Abstract
Lactobacillus rhamnosus GG is of general interest as a probiotic. Although L. rhamnosus GG is often used in clinical trials, there are few genetic tools to further determine its mode of action or to develop it as a vehicle for heterologous gene expression in therapy. Therefore, we developed a reproducible, efficient electroporation procedure for L. rhamnosus GG. The best transformation efficiency obtained was 104 transformants per μg of DNA. We validated this protocol by tagging L. rhamnosus GG with green fluorescent protein (GFP) using the nisin-controlled expression (NICE) system. Parameters for overexpression were optimized, which allowed expression of gfp in L. rhamnosus GG upon induction with nisin. The GFP+ strain can be used to monitor the survival and behavior of L. rhamnosus GG in vivo. Moreover, implementation of the NICE system as a gene expression switch in L. rhamnosus GG opens up possibilities for improving and expanding the performance of this strain. The GFP-labeled strain was used to demonstrate that L. rhamnosus GG is sensitive to human beta-defensin-2 but not to human beta-defensin-1.
Increasing consumer awareness of health-promoting intestinal bacteria has increased the use of probiotic bacteria as functional ingredients in certain foods, most commonly dairy products (19). Probiotics are defined as live microorganisms which, when adequate amounts are administered, provide a health benefit to the host beyond basic nutrition (11, 12). Although the concept of probiotics is becoming established in medicine and the food and feed industry, the underlying scientific framework needs to be strengthened (44). A great number of trials have suggested beneficial effects of probiotics (10, 22, 24, 32, 41, 45). Nevertheless, the basic molecular mechanisms underlying the observations have not been completely determined yet (40, 44). A prerequisite for thoroughly addressing the mode of action of a probiotic strain is amenability to genetic analysis (46).
Lactobacillus rhamnosus GG (= ATCC 53103) (53), which was isolated from a naturally occurring healthy human gut flora, is one of the most extensively studied probiotic organisms (16, 23, 49). However, our understanding of the physiology and genetics of this bacterium is still limited, mainly due to the absence of a reliable transformation procedure. In the past decade electroporation has become the method of choice for transforming lactobacilli (34). The efficiencies of electroporation between different Lactobacillus species and even between strains belonging to the same species can vary significantly (34). Another difficulty in the development of a transformation procedure is choosing the plasmid used. Indeed, the ability of a plasmid to replicate and to express its selectable marker in a given species cannot be predicted and influences the transformation frequency (52). The objective of the present study was to develop a reproducible electroporation procedure for L. rhamnosus GG. We adapted the previously described procedure for electrotransformation of L. rhamnosus 1/6 (56). The optimized protocol was used to increase the arsenal of genetic tools for L. rhamnosus GG with the green fluorescent protein (GFP). The first report describing GFP labeling of Lactobacillus was made by Geoffroy et al. in 2000 (15). Since then, other GFP-labeled lactic acid bacteria have been described (17, 25, 43). A GFP-labeled Lactobacillus plantarum strain was obtained by using the nisin-controlled expression (NICE) system initially developed for Lactococcus lactis (4). After addition of subinhibitory amounts of the signaling molecule nisin (7), the two-component NisRK system drives expression of GFP under control of the nisin-inducible nisA promoter (4). The NICE system has been used successfully for a variety of lactic acid bacteria (9, 26, 29, 42). In this study, we used the optimized electroporation protocol to implement the NICE system in L. rhamnosus GG in order to tag L. rhamnosus GG with the green fluorescent protein. The GFP-tagged strain was used to determine the sensitivity of L. rhamnosus GG to human beta-defensins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown at 37°C. L. rhamnosus GG was inoculated from a glycerol stock (−80°C) into Man-Rogosa-Sharpe (MRS) medium (Difco) and grown in static conditions. Escherichia coli was grown with aeration in Luria-Bertani broth (47). When appropriate, antibiotics were added to the culture medium. For Lactobacillus, erythromycin and chloramphenicol were used at final concentrations of 5 and 10 μg/ml, respectively. In case of E. coli, antibiotics were supplied at following final concentrations: ampicillin, 100 μg/ml; kanamycin, 30 μg/ml; erythromycin, 250 μg/ml; and chloramphenicol, 30 μg/ml.
TABLE 1.
Bacterial strains, plasmids, and primer sequences used for PCR
| Strain or plasmid | Relevant genotype and/or phenotypea | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10F′ | F′ [lacIq Tn10(Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli GM2163 | Dam−/Dcm−; F−ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL136(Strr) dam13::Tn9(Camr) xylA5 mtl-1 thi-1 mcrB1 hsdR2; Cmr | New England BioLabs Inc. |
| FAJ1905 | L. rhamnosus GG with chromosomal insertion of pMEC10 in the attB sequence of phage mv4; Eryr | This study |
| FAJ1906 | L. rhamnosus GG with chromosomal insertion of pFAJ1934 in the attB sequence of phage A2; Eryr | This study |
| L. casei ATCC 393 | Wild type; cheese isolate | American Type Culture Collection, Rockville, MD |
| L. rhamnosus 1/6 | Wild type; cheese isolate | Valio Ltd. |
| L. rhamnosus GG | Wild type; human isolate | ATCC 53103 (53) |
| Plasmids | ||
| pEM40 | pUC19E-derived integration vector (attB located at the 3′ end of the tRNALeu locus) containing a 1.6-kb int-attP cassette of phage A2; Eryr Ampr | 1 |
| pFAJ1934 | 2,634-bp PCR fragment (flanking EcoRI sites) carrying the nisRK genes of pMEC10 cloned into the EcoRI site of pEM40 downstream of the ery gene in the opposite orientation; Eryr Ampr | This study |
| pGK13 | Shuttle vector between E. coli and Lactococcus lactis; pWV01 replicon; Eryr Cmr | 27 |
| pLAB1301 | L. hilgardii pLAB1000 replicon; shuttle vector between E. coli and L. plantarum; Eryr Cmr | 21 |
| pMEC10 | Integration plasmid (attB located at the 3′ end of the tRNASer locus) containing int-attP cassette (of phage mv4) of pMC1 cloned into pNZ9500 (pUC19 derivative carrying a 2.7-kb chromosomal DNA fragment of L. lactis NZ9700 containing the 3′ end of nisP and nisRK); expression driven by ery read-through; Eryr | 42 |
| pMEC30 | L. hilgardii pLAB100 replicon; gfpuv cloned in expression vector containing the constitutive ldhL promoter from L. plantarum; Eryr Ampr | 15 |
| pMEC45 | L. lactis pSH71 replicon; pNZ8037 derivative with gfpuv cloned downstream of the nisA promoter from L. lactis NZ9800; Cmr | 15 |
| pNZ8008 | pNZ273 (pSH71 replicon) carrying the E. coli gusA reporter gene transcriptionally fused to the nisA promoter; Cmr | 28, N1Z0 |
| pRV85 | gfpuv cloned downstream of the L. sakei ldhL promoter in the replicative plasmid pG+host5; Eryr | 17 |
gfpuv, synthetic GFP gene (with improved codon usage) that was subsequently subjected to recursive cycles of DNA shuffling, thereby selecting for bright fluorescence that could be observed under UV light.
