Abstract
Gibberellin A1 (GA1) levels drop significantly in wild-type pea (Pisum sativum) plants within 4 h of exposure to red, blue, or far-red light. This response is controlled by phytochrome A (phyA) (and not phyB) and a blue light receptor. GA8 levels are increased in response to 4 h of red light, whereas the levels of GA19, GA20, and GA29 do not vary substantially. Red light appears to control GA1 levels by down-regulating the expression of Mendel's LE (PsGA3ox1) gene that controls the conversion of GA20 to GA1, and by up-regulating PsGA2ox2, which codes for a GA 2-oxidase that converts GA1 to GA8. This occurs within 0.5 to 1 h of exposure to red light. Similar responses occur in blue light. The major GA 20-oxidase gene expressed in shoots, PsGA20ox1, does not show substantial light regulation, but does show up-regulation after 4 h of red light, probably as a result of feedback regulation. Expression of PsGA3ox1 shows a similar feedback response, whereas PsGA2ox2 shows a feed-forward response. These results add to our understanding of how light reduces shoot elongation during de-etiolation.
The involvement of gibberellins (GA) and phytochrome (phy) in the de-etiolation of pea (Pisum sativum) has been discussed for over 40 years (Lockhart, 1956; Kende and Lang, 1964; Reid, 1983). However, it is only in the last few years that it has been firmly established that the level of the major bioactive GA in peas, GA1, drops during the first 24 h of exposure to light (Ait-Ali et al., 1999; Gil and Garcia-Martinez, 2000; O'Neill et al., 2000). The reason for the previous lack of clarity is probably that the GA1 level in the elongating stem rises after the first 24 h of exposure to light, to levels similar to those found in dark-grown plants (O'Neill et al., 2000). Therefore, several earlier studies had failed to show any major difference in GA1 levels between continuously light- and dark-grown plants (e.g. Ross et al., 1992; Weller et al., 1994). The continuing difference in elongation between light- and dark-grown plants appears to be attributable to a reduction in the responsiveness of elongating stem tissue to GA1 in light-grown shoots compared with dark-grown shoots (Reid, 1988; O'Neill et al., 2000).
There have been several studies showing that light controls GA synthesis, and hence development, via the photoreceptor phy (Kamiya and Garcia-Martinez, 1999). Perhaps the best studied examples are the light-regulated control of seed germination in lettuce (Lactuca sativa) and Arabidopsis. In lettuce, the Ls3h1 gene is dramatically up-regulated by red light, which leads to increased GA1 levels (Toyomasu et al., 1998). In Arabidopsis, two genes encoding GA 3β-hydroxylases, GA4 and GA4H, are induced by red light (Yamaguchi et al., 1998). Through the use of a phyB mutant, it was shown that GA4H was regulated by phyB, but that some other member of the phy gene family presumably regulates the GA4 gene (Yamaguchi et al., 1998). However, bioactive GA levels were not directly determined in this study. Regulation of GA levels has also been shown by photoperiod in long-day rosette plants such as spinach (Spinacia oleracea; Talon et al., 1991) and during tuberization in potato (Solanum tuberosum; Xu et al., 1998). GA 20-oxidase mRNA levels are regulated by light in spinach (Wu et al., 1996), whereas in potato, phyB mediates the tuberization response (Jackson et al., 2000).
In this study, we have used mutants deficient in phyA and/or phyB and a range of light conditions to determine the photoreceptor(s) involved in regulating GA levels during de-etiolation in pea. We have also determined the timing of changes in GA levels and the expression of genes controlling the later steps of GA metabolism (Fig. 1) under a range of light conditions. The results suggest that phyA and a blue light receptor are involved in regulating GA levels. The changes in GA levels probably result from direct light regulation of mRNA levels of specific GA 3-oxidase and GA 2-oxidase genes. In addition, feedback regulation of GA 20-oxidase and GA 3-oxidase gene expression, and feed-forward regulation of GA 2-oxidase expression are also important in regulating bioactive GA1 levels during the de-etiolation process.
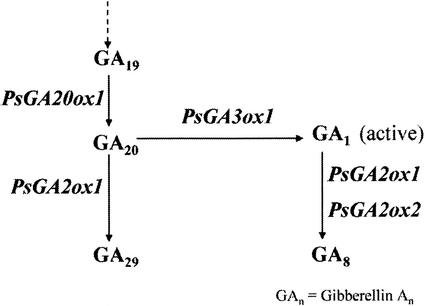
Figure 1.
The later stages of GA biosynthesis in pea, and the site of action of the major genes. PsGA3ox1 is Mendel's LE gene and PsGA2ox1 is SLN. PsGA2ox2 may have a very minor effect on the conversion of GA20 to GA29 based on metabolism studies of Lester et al. (1999).
