DBT were shaken separately with cell suspensions of P. putida A4 in M9 minimal medium supplemented with 10% p-xylene to investigate the ability of P. putida A4 to desulfurize different heterocyclic sulfur compounds in an organic solvent. P. putida A4 was able to degrade 97% of the DBT, 54% of the 4-M-DBT, 71% of the 4,6-DM-DBT, and 53% of the 3-M-BT (Fig. 4). The specific rates of degradation of DBT, 4-M-DBT, 4,6-DM-DBT, and 3-M-BT in the first 2 h were 1.29 mM g (dry weight) of cells−1 h−1, 0.72 mM g (dry weight) of cells−1 h−1, 0.69 mM g (dry weight) of cells−1 h−1, and 0.54 mM g (dry weight) of cells−1 h−1, respectively.
FIG. 4.
Degradation of heterocyclic sulfur compounds. Experiments were performed in M9 minimal medium containing 10% (vol/vol) p-xylene. (A) Degradation of DBT. □, control; ▪, P. putida A4. (B) Degradation of 4-M-DBT. ○, control; •, P. putida A4. (C) Degradation of 4,6-DM-DBT. ▾, control; ▴, P. putida A4. (D) Degradation of 3-M-BT. ▸, control; ◂, P. putida A4. The values are means of at least three replicates, and the error bars indicate standard deviations.
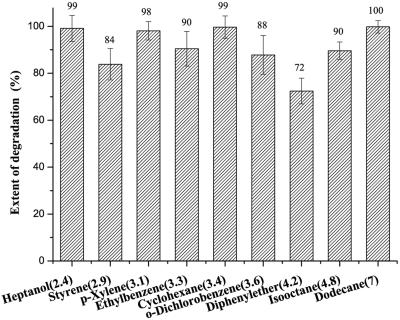
Organic solvents with different values for the common logarithm of the partition coefficient of a solvent in the n-octane and aqueous phases (log P) were used to demonstrate the effects of different organic solvents on the desulfurizing activity. Heptanol (log P, 2.4), styrene (log P, 2.9), p-xylene (log P, 3.1), ethylbenzene (log P, 3.3), cyclohexane (log P, 3.4), o-dichlorobenzene (log P, 3.6), diphenylether (log P, 4.2), isooctane (log P, 4.8), and n-dodecane (log P, 7.0) were added to cell suspensions (in M9 minimal medium) of induced P. putida A4 cells at a concentration of 10% (vol/vol), and the total volume in every 250 ml seal-capped flask was 20 ml. Then 0.5 mM DBT was added to each flask, and the flasks were incubated for 12 h with shaking at 30°C at 180 rpm on a rotary shaker. P. putida A4 was capable of degrading DBT with high activity in the presence of organic solvents (Fig. 5).
FIG. 5.
Extents of degradation in different organic solvents. The numbers in parentheses are the log P values of the organic solvents. Experiments were performed in 300-ml flasks, each of which was filled with 20 ml of a reaction mixture composed of a cell suspension and 10% organic solvent. After incubation the organic phase was sampled and analyzed by gas chromatography with flame ionization detection. The extent of degradation was determined by comparison with the control samples. The control samples were samples without any bacteria incubated under the same conditions. The values are means of at least three replicates, and the error bars indicate standard deviations.
Additionally, since P. putida A4, as well as the host strain P. putida Idaho, was able to tolerate up to 50% p-xylene, the biodesulfurization activities in the presence of different concentrations of p-xylene were examined. When 20% (vol/vol), 30%(vol/vol), 40%(vol/vol), and 50%(vol/vol) p-xylene were added to P. putida A4 cell suspensions in M9 minimal medium in 300-ml seal-capped flasks and shaken at 30°C for 12 h, the extents of DBT degradation were 70%, 58%, 48%, and 48%, respectively.
DISCUSSION
Pseudomonas sp. is considered the ideal host for biodesulfurization because of its high growth rate, its metabolic diversity, its well-documented ability to carry out biotransformation in biphasic systems, and the availability of genetic techniques (29). In addition, biodesulfurization is an energetically expensive multistep process that consumes flavin mononucleotide as reducing equivalents (11). Cell integrity and viability are necessary for this type of reaction (14, 21, 34). Therefore, solvent-tolerant Pseudomonas strains may be efficient hosts for the desulfurizing enzyme system, as they are able to supply energy and the necessary reaction conditions in the presence of organic solvents and thereby maintain good desulfurizing activity in the oil desulfurization process. In this study, the dszABCD genes were introduced into P. putida Idaho in order to construct a solvent-tolerant desulfurizing bacterium which could work well with organic solvents. This study is a significant step in the exploration of the biotechnological potential of novel biocatalysts for developing an efficient biodesulfurization process.