Molecular methods.
Standard protocols were used for buffer preparation, cloning, plasmid isolation, preparation of E. coli competent cells, and transformation (47). Restriction enzymes were used according to the manufacturer's instructions. Southern hybridization was carried out using a nonradioactive labeling and detection system, as previously described (47). DNA sequences were determined by the dideoxynucleoside triphosphate chain termination method (48) with an AutoRead sequencing kit (Pharmacia-LBK) and an automated sequencer (ALX; Pharmacia-LBK). PCR primers were purchased from Eurogentec. PRO-12 (5′-ATGAATTCGGTCAGGATGATGCTATTAACC-3′) and PRO-13 (5′-ATGAATTCGCACCGTTAACTCCTAATAAACC-3′) (EcoRI recognition sites are underlined) were used for construction of pFAJ1934 (see below); RHI-144 (5′-ATTAGAAATGAGAAACTATGAAGTTGC-3′) and RHI-145 (5′-TACTCCTAATCTGATAAATATACTCCG-3′) are homologous to the 5′ and 3′ regions of nisR; RHI-146 (5′-AATTTTCAAGCCGTTCAAAAGATTGC-3′) and RHI-147 (5′-ATTAACAAAGATATTGATAAGTGCTCG-3′) are homologous to the 5′ and 3′ regions of nisK; and RHI-148 (5′-AACTTTTCACTGGAGTTGTCC-3′) and RHI-149 (5′-TAATGGTCTGCTAGTTGAACG-3′) are homologous to the 5′ and 3′ regions of gfpuv. If single colonies of L. rhamnosus GG were used, the colony smeared in a PCR tube was heated in a microwave oven at full power for 3 min before the PCR mixture was added. The PCR was carried out with a Personal Mastercycler (Eppendorf) used according to the manufacturer's instructions. Integration of pMEC10 at the tRNASer locus and integration of pFAJ1934 at the tRNALeu locus were confirmed by PCR and Southern blotting, as previously described (1, 8). The following primers were used: PRO-567 (5′-GTCGACACAGGATTTGAACC-3′; corresponding to primer T2 [8]) and PRO-570 (5′-CAAGCCAACAGACGTGCAAGCA-3′; binding to the 3′ end of the int gene [8]) for tRNASer integration and PRO-564 (5′-AGCAGGACGAGAAAGCAATGAATGT-3′; corresponding to primer att1 [1]) and PRO-565 (5′-GCCGGTGTGGCGGAATTGGCAG-3′; corresponding to primer att7 [1]) for tRNALeu integration.
Construction of pFAJ1934.
The nisRK genes were amplified by PCR with the proofreading Pfx enzyme (Invitrogen) used according to the manufacturer's instructions and primers PRO-12 and PRO-13, using pMEC10 as the template DNA. The 2,634-bp fragment obtained, containing the nisR promoter, was digested with EcoRI and ligated into the corresponding unique site of pEM40. PCR, restriction, and sequence analysis confirmed the localization of nisRK downstream of the erythromycin resistance (ery) gene. The plasmid was designated pFAJ1934 and introduced into E. coli for propagation and subsequent plasmid DNA isolation. In pFAJ1934, the nisRK genes are localized downstream of the ery gene of pEM40, in the opposite orientation. This orientation of the nisRK genes prevents read-through transcription of ery, which lacks a functional transcription terminator (42) and allows low levels of expression of nisRK, which is required for a functional inducible expression system. Indeed, in order to retain the inducibility to nisin without a significant increase of basal activity, low levels of expression of nisRK are necessary (42).
Preparation of L. rhamnosus GG electrocompetent cells.
An overnight culture of L. rhamnosus GG was serially diluted (102- to 106-fold) into freshly prepared prewarmed MRS medium supplemented with 2% glycine and was incubated without agitation at 37°C. After overnight growth, 5 ml of the culture in the exponential growth phase (optical density at 600 nm [OD600], 0.8 to 1) was inoculated into 100 ml of freshly prepared, prewarmed MRS medium supplemented with 2% glycine. The resulting culture, which was kept in a tightly closed 100-ml Duran bottle (to minimize oxygen transfer), was incubated without agitation at 37°C. When the OD600 was 0.2 to 0.3, ampicillin was added to obtain a final concentration of 10 μg/ml (57). Then the cells were incubated until the OD600 was 0.4 to 0.5. This optical density should have been reached within 3 to 4 h after the initial inoculation of L. rhamnosus GG in order to obtain efficient competent cells. Cells were harvested by centrifugation at room temperature (10 to 15 min, 6,000 × g). The cells were washed twice at room temperature with electroporation buffer (0.5 M sucrose, 7 mM potassium phosphate [pH 7.4], 1 mM MgCl2) (2), resuspended in 1 ml of the same buffer, and placed on ice. The electrocompetent cells were used immediately for electroporation. After 16 h of freezing at −80°C, a slight decrease in the electrocompetency of the cells was noticed, but after 1 week of storage, no transformants could be obtained.