RESULTS
Photoreceptors Involved in the Regulation of GA Levels by Light
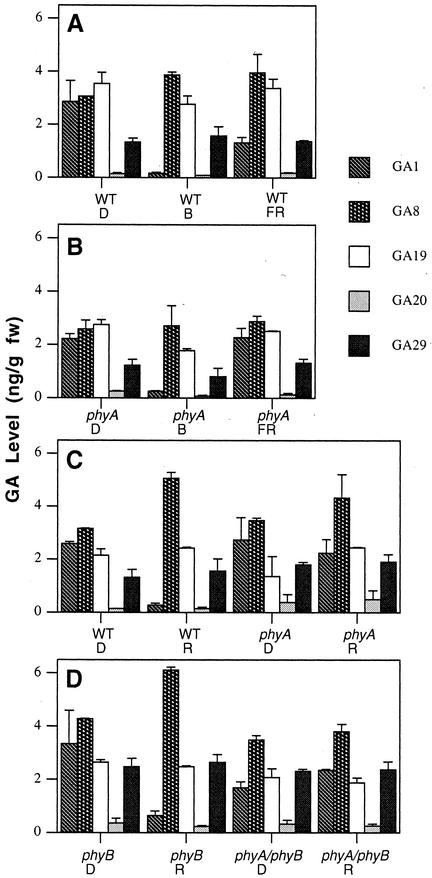
White light has been shown to dramatically reduce the level of GA1 during the first 24 h of de-etiolation (Ait-Ali et al., 1999; Gil and Martinez, 2000; O'Neill et al., 2000), with an associated increase in its inactive 2β-hydroxylated product, GA8 (Gil and Martinez, 2000; O'Neill et al., 2000). The results show clearly that 4 h of red light (P < 0.01; Fig. 2C), blue light (P < 0.01; Fig. 2A) and, to a lesser extent, far-red light (P < 0.05; Fig. 2A) also lead to significant reductions in GA1 levels in wild-type (WT) seedlings. A concomitant increase in GA8 levels occurs in red light (P < 0.01; Fig. 2C), whereas the levels of GA20, GA19, and GA29 show little variation. In a similar manner, significant changes in GA1 and GA8 levels are seen between phyB mutants grown in the dark and plants exposed to 4 h of red light (P < 0.01; Fig. 2D), suggesting that phyB is not involved in the regulation by red light of GA1 levels in this system. However, a quite different picture is seen in phyA mutants and in the double mutant phyA phyB. No significant change in GA1 or GA8 levels are seen when phyA (Fig. 2C) or phyA phyB (Fig. 2D) seedlings exposed to 4 h of red light are compared with dark-grown seedlings. The levels of GA19, GA20, and GA29 also do not vary substantially. This suggests phyA controls the change in GA1 and GA8 levels under red light. To confirm that phyA was responsible for regulating GA1 and GA8 levels, phyA seedlings were exposed for 4 h to far-red light. Again, there was no significant change in GA1 or GA8 levels (Fig. 2B). However, after 4 h exposure of phyA seedlings to blue light, a 10-fold decrease in GA1 level occurred (P < 0.01; Fig. 2B), similar to the change seen in WT seedlings (Fig. 2A). This suggests that although phyA regulates bioactive GA1 levels under red and far-red light in pea, a separate blue light receptor(s) is involved in the response to blue light.
Figure 2.
Comparison of the level of 13-hydroxylated GAs in WT (line 107) and phyA-deficient (phyA), phyB-deficient (phyB), and phyA phyB-deficient (phyA/phyB) mutant lines of pea transferred to red (R), far-red (FR), or blue (B) light for 4 h. Seedlings were grown in continuous dark (D) for 7 d prior to transfer. A and B were part of one experiment and C and D were part of another experiment.
Effect of phyA on GA Responsiveness
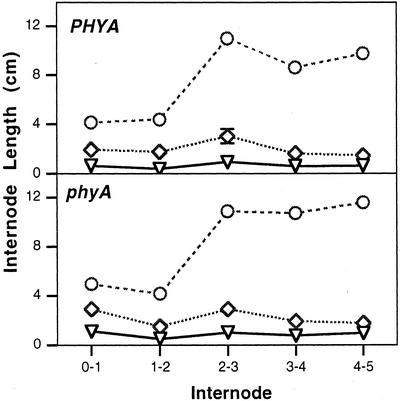
WT seedlings of pea show reduced elongation under continuous red, far-red, blue, and white light compared with dark-grown plants (Behringer et al., 1992; Weller et al., 2001). The phyB mutants show a reduced inhibition in response to red, blue, and white light (Behringer et al., 1992; Weller et al., 2001), whereas phyA mutants are effectively blind to far-red light (Weller et al., 2001). The phyA phyB double mutant is effectively blind to far-red and red light (Weller et al., 2001). The phyB plants have been shown to be more responsive to GA1 under white light than comparable WT plants (Reid and Ross, 1988). This presumably explains why phyB plants are longer than WT plants under red light even though the GA1 levels respond similarly to exposure to red light (Fig. 2, C and D). However, the responsiveness of phyA seedlings to GA1 in red light has not been previously examined. The results in Figure 3 show that regardless of the level of phyA, seedlings dwarfed with paclobutrazol (PP333) and on a GA1-deficient le-3 genetic background respond similarly to 1- and 10-μg doses of GA1. In fact, the response to GA1 was marginally less between nodes 1 and 4 in phyA plants than in WT plants (277% versus 330% increase with 1 μg of GA1 and 1,120% versus 1,230% increase with 10 μg of GA1). This clearly suggests that phyA does not significantly influence the response of red light-grown plants to GA1 (Fig. 3), although phyA does regulate GA1 levels during the early stages of de-etiolation (Fig. 2C).