There are factors about a host strain, such as the pH of the cytosol, the penetrability of the cell wall and cell membrane, and the respiratory activity, that can affect the efficiency and specificity of the enzyme system introduced into the host strain (3, 33). Thus, different PASHs were used to determine the effect of host strain P. putida Idaho on the specificity of the desulfurizing enzyme system. It should be noted that the metabolite(s) of each PASH produced by P. putida A4 was the same as the metabolite(s) produced by R. erythropolis XP (38). This result indicated that host strain P. putida Idaho did not affect the specificity of the desulfurizing enzyme system from R. erythropolis XP and that the desulfurizing enzyme system worked well in the host strain.
In general, solvents with log P values between 1 and 4 are considered extremely toxic to microorganisms, as the degree of partition into the cell membrane is high, and most microorganisms are not able to survive in organic solvents present in the environment (1, 13, 32). This is a shortcoming for the use of many bacteria in biodesulfurization and biphasic reactions (12). p-Xylene is one of these extremely toxic organic solvents and has log P value of 3.1. P. putida A4 was able to grow well in M187 supplemented with 10% p-xylene, although a transient decrease in biomass and a lag phase were observed after addition of 10% p-xylene (Fig. 2). It is reasonable to suggest that the transient decrease in biomass and the lag phase were due to the organic solvent shock (4). It is notable that there was no growth lag after IPTG was added at 11 h, which indicated that gene expression did not affect the growth of P. putida A4 in organic solvent. It is also notable that P. putida A4 was able to degrade various PASHs, such as DBT, 4-M-DBT, 4,6-DM-DBT, and, 3-M-BT, in the presence of 10% p-xylene and could maintain desulfurizing activity even in the presence of 50% p-xylene. Desulfurization of oils may be more successful with the solvent-tolerant strain P. putida A4.
There are many different organic solvents in oil, polluted environments, and biocatalytic media, and the toxic effects of them on microorganisms correlate with the hydrophobicity, expressed as log P. Thus, solvents with different log P values were examined to determine their effects on desulfurizing activity. P. putida A4 was able to tolerate all the organic solvents used in our study and to maintain the desulfurizing activity of the desulfurizing enzyme system (Fig. 5). de Carvalho et al. investigated the toxicities of dimethylformamide, ethanol, and butanol for Mycobacterium sp., R. erythropolis, and P. putida using fluorescence microscopy technology and found that the toxicities of organic solvents did not correspond to the log P values of solvents (6). Similar results were obtained in this study. It is reasonable to suggest that the unique mechanism of solvent tolerance of P. putida A4 and the unique chemical characteristics of different solvents led to these unexpected results. Cruden et al. suggested that the resistance of P. putida Idaho was due to the ability of this organism to synthesize membranes rapidly to compensate for the membranes damaged by solvents or due to some biochemical difference in the cytoplasmic membrane which makes it more stable in the presence of solvent (4, 30). This unique mechanism of solvent tolerance may be the reason for the broad range of substrates which P. putida A4 can tolerate.
In conclusion, our results suggested that P. putida A4, constructed by introducing dszABCD into P. putida Idaho, could remain viable and exhibit desulfurizing activity with a variety of organic solvents that were present separately. This is the first example of an organism that can efficiently desulfurize in organic solvent-based biphasic media, and this implies that this technology has a future. Currently, the expression of dsz genes is directed by the tac promoter, which makes commercial application of this strain less advisable. For practical application, promoter substitution is necessary, and such work is being performed by members of our group.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 20590368 and 29977011).