Electroporation of L. rhamnosus GG.
A mixture containing 100 μl of a cooled cell suspension and 400 ng of DNA (maximum volume, 4 to 5 μl) was transferred into a precooled electroporation cuvette (Eurogentec) with a 0.2-cm electrode gap and immediately electroporated (Gene Pulser; Bio-Rad Laboratories) by using the following settings: peak voltage, 1.7 kV; capacitance, 25 μF; and parallel resistance, 200 Ω. Following the pulse, the cells were immediately diluted with 5 ml of MRS medium containing 2 mM CaCl2 and 20 mM MgCl2 and incubated (without agitation) at 37°C for 3 h before they were plated onto MRS agar containing the appropriate antibiotic. The transformation efficiency was assessed by determining the number of transformants per microgram of plasmid DNA after anaerobic incubation (BBL GasPak system) at 37°C for 48 to 72 h. Transformants were validated either by restriction digestion (after plasmid isolation and back-transformation to E. coli) or by colony PCR.
Isolation of L. rhamnosus GG DNA.
Total DNA of L. rhamnosus GG was isolated as previously described for Rhodococcus (36), with following modifications. Prewarmed MRS medium (freshly prepared) was inoculated with ca. 2 × 108 CFU of an overnight culture of L. rhamnosus GG. After 2 to 3 h of growth without agitation at 37°C, 600 μg/ml ampicillin was added to the culture, which was then incubated for another 2 to 3 h. Bacterial cells were washed with Tris-EDTA buffer (10 mM Tris, 1 mM EDTA; pH 8.0) and treated with lysozyme (2 mg/ml; Sigma) at 37°C for 30 min. The cells were lysed further by addition of sodium dodecyl sulfate (final concentration, 1%) and NaCl (final concentration, 1 M). RNase A (final concentration, 35 μg/ml; Sigma), prepared as described by Sambrook et al. (47), was added, and this was followed by 15 min of incubation at 55°C. The remainder of the protocol has been described previously (36). The precipitated DNA was resuspended at 4°C for 48 to 72 h. Total DNA was stored at 4°C. For isolation of plasmid DNA from L. rhamnosus GG, the protocol used for isolation of total DNA was used, with some minor adaptations. After sodium dodecyl sulfate and NaCl were added, the mixture was incubated at −20°C for 1 h. Cell wall components and degraded total DNA were removed by centrifugation, after which the procedure used for isolation of total DNA was used.
Microtiter plate β-glucuronidase assay to optimize nisin induction.
Nisin-induced expression of gusA (pNZ8008) (28) was optimized essentially as described by Pavan et al. (42), except that strain L. plantarum NCIMB8826 was replaced by L. rhamnosus GG FAJ1905. pNZ8008 (28) was introduced into FAJ1905 by using the optimized electroporation protocol described above. β-Glucuronidase assays were performed by the method described by de Ruyter et al. (4), modified as described by Miller (35), which resulted in the following optimized microtiter plate-based protocol. After appropriate induction, 1 ml of cells was harvested by centrifugation (5 min, 5,000 × g) and resuspended in 500 μl of NaPi buffer (50 mM NaHPO4, pH 7.0). After the OD595 of 135 μl was determined, cell suspensions were frozen at −80°C in 10-μl aliquots in 96-well plates until they were assayed for β-glucuronidase activity. To each 10-μl cell suspension, 90 μl of GUS buffer (50 mM NaHPO4 [pH 7.0], 14.3 mM β-mercaptoethanol, 1 mM Na2EDTA, 0.1% Triton X-100, 0.1% sodium laurylsarcosine, 25 mM p-nitrophenyl-β-d-glucuronic acid [PNPG] [Sigma]) was added. The mixture was incubated at 37°C. The reaction was stopped by adding 35 μl of a 1 M Na2CO3 solution after sufficient yellow color had developed. The reaction was stopped at at least three different times (replicates) to ascertain that enzyme activity was still linearly increasing with incubation time. The reaction time was recorded. Both OD415 and OD595 were determined. Identical treatments were performed with NaPi buffer but without cells as controls to correct measured sample values. Miller units (MU) of β-glucuronidase activity were defined as 1,000 times the increase in absorbance at 415 nm per minute per unit of optical density at 595 nm of the cell suspension, as follows: MU = 1,000 × {[(OD415,PNPG − 1.75 × OD595,PNPG) × v1]/(t × vt × OD595)}, where t is the time of the reaction (in minutes), OD595 is a measure of cell density just before the assay, OD415,PNPG is the absorbance by PNPG measured after the reaction, OD595,PNPG is a measure of the cell density after the β-glucuronidase reaction (used as a correction for light scattering by cell debris), 1.75 is the corresponding correction factor, v1 is the volume (in μl) of cells used in the reaction mixture, and vt is the total volume (in μl) of the reaction mixture. The statistical analysis was based on at least three independent repetitions, as designed in the optimization experiment. Optical densities were determined with a microtiter plate reader (VERSAmax; Molecular Devices).
Expression of gfp under control of the nisin promoter.
Expression of gfp driven by the nisA promoter (pMEC45) in L. rhamnosus GG was optimally induced as follows. An overnight culture of L. rhamnosus GG FAJ1905/pMEC45 or FAJ1906/pMEC45 was used to inoculate fresh, prewarmed MRS medium diluted 1:50. After 30 min of incubation at 37°C without shaking, 500 ng/ml nisin was added to the culture, which was then incubated for 2.5 to 3 h. Nisin was prepared as previously described (9).
Epifluorescence microscopy and flow cytometric analysis.
Fluorescence microscopy was performed as previously described (58). Flow cytometric analysis was performed with a FACSCalibur (Becton Dickinson, Erembodegem, Belgium) equipped with a 15-mW, air-cooled argon ion laser excitation light source (488 nm), as previously described (31, 55). For two-color analysis, fluorescence compensation was used to correct for spectral overlap of the green and red signals. Data were analyzed using CellQuestPro (Becton Dickinson).
Human beta-defensin sensitivity assay.