Figure 3.
The mean length of internodes 0 through 5 in PHYA and phyA plants (on a dwarf le-3 background) treated with 5 μg of the GA biosynthesis inhibitor PP333 (▿), 5 μg of PP333 + 1 μg of GA1 (⋄), or 5 μg of PP333 + 10 μg of GA1 (○). Plants were grown under continuous red light. Where se bars are not visible, they lie within the dimensions of the symbol. n = 10. Node 0 is the cotyledonary node.
Changes in GA Levels during De-Etiolation
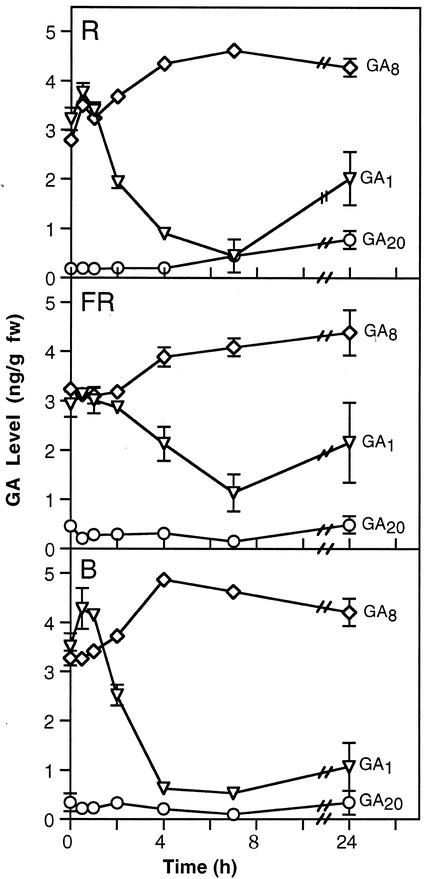
Ait-Ali et al. (1999) have suggested that changes in GA levels during de-etiolation may occur prior to changes in GA 20-oxidase and GA 3-oxidase gene expression in white light. To examine this apparent paradox, the timing of changes in GA levels was examined during the first 24 h of de-etiolation of WT pea seedlings under red, blue, and far-red light (Fig. 4). Under red and blue light, there was no significant change in the levels of the bioactive GA, GA1, after 0.5 or 1 h of light exposure. A significant reduction was apparent at 2 h in red and blue light (P < 0.05), in agreement with results under white light reported by Ait-Ali et al. (1999) and Gil and Garcia-Martinez (2000). A further reduction was seen at 4 and 7 h with the level beginning to rise by 24 h, although still below that seen in continuous dark. This rise is consistent with results of O'Neill et al. (2000) who showed that GA1 levels initially drop in de-etiolating seedlings before recovering to reach levels seen in continuously light-grown plants after 5 d. This rise explains why comparisons of GA1 levels in continuously dark-grown versus light-grown seedlings can be misleading regarding the control of de-etiolation by GAs.
Figure 4.
Comparison of mean GA1(▿), GA8(⋄), and GA20(○) levels in 7-d-old seedlings on transfer from continuous darkness (D) to continuous red (R), far-red (FR), or blue (B) light. Where se bars are not visible they lie within the dimensions of the symbol; n = 2.
Under far-red light, a significant drop in GA1 levels is not seen until 4 h (P < 0.05) after the start of illumination, and even at 7 h (P < 0.001) the reduction is only 3-fold rather than the approximately 7-fold change seen under red and blue light. These results are correlated with a smaller reduction in internode length caused by far-red light compared with red and blue light (Weller et al., 2001) and with the extended timing of phyA movement from the cytoplasm to the nucleus under far-red compared with red light (Hisada et al., 2000).
Under all three light treatments, a significant increase in GA8 levels occurred by 7 h (P < 0.05). At the same time there was a significant drop in GA1 (P < 0.01) levels. This supports the results of Gil and Garcia-Martinez (2000) and O'Neill et al. (2000) under white light, and suggests that the 2β-hydroxylation of GA1 (Fig. 1) may be an important step in the regulation of the level of bioactive GA1 during de-etiolation in pea.
The levels of GA19, and GA29, the 2β-hydroxylation product of GA20 (Fig. 1), do not differ significantly under red, blue, or far-red light (data not shown). The level of GA20 shows a tendency to start to rise after 24 h of exposure to red, far-red, or blue light (Fig. 4), again consistent with the results of O'Neill et al. (2000), who showed that after 5 d of de-etiolation under white light, GA20 levels rose to the levels found in continuously light-grown plants.