We thank David T. Gibson for supplying P. putida Idaho, Erich Lanka for supplying plasmid pMMB66EH, David H. Figurski for supplying plasmid pRK2013, and Juan L. Ramos for supplying important literature.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aono, R., and H. Kobayashi. 1997. Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl. Environ. Microbiol. 63:3637-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgne, S. L., and R. Quintero. 2003. Biotechnological processes for the refining of petroleum. Fuel Process. Technol. 81:155-169. [Google Scholar]
- 3.Bouchez-Naïtali, M., S. Abbad-Andaloussi, M. Warzywoda, and F. Monot. 2004. Relation between bacterial strain resistance to solvents and biodesulfurization activity in organic medium. Appl. Microbiol. Biotechnol. 65:440-445. [DOI] [PubMed] [Google Scholar]
- 4.Cruden, D. L., J. H. Wolfram, R. D. Rogers, and D. T. Gibson. 1992. Physiological properties of a Pseudomonas strain which grows with p-xylene in a two-phase organic-aqueous medium. Appl. Environ. Microbiol. 58:2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bont, J. A. M. 1998. Solvent-tolerant bacteria in biocatalysis. Trends Biotechnol. 16:493-499. [Google Scholar]
- 6.de Carvalho, C. C. C. R., A. A. R. L. Da Cruz, M. N. Pöns, H. M. R. V. Pinheiro, J. M. S. Cabral, M. M. R. Da Fonseca, B. S. Ferreira, and F. Fernandes. 2004. Mycobacterium sp., Rhodococcus erythropolis, and Pseudomonas putida behavior in the presence of organic solvents. Microsc. Res. Tech. 64:215-222. [DOI] [PubMed] [Google Scholar]
- 7.Früste, J. P., W. Pansegrau, R. Frank, H. Blöcker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 8.Galán, B., E. Díaz, and J. L. García. 2000. Enhancing desulfurization by engineering a flavin reductase-encoding gene cassette in recombinant biocatalysts. Environ. Microbiol. 2:687-694. [DOI] [PubMed] [Google Scholar]
- 9.Gallardo, M. E., A. Ferrández, V. de Lorenzo, J. L. García, and E. Díaz. 1997. Designing recombinant Pseudomonas strains to enhance biodesulfurization. J. Bacteriol. 179:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, K. A., G. T. Mrachkoyz, and C. H. Squiresy. 2003. Biodesulfurization of fossil fuels. Curr. Opin. Microbiol. 6:229-235. [DOI] [PubMed] [Google Scholar]
- 11.Gray, K. A., O. S. Pogrebinsky, G. T. Mrachko, X. Lei, D. J. Monticello, and C. H. Squires. 1996. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 14:1705-1709. [DOI] [PubMed] [Google Scholar]
- 12.Gupta, N., P. K. Roychoudhury, and J. K. Deb. 2005. Biotechnology of desulfurization of diesel: prospects and challenges. Appl. Microbiol. Biotechnol. 66:356-366. [DOI] [PubMed] [Google Scholar]
- 13.Inoue, A., and K. Horikoshi. 1989. A Pseudomonas thrives in high concentrations of toluene. Nature 338:264-266. [Google Scholar]
- 14.Ishige, T., K. Honda, and S. Shimizu. 2005. Whole organism biocatalysis. Curr. Opin. Chem. Biol. 9:174-180. [DOI] [PubMed] [Google Scholar]
- 15.Isken, S., and J. A. M. de Bont. 1998. Bacteria tolerant to organic solvents. Extremophiles 2:229-238. [DOI] [PubMed] [Google Scholar]
- 16.Jonathan, D., V. Hamme, A. Singh, and O. P. Ward. 2003. Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67:503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabe, T., A. Ishihara, and H. Tajima. 1992. Hydrodesulfurization of sulfur containing polyaromatic compounds in light oil. Ind. Eng. Chem. Res. 31:1577-1580. [Google Scholar]
- 18.Kilbane, J. J. 1989. Desulfurization of coal: the microbial solution. Trends Biotechnol. 7:97-101. [Google Scholar]
- 19.Kobayashi, M., T. Onaka, Y. Ishii, J. Konishi, M. Takaki, H. Okada, Y. Ohta, K. Koizumi, and M. Suzuki. 2000. Desulfurization of alkylated forms of both dibenzothiophene and benzothiophene by a single bacterial strain. FEMS Microbiol. Lett. 187:123-126. [DOI] [PubMed] [Google Scholar]
- 20.Konishi, J., T. Onaka, Y. Ishii, and M. Suzuki. 2000. Demonstration of the carbon-sulfur bond targeted desulfurization of benzothiophene by thermophilic Paenibacillus sp. strain A11-2 capable of desulfurizing dibenzothiophene. FEMS Microbiol. Lett. 187:151-154. [DOI] [PubMed] [Google Scholar]