L. rhamnosus GG cells were induced with nisin as described above and collected by centrifugation, and ca. 106 CFU was resuspended in 45 μl of sixfold-diluted Dulbecco's modified Eagle's medium (Gibco, Invitrogen) supplemented with 16.67 mM glucose. Subsequently, 5 μl of human beta-defensins (Peptides International, Inc., Louisville, KY) dissolved in 10 mM acetic acid was added. For human beta-defensin-2 (hBD2), final concentrations ranging from 0 to 12 μg/ml were tested. For hBD1, a final concentration of 12 μg/ml was used. After 3 h of incubation at 37°C, bacterial viability was measured either by plating serial dilutions on MRS agar or by flow cytometry using the green fluorescent protein-propidium iodide (PI) assay as previously described (31). Each experiment was performed in triplicate.
RESULTS AND DISCUSSION
Optimization of the electroporation protocol.
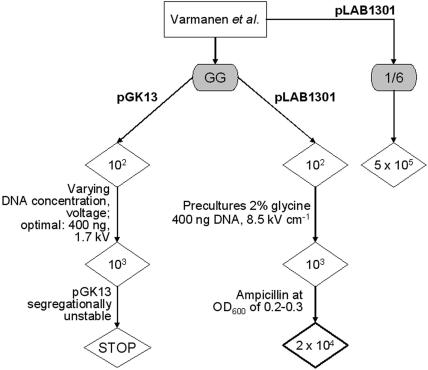
The optimization strategy used for electroporation is shown in Fig. 1. Initially, pGK13 (27) was used as the DNA source, which resulted in an efficiency of 103 CFU/μg DNA after some optimization (Fig. 1). The optimal pulse strength was 8.5 kV cm−1, which is higher than the optimal pulse strength in the protocol described by Varmanen et al. (7.5 kV cm−1) (56) but lower than the optimal pulse strength described by Wei et al. (12.5 kV cm−1) (57), indicating that optimization of conditions for electrotransformation is necessary for each species or even for each strain of lactic acid bacteria. However, pGK13 seemed to be segregationally unstable in L. rhamnosus GG, and the results were not reproducible (data not shown). Plasmid pLAB1301 (21), which contains the replicon of a native plasmid isolated from Lactobacillus hilgardii, was used subsequently. Different electrocompetence-promoting protocols were tried, as shown in Fig. 1. Adding the cell wall weakener glycine (25) at a concentration of 2% to the preculture medium increased the transformation efficiency 10-fold. Addition of ampicillin, also a cell wall weakener (57), during the early exponential growth phase of L. rhamnosus GG led to a 20-fold increase in the efficiency compared to the efficiency of an identical protocol without addition of ampicillin. Addition of ampicillin was first described by Wei et al. (57) for electroporation of Lactobacillus isolates, and this protocol was also used by McCracken et al. (33) for electroporation of L. rhamnosus GG. However, in our hands, the protocol described by Wei et al. (57) did not result in efficient electroporation of L. rhamnosus GG (data not shown). Compared to the protocol of Wei et al. (57), we included in our optimization protocol an extra cell wall weakener, glycine, during the two culture phases, and we also changed the composition of the washing and electroporation buffer, the temperature at which the washing step was performed, and the electrical parameters used for electroporation (33, 57).
FIG. 1.
Optimization of the electroporation protocol. The protocol described by Varmanen et al. (56) for electrotransformation of L. rhamnosus 1/6, a cheese isolate, was used as a starting point. Using the originally described protocol with L. rhamnosus GG resulted in an efficiency of only 102 CFU/μg DNA. By gradually varying the DNA concentration (0.2 to 3 μg DNA) and the voltage applied (1.4 to 2.0 kV) we increased the efficiency 10-fold, and the optimal parameter combination was 400 ng DNA and 1.7 kV. However, pGK13 seemed to be segregationally unstable in L. rhamnosus GG and was not used further. Electroporation of L. rhamnosus GG and L. rhamnosus 1/6 using pLAB1301 resulted in reproducible efficiencies of 102 and 5 × 105 CFU/μg DNA, respectively. The resilience of the peptidoglycan layer in gram-positive bacteria complicates the development of electroporation protocols (54). The composition of the growth medium directly affects the cell wall density and thickness. Therefore, weakening the cell wall by addition of cell wall weakeners, such as glycine (18), can improve the transformation efficiency. We included two culture steps with 2% glycine in the growth medium. When 2% glycine was added to the serial dilutions of an overnight culture of L. rhamnosus GG, the electroporation efficiency increased 10-fold compared to the electroporation efficiency of an identical protocol without an overnight preculture supplemented with glycine. Addition of ampicillin, also a cell wall weakener (57), during the early exponential growth phase of L. rhamnosus GG led to a 20-fold increase in the efficiency compared to the efficiency of an identical protocol without addition of ampicillin.
With this optimized protocol, a transformation efficiency of 104 L. rhamnosus GG transformants per μg of pLAB1301 DNA was obtained. A similar efficiency was obtained for Lactobacillus delbrueckii subsp. bulgaricus (52), but this efficiency was still less than the efficiency obtained for L. plantarum (106 to 107 CFU/μg) (14). Perhaps the efficiency can still be improved, but 104 CFU/μg DNA is sufficient to select for double homologous recombination events, as described by Varmanen et al. (56) for the construction of a L. rhamnosus 1/6 pepR mutant. Here, we validated the protocol by overexpressing gfp under control of the NICE system, resulting in GFP-labeled L. rhamnosus GG that could be sorted by fluorescence-activated cell sorting and be used in other applications.
Expression of gfp in L. rhamnosus GG driven by constitutive promoters.