Changes in the Expression of GA Biosynthesis Genes during De-Etiolation
The later steps in GA metabolism are controlled by 2-oxoglutarate-dependent dioxygenases (Fig. 1; Hedden and Phillips, 2000). Each step is controlled by a small gene family with the various members being expressed in different tissues and/or different environmental conditions (Hedden and Phillips, 2000). We examined the mRNA levels of the members of these families that are expressed in the young shoot tissue of pea. Hence, we examined the GA 20-oxidase gene, PsGA20ox1 (Garcia-Martinez et al., 1997), Mendel's GA 3-oxidase gene, PsGA3ox1 (LE; Lester et al., 1997), and two GA 2-oxidase genes, PsGA2ox1 (SLN; Lester et al., 1999) and PsGA2ox2 (Lester et al., 1999; Fig. 1). In each case, we examined the mRNA level from a sample of each replicate experiment used to determine GA levels in Figure 4.
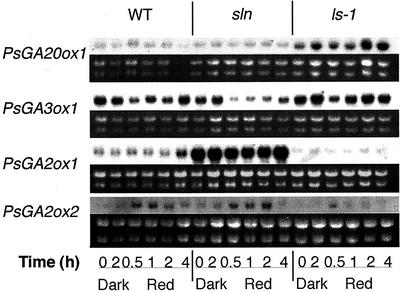
Mendel's PsGA3ox1 gene, which regulates the conversion of GA20 to GA1 in shoots (Fig. 1), showed clear down-regulation by 0.5 h of red, far-red, or blue light (Fig. 5B). This down-regulation continued until 2 h, but had returned to near the levels seen in dark-grown plants by 7 h. These results suggest that Mendel's PsGA3ox1 gene is light-regulated during the early stages of de-etiolation and they also suggest that feedback regulation by the reduced GA1 levels at 2 and 4 h may then lead to renewed gene expression. To examine this proposed feedback regulation, a dwarf GA1-deficient mutant, ls-1, and a slender, GA1-overproducing mutant, sln (mutation in gene PsGA2ox1), were also analyzed during de-etiolation under red light (Fig. 6). Clear light regulation of the PsGA3ox1 gene was apparent in WT, sln, and ls-1 plants. However, in sln plants, it was much further down-regulated and for a longer period than in ls-1 plants, which only showed strong down-regulation at the 0.5-h time point. These results suggest an interesting interaction between light regulation and feedback regulation.
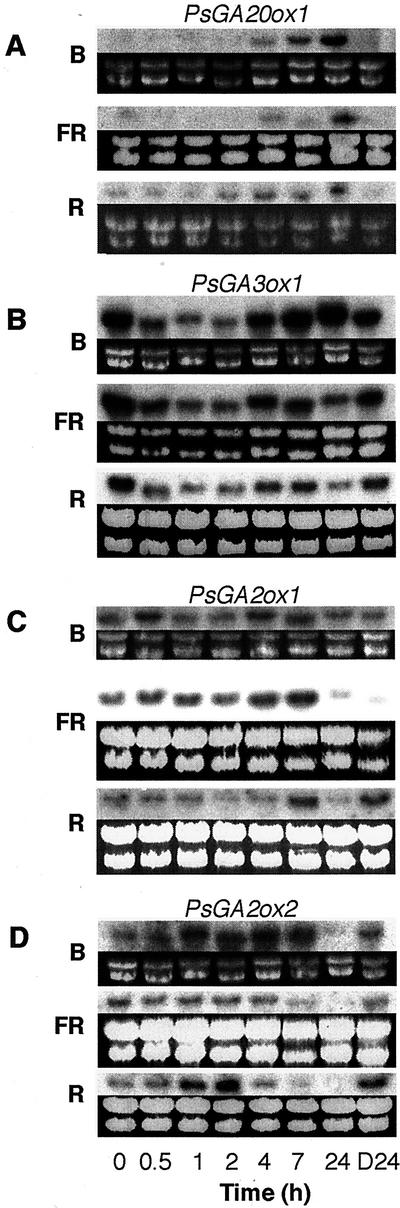
Figure 5.
PsGA20ox1 (A), PsGA3ox1 (B), PsGA2ox1 (C), and PsGA2ox2 (D) mRNA levels in 7-d-old pea seedlings transferred to blue (B), far-red (FR), or red (R) light as shown by northern analysis. The plants were transferred from darkness (D) to the relevant light treatment for the time (h) indicated. The dark 24-h sample (D24) represents the mRNA level in continuous darkness 24 h after the initial transfer. The corresponding gels showing the ribosomal RNAs stained with ethidium bromide are shown immediately below each northern blot to indicate the loading of lanes.
Figure 6.
PsGA20ox1, PsGA3ox1, PsGA2ox1, and PsGA2ox2 mRNA levels after transfer to red light in WT, GA1-overproducing (sln), and GA1-deficient (ls-1) mutant lines of pea, as shown by northern analysis. Seedlings were grown in continuous dark (D) for 7 d prior to transfer. The dark 2-h sample (D2) represents the relevant mRNA level in continuous darkness at the time of sampling of seedlings transferred to R for 2 h. Immediately below each northern blot is the gel showing the corresponding ribosomal RNAs (stained with ethidium bromide) to indicate the loading of lanes.