- 21.León, R., P. Fernandes, H. M. Pinheiro, and J. M. S. Cabral. 1998. Whole-cell biocatalysis in organic media. Enzyme Microb. Technol. 23:483-500. [Google Scholar]
- 22.Li, F. L., P. Xu, C. Q. Ma, L. L. Luo, and X. S. Wang. 2003. Deep desulfurization of hydrodesulfurization-treated diesel oil by a facultative thermophilic bacterium, Mycobacterium sp. X7B. FEMS Microbiol. Lett. 223:301-307. [DOI] [PubMed] [Google Scholar]
- 23.Li, F. L., P. Xu, J. H. Feng, L. Meng, Y. Zheng, L. L. Luo, and C. Q. Ma. 2005. Microbial desulfurization of gasoline in a Mycobacterium goodii X7B immobilized-cell system. Appl. Environ. Microbiol. 71:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Matsui, T., K. Hirasawa, J. Konishi, Y. Tanaka, K. Maruhashi, and R. Kurane. 2001. Microbial desulfurization of alkylated dibenzothiophene and alkylated benzothiophene by recombinant Rhodococcus sp. strain T09. Appl. Microbiol. Biotechnol. 56:196-200. [DOI] [PubMed] [Google Scholar]
- 26.Miller, L. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Oldfield, C., O. Pogrebinsky, J. Simmonds, E. S. Olson, and C. F. Kulpa. 1997. Elucidation of the metabolic pathway for dibenzothiophene desulfurization by Rhodococcus sp. strain IGTS8 (ATCC 53968). Microbiology 143:2961-2973. [DOI] [PubMed] [Google Scholar]
- 28.Piddington, C. S., B. R. Kovaceich, and J. Rambosek. 1995. Sequence and molecular characterization of a DNA region encoding the dibenzothiophene desulfurization operon of Rhodococcus sp. strain IGTS8. Appl. Environ. Microbiol. 61:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pienkos, P. T. 1999. Choosing the best platform for the biotransformation of hydrophobic molecules, p. 875-888. In C. R. Bell, M. Brylinsky, and P. Johnson-Green (ed.), Proceedings of the 8th International Symposium on Microbial Ecology. Atlantic Canada Society for Microbial Ecology, Halifax, Canada.
- 30.Pinkart, H. C., and D. C. White. 1997. Phospholipid biosynthesis and solvent tolerance in Pseudomonas putida strains. J. Bacteriol. 179:4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Sardessai, Y. N., and S. Bhosle. 2004. Industrial potential of organic solvent tolerant bacteria. Biotechnol. Prog. 20:655-660. [DOI] [PubMed] [Google Scholar]
- 33.Sarthiy, A. V., B. L. Mcconaughy, Z. Lobo, and J. A. Sundstrom. 1987. Expression of the Escherichia coli xylose isomerase gene in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 53:1996-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid, A., J. S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409:258-268. [DOI] [PubMed] [Google Scholar]
- 35.Sepulveda-Torres, L. D. C., N. Rajendran, M. J. Dybas, and C. S. Criddle. 1999. Generation and initial characterization of Pseudomonas stutzeri KC mutants with impaired ability to degrade carbon tetrachloride. Arch. Microbiol. 171:424-429. [DOI] [PubMed] [Google Scholar]
- 36.Shan, G. B., J. M. Xing, H. Y. Zhang, and H. Z. Liu. 2005. Biodesulfurization of dibenzothiophene by microbial cells coated with magnetite nanoparticles. Appl. Environ. Microbiol. 71:4497-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi, L., C. H. Squires, D. J. Monticello, and J. D. Childs. 1997. A flavin reductase stimulates DszA and DszC proteins of Rhodococcus erythropolis IGTS8 in vitro. Biochem. Biophys. Res. Commun. 230:73-75. [DOI] [PubMed] [Google Scholar]
- 38.Yu, B., P. Xu, Q. Shi, and C. Q. Ma. 2006. Deep desulfurization of diesel oil and crude oils by a newly isolated Rhodococcus erythropolis strain. Appl. Environ. Microbiol. 72:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, B., P. Xu, S. S. Zhu, X. F. Cai, Y. Wang, L. Li, F. L. Li, X. Y. Liu, and C. Q. Ma. 2006. Selective biodegradation of S and N heterocycles by a recombinant Rhodococcus erythropolis strain containing carbazole dioxygenase. Appl. Environ. Microbiol. 72:2235-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.