Using the optimized transformation protocol, we introduced pMEC30 (15) (a pLAB1301 derivative containing gfpuv under control of the constitutive ldhL promoter from L. plantarum) into L. rhamnosus GG (transformation efficiency, 104 CFU/μg DNA). pMEC30 could be successfully maintained in L. rhamnosus GG, as verified by PCR and plasmid isolation, and subsequently reelectroporated into E. coli. Although pMEC30 was functional in E. coli, no fluorescent phenotype was observed in L. rhamnosus GG.
pRV85 was used previously to tag Lactobacillus sakei with GFP (17). pRV85 (17) could be successfully transferred to L. rhamnosus GG, as confirmed by PCR and Southern analysis (data not shown). L. rhamnosus GG cells bearing pRV85 appeared as green fluorescent bacteria in a fluorescence microscopic analysis, albeit with a low intensity (data not shown). However, pRV85 is a replication-thermosensitive derivative of pWV01 and replicates in gram-positive bacteria at 28°C and is lost at temperatures above 37°C (3). Expression of gfp from pldhL in L. sakei was reported to increase when the culture temperature was decreased (17). In view of future applications, optimization of gfp expression by altering the growth temperature of L. rhamnosus GG, which prefers 37°C for growth, was not pursued.
Also, in L. plantarum gfp expression from a constitutive promoter appeared not to be feasible (15). We attempted to express gfp under control of a nisin-inducible promoter, which was previously reported to be successful in L. plantarum (15).
Expression of gfp in L. rhamnosus GG through implementation of the NICE system.
The nisRK genes with their own promoter were integrated into the chromosome of L. rhamnosus GG. Site-specific integration was accomplished by electroporating the suicide plasmid pMEC10 (42) into L. rhamnosus GG. It is noteworthy that successful integration was obtained only with pMEC10 DNA isolated from E. coli GM2163, a Dam− Dcm− strain (unmethylated DNA). Putative integrants, which were able to grow on erythromycin, were analyzed by PCR and Southern hybridization (data not shown). No plasmid DNA containing nisRK could be reisolated. One integrant, FAJ1905, exhibited the expected tRNASer integration patterns and was used for further manipulation.
In parallel, we integrated nisRK into the L. rhamnosus GG chromosome by an alternative method using plasmid pFAJ1934. The nisRK genes were cloned in pEM40, a nonreplicative integration vector containing the A2-specific recombination cassette int-attP (1). Bacteriophage A2 integrates at the 3′ end of the tRNALeu locus. This attachment site allowed integration of A2 attP-containing vectors into the genomes of Lactobacillus casei and Lactobacillus paracasei (1), both of which are closely related to L. rhamnosus GG. Transformation of L. rhamnosus GG with pFAJ1934 and selection for erythromycin-resistant colonies yielded putative integrants. PCR and Southern hybridization confirmed that there was chromosomal integration of pFAJ1934 at the corresponding tRNA loci in these clones (data not shown). One integrant that had the correct structure was selected and designated FAJ1906.
The two integration plasmids supported successful chromosomal insertion at permissive sites based on the integration properties of a bacteriophage. This method provides an efficient way to generate stable heterologous DNA integration in L. rhamnosus GG.
Subsequently, pMEC45 (15), the expression vector containing gfp under control of the nisA promoter, was electroporated into FAJ1905 and FAJ1906. Transformants were verified by PCR analysis (data not shown).
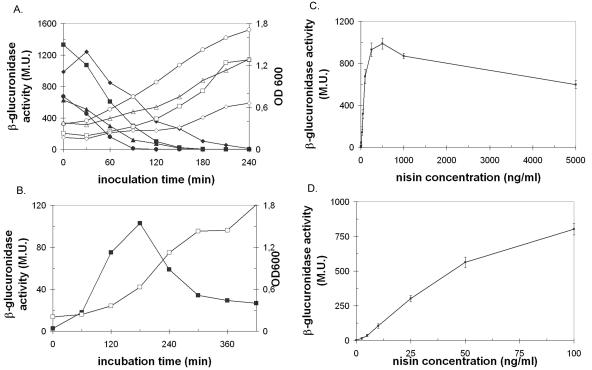
In previous applications of the NICE system in different species the workers stressed the need for optimization of nisin induction conditions in each host separately (5, 9, 42). The optimal growth phase for nisin induction, the optimal contact time with the inducer, and the optimal nisin concentration for induction were determined as described previously (42). To do this, we used the β-glucuronidase activity of FAJ1905/pNZ8008 as a reporter for nisin induction, concomitantly optimizing a user-friendly protocol for analysis of gusA expression in L. rhamnosus GG (see Materials and Methods). The results are summarized in Fig. 2. As Fig. 2 shows, the maximal levels of β-glucuronidase activity were obtained when the inducer was added early in the exponential phase, 30 min after inoculation (1/50 inoculum; 37°C), after 2.5 to 3 h of induction. Similar to the situation in L. plantarum (42), L. rhamnosus GG was most receptive to nisin when it was added very early in the exponential growth phase; in contrast, in L. lactis addition in the middle of the exponential growth phase was more effective (4).
FIG. 2.
β-Glucuronidase activity of FAJ1905/pNZ8008 cell extracts. (A) β-Glucuronidase activity (solid symbols) was determined during growth (open symbols) in different conditions (squares, 1/50 inoculum and incubation at 37°C; triangles, 1/20 inoculum and incubation at 30°C; circles, 1/20 inoculum and incubation at 37°C; diamonds, 1/20 inoculum and incubation at 30°C) after induction with 50 ng of nisin/ml at different times after inoculation. The β-glucuronidase activity was measured after 2.5 h of contact with nisin. (B) β-Glucuronidase activity (▪) after different periods of contact with 50 ng nisin/ml. The OD600 (□) was also determined. (C) Dose-response curve for gusA expression in FAJ1905 harboring pNZ8008 induced with different concentrations of nisin (0 to 5,000 ng/ml). (D) β-Glucuronidase activity with nisin concentrations ranging from 0 to 100 ng/ml. β-Glucuronidase activity is expressed in Miller units (M.U.), as described in Materials and Methods. The error bars indicate standard deviations.