The shoot-expressed GA 20-oxidase gene, PsGA20ox1, showed low expression over the first 2 h of red, far-red, or blue light (Fig. 5A). Under all three light treatments, its expression then increased (after 4 or 7 h) until 24 h of exposure. This change probably reflects feedback regulation rather than light up-regulation because it occurs much later than the light regulation seen for PsGA3ox1 (Fig. 5B) and occurs after the observed drop in bioactive GA1 (Fig. 4). Feedback regulation of this gene is well established in pea (Martin et al., 1996; Ross et al., 1999). This appeared to be confirmed when the expression of PsGA20ox1 was examined in the sln and ls-1 mutants (Fig. 6). Only small changes in expression were evident over the first 4 h of red light, but the expression is markedly higher in ls-1 plants compared with sln plants at all time points and in the dark.
The two GA 2-oxidase genes, PsGA2ox1 and PsGA2ox2, cloned from pea (Lester et al., 1999) appear to be expressed in expanding internodes (Elliott et al., 2001). The PsGA2ox1 (SLN) product clearly regulates the 2-oxidation of GA20 to GA29 to GA29-catabolite, especially in developing seed because sln plants have a clear morphological phenotype (Reid et al., 1992; Lester et al., 1999). However, the sln mutation did not substantially affect the metabolism of GA1 to GA8 in mature shoot tissues, although GA1 levels were elevated to a small extent in mature sln plants (Ross et al., 1995). However, PsGA2ox1 gene product can convert GA1 to GA8 in vitro (Lester et al., 1999). The PsGA2ox2 gene product has a strong preference for GA1 rather than GA20 as a substrate (Lester et al., 1999). On this evidence, it has been suggested that it is a good candidate for the enzyme that deactivates GA1 in the shoot. The results show that PsGA2ox2 expression is up-regulated by 1 or 2 h of exposure to red light (Fig. 5D). A small but similar response is evident for blue light (Fig. 5D). A similar pattern is shown in sln and ls-1 plants in red light (Fig. 6). However, due to feed-forward regulation of this gene (Elliott et al., 2001), the effect is more pronounced in sln plants than ls-1 plants. The single base substitution in the sln (PsGA2ox1) gene may also have affected transcript abundance due to altered mRNA stability. The down-regulation of PsGA2ox2 expression seen (depending on light treatment) between 4 and 24 h exposure to light (Fig. 5D) probably results from this feed-forward mechanism because GA1 levels have dropped dramatically by 4 h (Fig. 4).
The regulation of PsGA2ox1 expression is less clear. Strong feed-forward regulation for this gene is seen when the expression is compared between WT, sln, and ls-1 seedlings (Fig. 6), consistent with the results of Elliott et al. (2001). However, this gene shows little photoregulation of expression after 2 h of red light, but it does show a small increase after 4 h of exposure (Fig. 6). This is confirmed by the results in Figure 5C when exposure to red, blue, and far-red light over the first 24 h of de-etiolation also suggests some transient up-regulation of expression between 4 and 7 h of light.
DISCUSSION
O'Neill et al. (2000) showed that during de-etiolation in pea there is first a rapid reduction in GA1 levels, which is followed by a reduction in the response to GA1. This reduced response allows continued inhibition of shoot elongation even though GA1 levels return to homeostatic levels. The present results show that phyA and a blue light receptor(s) regulate the level of bioactive GA1 during the initial stages of de-etiolation. PhyB does not appear to be involved in this process. However, Reid and Ross (1988) showed that the phyB mutant is more responsive to GA1, suggesting that phyB controls the responsiveness of the shoot to GA1. The results in Figure 3 indicate that phyA does not influence this process in red light.
In Arabidopsis, phyA causes a transient decrease in hypocotyl elongation during the first 3 h of exposure to continuous red light (Parks and Spalding, 1999), whereas phyB regulates inhibition after 3 h (Parks and Spalding, 1999). In pea, phyB seedlings show a similar inhibition of growth to WT plants during the first 2 to 3 h of exposure to continuous red or white light, but reduced inhibition thereafter (Behringer et al., 1992), similar to the results in Arabidopsis. Behringer et al. (1992) suggested that this may indicate the transition of the primary photocontrol of stem elongation from a light-labile to a light-stable phy. This suggests an interesting parallel between the system in pea and Arabidopsis. Further, it is tempting to suggest that the transient phyA-regulated change in elongation seen in Arabidopsis may be caused by a rapid change in bioactive GA levels similar to that shown in pea. The longer-term response may be the result of a change in GA responsiveness because Reed et al. (1994) have shown that phyB in Arabidopsis does enhance the responsiveness of shoot tissue to applied GA3. The similarity in the responses to red and blue light in pea suggests that although distinct photoreceptors (phyA and a blue light receptor) are involved, they may share a common transduction pathway that leads to similar changes in the expression of the GA biosynthesis genes and GA levels.
Previous studies have suggested that the changes in GA1 levels during de-etiolation precede changes in the expression of genes in the GA biosynthetic pathway. Further, the expression response is in the opposite direction to that expected (Ait-Ali et al., 1999; Gil and Garcia-Martinez, 2000). In both cases, feedback was suggested as the cause of the change in gene expression. In these studies only, the expression of the genes PsGA20ox1 and PsGA3ox1 was monitored (Fig. 1). The combination of higher growth temperatures and the different time of sampling suggests that the response in gene expression monitored was the increase in the expression of these genes following the initial rapid drop in GA1 levels apparent in the current studies at around 4 to 7 h of light exposure (Fig. 4). The low level of gene expression seen in the dark controls in the studies of Ait-Ali et al. (1999) and Gil and Martinez (2000) may have been attributable to the use of green safelights. Our work clearly shows that a 15-min exposure to our green safelight can cause substantial down-regulation of PsGA3ox1 expression 2 h later (data not shown), and a small down-regulation is also apparent in the data of Ait-Ali et al. (1999). This may not be surprising because phyA is the photoreceptor involved and this is thought to be the photoreceptor responsible for the very low fluence response to red light in pea (Weller et al., 1995). Ait-Ali et al. (1999) and Gil and Garcia-Martinez (2000) appear to have missed the rapid (as early as 0.25 h after commencement of red light, data not shown) down-regulation of PsGA3ox1 expression by light (Fig. 5).
To our knowledge, this study is the first to examine the photoregulation of GA-deactivating genes. A rapid (by 0.5 h) and clear photoregulation of PsGA2ox2 is apparent (Fig. 6) and certainly occurs sufficiently early to explain the significant increase in GA8 levels seen after light exposure (Fig. 4). This gene has been proposed as a likely candidate to regulate deactivation of GA1 to GA8 in the shoot (Lester et al., 1999). This is consistent with a light-mediated increase in 2β-hydroxylation of GA1 to GA8 after feeds of labeled GA20 during de-etiolation (O'Neill et al., 2000).
In conclusion, the results suggest that phyA and a blue light photoreceptor(s) regulate the levels of bioactive GA1 during de-etiolation. This is achieved at least partly by regulating the deactivation of GA1 to GA8 by PsGA2ox2. Photoreceptors also regulate the expression of PsGA3ox1 (Mendel's LE gene), the gene responsible for controlling the 3β-hydroxylation of GA20 to GA1 in the shoot during the first 4 h of de-etiolation. PhyB does not appear to be involved in regulating GA levels during de-etiolation (Fig. 2D), but it does appear to control the change in responsiveness of stem tissue to GA1 during de-etiolation (Reid and Ross, 1988). Feedback up-regulation of PsGA20ox1 and PsGA3ox1 gene expression occurs after the first 4 h of de-etiolation, whereas feed-forward down-regulation of PsGA2ox1 and PsGA2ox2 gene expression is also apparent. The results clearly demonstrate a complex regulation of the GA biosynthesis genes at the level of transcript accumulation, which results in reduced GA1 levels and hence elongation during the first 4 h of de-etiolation.
MATERIALS AND METHODS
Plant Material
The pure pea (Pisum sativum) lines used were from the collection held at the University of Tasmania (Hobart, Tasmania, Australia). The WT line 107 (derived from cv Torsdag), the phyA-deficient mutant phyA-1 (Weller et al., 1997; Weller et al., 2001; formerly fun1-1), the phyB-deficient mutant phyB-5 (Weller et al., 2001; formerly lv-5), the double mutant phyA-1 phyB-5, the GA-deficient mutant ls-1 (Ait-Ali et al., 1997), and the GA-accumulating mutant sln (Reid et al., 1992) were used in the gene expression and GA level experiments. The GA1-deficient mutant le-3 (Ross et al., 1995) and the double mutant phyA-1 le-3 were used in the determination of GA responsiveness.
Growing Conditions
Plants were grown in plastic tote boxes, 50 plants per box, in a 1:1 (v/v) mixture of dolerite chips:vermiculite, topped with 2 to 3 cm of potting soil. Testae were nicked with a razor blade prior to planting. Plants were grown in complete darkness for 7 d at 20°C and were then transferred to blue, red, or far-red light. Control boxes were left in the dark. The light cabinets were at 20°C with light intensities of 23 μmol m−2 s−1. Red light was obtained by using TLD 36 W/15 Red (internally coated) fluorescent tubes (Philips, Eindhoven, Holland), blue light was obtained by using TLD 36 W/18 Blue (internally coated) fluorescent tubes (Philips) wrapped in two layers of blue plastic film (cutting sheet 521C; Nakagawa Chemical, Tokyo), and far-red light was obtained by using 20 W long-wavelength fluorescent tubes (FL20S/FR-74; Toshiba, Tokyo) filtered through far-red plastic (Westlakes Plastic Company, Lenni, PA). The green safelight was L40 W/20S cool-white fluorescent tubes (Osram, Munich, Germany), covered in alternate layers of blue, yellow, and green plastic film (cutting sheets 521C, 321C, and 431C, respectively; Nakagawa Chemical).