The inoculation and incubation times were optimized as described by Pavan et al. (42), using 50 ng/ml nisin. However, the maximum levels of β-glucuronidase activity were observed after induction with 500 ng/ml (final concentration) nisin. In retrospect, this concentration may have been more appropriate in our study for optimizing the inoculation and incubation times. Remarkably higher doses of the inducer are necessary for L. rhamnosus GG (250 to 500 ng/ml) than for L. lactis (1 to 5 ng/ml) (5) or L. plantarum (20 to 25 ng/ml) (42). The optimal nisin concentrations for L. rhamnosus GG are in the same range as the concentrations used for induction in Streptococcus species (1 to 5 μg/ml) (9). In Bacillus subtilis and Enterococcus faecalis, concentrations as high as 20 μg/ml are necessary, and these concentrations are just below the inhibitory level (9). In addition, the optimal contact times with the inducer are different for L. plantarum and L. rhamnosus GG and are 5 h (42) and 2.5 h (this study), respectively. It has been reported previously that the optimal contact time with the inducer, leading to maximal protein production, can vary greatly with the host (26, 42). The induction factor, defined as the ratio of the β-glucuronidase activity observed after maximal induction to the basal activity with no inducer, is 500. This value is similar to values reported for L. plantarum (42), which in turn were only two- to threefold lower than the values reported for L. lactis (4).
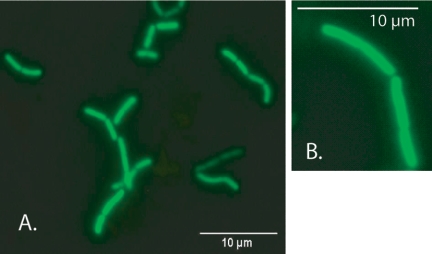
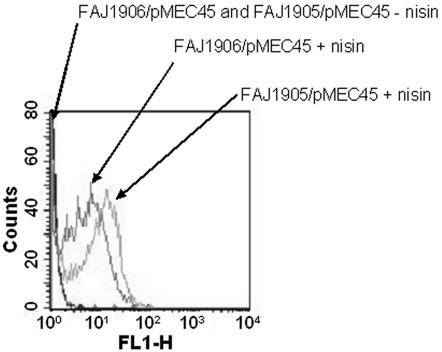
Using these inducing conditions with FAJ1905/pMEC45 and FAJ1906/pMEC45 resulted in bright green fluorescent bacteria, as determined by fluorescence microscopy (Fig. 3) and flow cytometric analysis (Fig. 4). FAJ1905/pMEC45 exhibited slightly enhanced fluorescence intensity compared to FAJ1906/pMEC45 (Fig. 4). This could have been due to the two different locations of nisRK (i.e., in the tRNASer and tRNALeu loci, respectively).
FIG. 3.
GFP-tagged L. rhamnosus GG, based on implementation of the NICE system: epifluorescence microscopic analysis of bacterial cells grown in liquid broth. (A) FAJ1905/pMEC45 induced with nisin as described in Materials and Methods. (B) FAJ1906/pMEC45 induced with nisin as described in Materials and Methods.
FIG. 4.
Flow cytometric analysis of induced (GFP+) and uninduced (GFP−) L. rhamnosus GG. FAJ1905/pMEC45 and FAJ1906/pMEC45 were induced with nisin as described in Materials and Methods (FAJ1905/pMEC45 + nisin and FAJ1906/pMEC45 + nisin). Cultures without nisin were used as negative controls (FAJ1906/pMEC45 and FAJ1905/pMEC45 − nisin). Green fluorescence intensities (FL1-H) are indicated on the x axis, and cell counts are indicated on the y axis. The cytometric analysis was performed for 2 × 104 events.
Use of GFP-tagged L. rhamnosus GG to determine sensitivity to human beta-defensins.
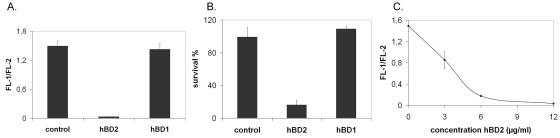
The combination of GFP and PI has been shown to have some advantages for analysis of bacterial viability compared to the frequently used LIVE/DEAD BacLight bacterial viability kit (31). Here we used the GFP-PI-based assay for flow cytometric measurement of the viability of L. rhamnosus GG after contact with human beta-defensins. Human beta-defensins are antimicrobial peptides expressed by epithelial cells (30, 50). It is now recognized that beta-defensins are integral components of the constitutive and regulated innate host defense; hBD1 is a constitutively expressed molecule, while hBD2, hBD3, and hBD4 are upregulated by bacterial challenge and proinflammatory cytokines (38, 39). It has been reported that constitutively expressed hBD1 can mediate epithelial interactions with the commensal flora, whereas hBD2 may participate in the host defense response to enteric microbes that can breach the epithelial barrier. Human beta-defensins are antimicrobial peptides that are particularly efficient in destroying gram-negative bacteria (38), while it has been reported that a bifidobacterial probiotic strain is not sensitive to human beta-defensin-mediated killing by recombinant hBD1, hBD2, and hBD3 (13). We investigated the effect of hBD1 and hBD2 on GFP-labeled L. rhamnosus GG using flow cytometry (Fig. 5). While hBD1 did not kill L. rhamnosus GG (Fig. 5A), L. rhamnosus GG was very sensitive to hBD2 (Fig. 5A). These results were confirmed by plate counting (Fig. 5B). The sensitivity was directly related to the defensin concentration (Fig. 5C). Subsequently, a time course experiment was performed in order to determine the defensin contact time required to kill the bacterial cells. At an hBD2 concentration of 6 μg/ml, a decrease in cell viability was observed 2 h after addition of the defensin to the culture medium. However, at a concentration of 12 μg/ml, L. rhamnosus GG was killed almost immediately after addition of the defensin (data not shown). Hudault et al. (20) reported that the number of L. rhamnosus GG cells in the gut of L. rhamnosus GG-monoassociated mice was dramatically decreased after a challenge with Salmonella, but the mechanism behind this phenomenon is unknown. It could be speculated that Salmonella infection upregulates defensin production (37), which leads to a decrease in the number of viable L. rhamnosus GG cells and reduced protection against the pathogen. Therefore, incorporation of a screen for defensin resistance in the selection of probiotic strains could be appropriate.
FIG. 5.
Sensitivity of L. rhamnosus GG to human beta-defensins. (A) Bacterial viability of L. rhamnosus GG after 3 h of contact with 12 μg/ml hBD1 or hBD2, as determined by GFP-PI-based flow cytometric analysis. The values are the ratios of the mean intensities of the green (FL-1) and red (FL-2) signals. The error bars indicate standard deviations of three independent measurements. No human beta-defensin was added to the control. (B) Bacterial viability of L. rhamnosus GG after 3 h of contact with 12 μg/ml hBD1 or hBD2, as determined by plate counting. The values are percentages of survival of the initial number of L. rhamnosus GG cells. The error bars indicate standard deviations of three independent measurements. (C) Concentration-dependent sensitivity of L. rhamnosus GG to hBD2, as determined by GFP-PI-based flow cytometric analysis as described above.
Concluding remarks.
Although there have been many studies of L. rhamnosus GG and L. rhamnosus GG has been used in numerous clinical trials, molecular genetic analysis of L. rhamnosus GG is poorly developed. This fact can be explained in part by the lack of a reliable transformation procedure. In this paper, we describe a reproducible electroporation protocol for L. rhamnosus GG. This protocol enabled us to implement the NICE system in L. rhamnosus GG and subsequently label this strain with GFP. As previously shown (15), preloaded fluorescent bacteria can be used for a variety of in vitro and in vivo experiments, including studies of the interaction of L. rhamnosus GG with members of the microbiota and with specific members of the immune system. Here, we used preloaded fluorescent bacteria to show L. rhamnosus GG sensitivity to hBD2.
The flexibility of the L. rhamnosus GG NICE system should allow expression of other genes of interest under control of the nisA promoter. This could be used to increase the technological suitability of L. rhamnosus GG by food-grade (nisin is a food-grade, approved antibiotic peptide) genetic manipulation approaches (46), as shown by overexpression of the GroESL heat shock protein chaperones in Lactobacillus paracasei NFBC338 (6). Moreover, this system could be used for development and design of live recombinant vaccines (51) in which L. rhamnosus GG is used as a live delivery vehicle. Thus, the implementation of the NICE system in L. rhamnosus GG provides an approach for improving and expanding both the technical performance and the probiotic performance of this strain.
Acknowledgments
S.D.K. was a research associate of the Belgian Fund for Scientific Research (FWO-Vlaanderen) when this study was conducted. P.H. is a research associate at FNRS. This work was supported by the IWT through research projects STWW-00162 and GBOU-20160.
We gratefully acknowledge Valio Ltd., A. Mercenier, M. Alvarez, W. de Vos, and M. Zagorec for kindly providing strains and plasmids used in this study.
REFERENCES
- 1.Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in Gram-positive and Gram-negative bacteria. Virology 250:185-193. [DOI] [PubMed] [Google Scholar]
- 2.Aymerich, M. T., M. Hugas, M. Garriga, R. F. Vogel, and J. M. Monfort. 1995. Electrotransformation of meat lactobacilli. Effect of several parameters on their efficiency of transformation. J. Appl. Bacteriol. 75:320-325. [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desmond, C., G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2004. Improved stress tolerance of GroESL-overproducing Lactococcus lactis and probiotic Lactobacillus paracasei NFBC 338. Appl. Environ. Microbiol. 70:5929-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos, W. M. 1999. Gene expression systems for lactic acid bacteria. Curr. Opin. Microbiol. 2:289-295. [DOI] [PubMed] [Google Scholar]
- 8.Dupont, L., B. Boizet-Bonhoure, M. Coddeville, F. Auvray, and P. Ritzenthaler. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J. Bacteriol. 177:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezendam, J., and H. van Loveren. 2006. Probiotics: immunomodulation and evaluation of safety and efficacy. Nutr. Rev. 64:1-14. [DOI] [PubMed] [Google Scholar]
- 11.FAO/WHO. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report. FAO, Rome, Italy.
- 12.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 13.Furrie, E., S. MacFarlane, A. Kennedy, J. H. Cummings, S. V. Walsh, D. A. O'Neil, and G. T. MacFarlane. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasson, M. J., and G. F. Fitzgerald. 1994. Gene transfer and transposition, p. 1-51. In M. J. Gasson and W. M. de Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Chapmann & Hall, London, United Kingdom.
- 15.Geoffroy, M. C., C. Guyard, B. Quatannens, S. Pavan, M. Lange, and A. Mercenier. 2000. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl. Environ. Microbiol. 66:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbach, S. L. 2000. Probiotics and gastrointestinal health. Am. J. Gastroenterol. 95:S2-S4. [DOI] [PubMed] [Google Scholar]
- 17.Gory, L., M. C. Montel, and M. Zagorec. 2001. Use of green fluorescent protein to monitor Lactobacillus sakei in fermented meat products. FEMS Microbiol. Lett. 194:127-133. [DOI] [PubMed] [Google Scholar]
- 18.Hammes, W., K. H. Schleifer, and O. Kandler. 1973. Mode of action of glycine on the biosynthesis of peptidoglycan. J. Bacteriol. 116:1029-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heller, K. J. 2001. Probiotic bacteria in fermented foods: product characteristics and starter organisms. Am. J. Clin. Nutr. 73:374S-379S. [DOI] [PubMed] [Google Scholar]
- 20.Hudault, S., V. Lievin, M. F. Bernet-Camard, and A. L. Servin. 1997. Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella typhimurium C5 infection. Appl. Environ. Microbiol. 63:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a Gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid. 21:9-20. [DOI] [PubMed] [Google Scholar]
- 22.Kajander, K., K. Hatakka, T. Poussa, M. Farkkila, and R. Korpela. 2005. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment. Pharmacol. Ther. 22:387-394. [DOI] [PubMed] [Google Scholar]
- 23.Kalliomaki, M., S. Salminen, H. Arvilommi, P. Kero, P. Koskinen, and E. Isolauri. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076-1079. [DOI] [PubMed] [Google Scholar]
- 24.Kalliomaki, M., S. Salminen, T. Poussa, H. Arvilommi, and E. Isolauri. 2003. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 361:1869-1871. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y. H., K. S. Han, S. Oh, S. You, and S. H. Kim. 2005. Optimization of technical conditions for the transformation of Lactobacillus acidophilus strains by electroporation. J. Appl. Microbiol. 99:167-174. [DOI] [PubMed] [Google Scholar]
- 26.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kok, J., J. M. B. M. Van der Vossen, and G. Venema. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 29.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1997. Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol. 15:135-140. [DOI] [PubMed] [Google Scholar]
- 30.Lehrer, R. I., A. K. Lichtenstein, and T. Ganz. 1993. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu. Rev. Immunol. 11:105-128. [DOI] [PubMed] [Google Scholar]
- 31.Lehtinen, J., J. Nuutila, and E. M. Lilius. 2004. Green fluorescent protein-propidium iodide (GFP-PI) based assay for flow cytometric measurement of bacterial viability. Cytometry A. 60:165-172. [DOI] [PubMed] [Google Scholar]
- 32.Marteau, P. R. 2002. Probiotics in clinical conditions. Clin. Rev. Allergy Immunol. 22:255-273. [DOI] [PubMed] [Google Scholar]
- 33.McCracken, A., M. S. Turner, P. Giffard, L. M. Hafner, and P. Timms. 2000. Analysis of promoter sequences from Lactobacillus and Lactococcus and their activity in several Lactobacillus species. Arch. Microbiol. 173:383-389. [DOI] [PubMed] [Google Scholar]
- 34.Mercenier, A., P. H. Pouwels, and B. M. Chassy. 1994. Genetic engineering of lactobacilli, leuconostocs and Streptococcus thermophilus, p. 252-293. In M. J. Gasson and W. M. de Vos (ed.), Genetics and biotechnology of lactic acid bacteria. Chapmann & Hall, London, United Kingdom.