Harvest Procedure
Plants were excised at the soil surface, weighed, and counted. Plants were harvested at the following times after transfer to light: 0.5, 1, 2, 4, 7, and 24 h. Dark-grown control plants were harvested under a green safelight at time zero and at 24 h. Approximately 15 plants were harvested for hormone analysis and 10 were harvested for northern analysis. The plants for RNA extraction were wrapped in foil and were then immersed in liquid nitrogen. The plants used for hormone analysis were immersed in approximately 50 mL of cold (−20°C) 80% (v/v) methanol.
GA Responsiveness
The testae of all seeds were nicked. Seeds were treated with 5 μg of paclobutrazol (PP333) alone or with 5 μg of PP333 plus 1 μg of GA1 or 10 μg of GA1 in 10 μL of ethanol. Plants were grown under red light as previously described. Lengths of the first four internodes, the total height, and the number of nodes expanded were recorded at 19 d.
Northern Analysis
Total RNA was extracted from approximately 100 mg of ground tissue using a RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The RNA was quantified using a GBC UV/VIS 916 spectrophotometer (Dandenong, Victoria, Australia). Five micrograms of total RNA was run on a 1% agarose (w/v) denaturing formaldehyde gel and was blotted onto GeneScreen Plus (PerkinElmer Life Sciences, Boston). Hybridization was carried out in formamide prehybridization solution at 42°C or in DNA prehybridization solution at 65°C as described by Ausubel et al. (1994), previous experiments having shown either method to give the same results. DNA probes were labeled with 32P by random priming using a DecaLabel kit (MBI Fermentas, Progen, Queensland, Australia). Northern blots were washed at 65°C in 2× SSC and 0.1% (w/v) SDS, followed by a wash with 0.2× SSC and 0.1% (w/v) SDS. Blots were exposed to x-ray film (Biomax MS; Eastman-Kodak, Rochester, NY) at −70°C.
Hormone Analysis
The samples were kept at −20°C until extraction commenced. Internal standards of [17-2H2]GA1, [17-2H2]GA8, [17-2H2]GA19, [17-2H2]GA20, and [17-2H2]GA29 were added in amounts appropriate to the light treatment. Internal standards were provided by Prof. L.N. Mander (Research School of Chemistry, Australian National University, Canberra, Australian Capital Territory, Australia). Extraction and purification was carried out as described by O'Neill et al. (2000). GAs were separated by HPLC and were quantified by gas chromatography-selected ion monitoring as previously described (Ross et al., 1995). The ion pairs monitored for the quantification of GAs were as follows: 506 and 508 for GA1, 594 and 596 for GA8, 434 and 436 for GA19, 418 and 420 for GA20, and 506 and 508 for GA29.
ACKNOWLEDGMENTS
We thank Tracey Jackson, Ian Cummings, Annelli Binns, Bedrich Eckhart, Tracey Barker, Claire Hardman, Brit Bezemer, Felicity Chambers, Ben Boevink, Kate Brettingham-Moore, and Dr. Noel Davies for technical support, Dr. John Ross for helpful comments and assistance with the manuscript, and Prof. Lewis Mander for labeled GAs.
Footnotes
This work was supported by the Australian Research Council.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010607.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley Interscience; 1994. [Google Scholar]
- Ait-Ali T, Frances S, Weller JL, Reid JB, Kendrick RE, Kamiya Y. Regulation of gibberellin 20-oxidase and gibberellin 3β-hydroxylase transcript accumulation during de-etiolation of pea seedlings. Plant Physiol. 1999;121:783–791. doi: 10.1104/pp.121.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Ali T, Swain SM, Reid JB, Sun T, Kamiya Y. The LSlocus of pea encodes the gibberellin biosynthesis enzyme ent-kaurene synthase A. Plant J. 1997;11:443–454. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- Behringer FJ, Davies PJ, Reid JB. Phytochrome regulation of stem growth and indole-3-acetic acid levels in the lv and Lv genotypes of Pisum. Photochem Photobiol. 1992;56:677–684. [Google Scholar]
- Elliott RC, Ross JJ, Smith JJ, Lester DR, Reid JB. Feed-forward regulation of gibberellin deactivation in pea. J Plant Growth Regul. 2001;20:87–94. [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Gil J, Garcia-Martinez JL. Light regulation of gibberellin A(1) content and expression of genes coding for GA 20-oxidase and GA 3 β-hydroxylase in etiolated pea seedlings. Physiol Plant. 2000;180:223–229. [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Hisada A, Hanzawa H, Weller JL, Nagatani A, Reid JB, Furuya M. Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell. 2000;12:1063–1078. doi: 10.1105/tpc.12.7.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD, James PE, Carrera E, Prat S, Thomas B. Regulation of transcript levels of a potato gibberellin 20-oxidase gene by light and phytochrome B. Plant Physiol. 2000;124:423–430. doi: 10.1104/pp.124.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Garcia-Martinez JL. Regulation of gibberellin biosynthesis by light. Curr Opin Plant Biol. 1999;2:398–403. doi: 10.1016/s1369-5266(99)00012-6. [DOI] [PubMed] [Google Scholar]