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. de Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogushi, K., A. Wada, T. Niidome, N. Mori, K. Oishi, T. Nagatake, A. Takahashi, H. Asakura, S. Makino, H. Hojo, Y. Nakahara, M. Ohsaki, T. Hatakeyama, H. Aoyagi, H. Kurazono, J. Moss, and T. Hirayama. 2001. Salmonella enteritidis FliC (flagella filament protein) induces human beta-defensin-2 mRNA production by Caco-2 cells. J. Biol. Chem. 276:30521-30526. [DOI] [PubMed] [Google Scholar]
- 38.O'Neil, D. A. 2003. Regulation of expression of beta-defensins: endogenous enteric peptide antibiotics. Mol. Immunol. 40:445-450. [DOI] [PubMed] [Google Scholar]
- 39.O'Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 40.O'Sullivan, G. C., P. Kelly, S. O'Halloran, C. Collins, J. K. Collins, C. Dunne, and F. Shanahan. 2005. Probiotics: an emerging therapy. Curr. Pharm. Des. 11:3-10. [DOI] [PubMed] [Google Scholar]
- 41.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 42.Pavan, S., P. Hols, J. Delcour, M. C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Arellano, I., and G. Perez-Martinez. 2003. Optimization of the green fluorescent protein (GFP) expression from a lactose-inducible promoter in Lactobacillus casei. FEMS Microbiol. Lett. 222:123-127. [DOI] [PubMed] [Google Scholar]
- 44.Reid, G. 2005. The importance of guidelines in the development and application of probiotics. Curr. Pharm. Des. 11:11-16. [DOI] [PubMed] [Google Scholar]
- 45.Rinne, M., M. Kalliomaki, H. Arvilommi, S. Salminen, and E. Isolauri. 2005. Effect of probiotics and breastfeeding on the Bifidobacterium and Lactobacillus/Enterococcus microbiota and humoral immune responses. J. Pediatr. 147:186-191. [DOI] [PubMed] [Google Scholar]
- 46.Ross, R. P., C. Desmond, G. F. Fitzgerald, and C. Stanton. 2005. Overcoming the technological hurdles in the development of probiotic foods. J. Appl. Microbiol. 98:1410-1417. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxelin, M. 1997. Lactobacillus GG—a human probiotic strain with thorough clinical documentation. Food Rev. Int. 13:293-313. [Google Scholar]
- 50.Schneider, J. J., A. Unholzer, M. Schaller, M. Schafer-Korting, and H. C. Korting. 2005. Human defensins. J. Mol. Med. 83:587-595. [DOI] [PubMed] [Google Scholar]
- 51.Seegers, J. F. 2002. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20:508-515. [DOI] [PubMed] [Google Scholar]
- 52.Serror, P., T. Sasaki, S. D. Ehrlich, and E. Maguin. 2002. Electrotransformation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis with various plasmids. Appl. Environ. Microbiol. 68:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sherwood, L., and M. D. Gorbach. 1996. The discovery of Lactobacillus GG. Nutr. Today 31:2S-4S. [Google Scholar]
- 54.Trevors, J. T., B. M. Chassy, W. J. Dower, and H. P. Blaschek. 1992. Electrotransformation of bacteria by plasmid DNA, p. 265-290. In D. C. Chang, B. M. Chassy, J. A. Saunders, and A. E. Sowers (ed.), Guide to electroporation and electrofusion. Academic Press Inc., San Diego, CA.
- 55.Valdivia, R. H., A. E. Hromockyj, D. Monack, L. Ramakrishnan, and S. Falkow. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47-52. [DOI] [PubMed] [Google Scholar]
- 56.Varmanen, P., T. Rantanen, A. Palva, and S. Tynkkynen. 1998. Cloning and characterization of a prolinase gene (pepR) from Lactobacillus rhamnosus. Appl. Environ. Microbiol. 64:1831-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei, M., C. Rush, J. M. Norman, L. M. Hafner, R. J. Epping, and P. Timms. 1995. An improved method for the transformation of Lactobacillus strains using electroporation. J. Microbiol. Methods 21:97-109. [Google Scholar]
- 58.Zhu, G.-Y., S. Dobbelaere, and J. Vanderleyden. 2002. Use green fluorescent protein to visualize rice root colonization by Azospirillum irakense and A. brasilense. Funct. Plant Biol. 29:1-7. [DOI] [PubMed] [Google Scholar]