- Kende H, Lang A. Gibberellins and light inhibition of stem growth in peas. Plant Physiol. 1964;39:435–440. doi: 10.1104/pp.39.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Davies PJ, Reid JB. Mendel's stem length gene (Le) encodes a gibberellin 3β-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester DR, Ross JJ, Smith JJ, Elliott RC, Reid JB. Gibberellin 2-oxidation and the SLN gene of Pisum sativum. Plant J. 1999;19:1–9. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- Lockhart JA. The effect of light and the gibberellins on stem elongation in dwarf and normal pea seedlings. Plant Physiol. 1956;31:xii. [Google Scholar]
- Martin DN, Proebsting WM, Parks TD, Dougherty WG, Lange T, Lewis MJ, Gaskin P, Hedden P. Feed-back regulation of gibberellin biosynthesis and gene expression in Pisum sativumL. Planta. 1996;200:159–166. doi: 10.1007/BF00208304. [DOI] [PubMed] [Google Scholar]
- O'Neill DP, Ross JJ, Reid JB. Changes in gibberellin A1levels and response during de-etiolation of pea seedlings. Plant Physiol. 2000;124:805–812. doi: 10.1104/pp.124.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Spalding EP. Sequential and coordinated action of phytochromes A and B during Arabidopsisstem growth revealed by kinetic analysis. Proc Natl Acad Sci USA. 1999;96:14142–14146. doi: 10.1073/pnas.96.24.14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Chory J, Furuya M. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsisdevelopment. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB. Internode length in Pisum. Do the internode length genes effect growth in dark-grown plants. Plant Physiol. 1983;72:759–763. doi: 10.1104/pp.72.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB. Internode length in Pisum: comparison of genotypes in the light and dark. Physiol Plant. 1988;74:83–88. [Google Scholar]
- Reid JB, Ross JJ. A new gene, lv, conferring an enhanced response to gibberellin A1. Physiol Plant. 1988;72:595–604. [Google Scholar]
- Reid JB, Ross JJ, Swain SM. Internode length in Pisum: a new, slender mutant with elevated levels of C19gibberellins. Planta. 1992;188:462–467. doi: 10.1007/BF00197036. [DOI] [PubMed] [Google Scholar]
- Ross JJ, MacKenzie-Hose AK, Davies PJ, Lester DR, Twitchin B, Reid JB. Further evidence for feedback regulation of gibberellin biosynthesis in pea. Physiol Plant. 1999;105:532–538. [Google Scholar]
- Ross JJ, Reid JB, Swain SM, Hasan O, Poole AT, Hedden P, Willis CL. Genetic regulation of Gibberellin deactivation in Pisum. Plant J. 1995;7:513–523. [Google Scholar]
- Ross JJ, Willis CL, Gaskin P, Reid JB. Shoot elongation in Lathyrus odoratusL.: gibberellin levels in light and dark-grown tall and dwarf seedlings. Planta. 1992;187:10–13. doi: 10.1007/BF00201618. [DOI] [PubMed] [Google Scholar]
- Talon M, Zeevaart JAD, Gage DA. Identification of gibberellins in spinach and effects of light and darkness on their levels. Plant Physiol. 1991;97:1521–1526. doi: 10.1104/pp.97.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Mitsuhashi W, Inoue Y, Kamiya Y. Phytochrome regulates gibberellin biosynthesis during germination of photoblastic lettuce seeds. Plant Physiol. 1998;118:1517–1523. doi: 10.1104/pp.118.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Beauchamp N, Kerckhoffs LHJ, Platten JD, Reid JB. Interaction of phytochromes A and B in the control of de-etiolation and flowering in pea. Plant J. 2001;26:1–14. doi: 10.1046/j.1365-313x.2001.01027.x. [DOI] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB. Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol. 1997;114:1225–1236. doi: 10.1104/pp.114.4.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Nagatani A, Kendrick RE, Murfet IC, Reid JB. New lvmutants of pea are deficient in phytochrome B. Plant Physiol. 1995;108:525–532. doi: 10.1104/pp.108.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Ross JJ, Reid JB. Gibberellins and phytochrome regulation of stem elongation in pea. Planta. 1994;192:489–496. [Google Scholar]
- Wu K, Gage AD, Zeevaart JAD. Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from long-day plant spinach. Plant Physiol. 1996;110:547–554. doi: 10.1104/pp.110.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Van Lammeren AM, Vermeer E, Vreugdenhil D. The role of gibberellin, abscisic acid, and sucrose in the regulation of potato tuber formation in vitro. Plant Physiol. 1998;117:575–584. doi: 10.1104/pp.117.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RGS, Kamiya Y, Sun TP. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsisseeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]