Abstract
The environmental distribution and phylogeny of “Korarchaeota,” a proposed ancient archaeal division, was investigated by using the 16S rRNA gene framework. Korarchaeota-specific primers were designed based on previously published sequences and used to screen a variety of environments. Korarchaeota 16S rRNA genes were amplified exclusively from high temperature Yellowstone National Park hot springs and a 9°N East Pacific Rise deep-sea hydrothermal vent. Phylogenetic analyses of these and all available sequences suggest that Korarchaeota exhibit a high level of endemicity.
In 1994, 16S rRNA gene sequences PCR amplified from Yellowstone National Park hot spring Obsidian Pool were reported (3). These sequences were hypothesized to belong to an ancient division, the “Korarchaeota,” that diverged from the Archaea lineage before the separation of Crenarchaeota and Euryarchaeota (2). Other deeply branching clades have since been proposed, including the Nanoarchaeota and “Ancient Archaeal Group,” although phylogenetic support for any group having a basal position is not strong (2, 4, 29). What little is known about Korarchaeota comes from detection of their 16S rRNA genes during environmental diversity surveys that used “universal” Archaea primers. Nineteen such sequences which were believed to be in the Korarchaeota clade have been published (3, 12, 15, 17, 19, 22-24, 28, 31, 32). These genes were PCR amplified from fluid, sediment, sulfide chimneys, or microbial mats of high-temperature hot springs or submarine vents. Although attempts to grow Korarchaeota in pure culture have not been successful, Korarchaeota cells of the pJP27 phylotype originally detected in Obsidian Pool have now been highly enriched, and their (meta)genome is being sequenced (K. O. Stetter, J. G. Elkins, M. Keller, and the JGI-DOE [http://www.jgi.doe.gov/sequencing/why/CSP2006/korarchaeota.html]).
To further assess their diversity and distribution, we designed Korarchaeota-specific primers based on published sequences and used them to PCR amplify 16S rRNA genes from deep-sea hydrothermal vents, various Yellowstone National Park hydrothermal features, and a number of nonhydrothermal environments. PCR amplification products were cloned and sequenced, and the phylogeny of these 16S rRNA genes, as well as those previously named Korarchaeota or sharing identity to those previously named as such, was inferred. The relationships among Korarchaeota were compared to the properties of the environments from which they were amplified to gain insights into basic physiological properties of this enigmatic group.
Diverse deep-sea hydrothermal vent niches were screened for Korarchaeota. Samples, collected from the 9°N East Pacific Rise (EPR) 2,500-m-deep hydrothermal field in December 2002 using DSV Alvin on R/V Atlantis cruise AT-07, Leg 26, were from the following environments: the dark red surface of a black smoker sulfide chimney through which 370°C fluid flowed (P-vent; Dive 3851), a basalt settlement piece (FRIE 16.2) left by an active sulfide chimney for 1 week (Tica vent; Dives 3843 and 3849), and a 0.45-μm-pore-size, 142-mm-diameter mixed cellulose ester filter (Millipore) and a 1-μm-pore-size, 142-mm-diameter Petex prefilter (Sefar) through which 200 liters of water 1 m peripheral to a hydrothermal vent was filtered by using a McLane Research Laboratories, Inc., pump in situ (Tica vent). On board ship, sulfide chimney surfaces were scraped into 2-ml freezer tubes and dropped in liquid nitrogen, basalt pieces were placed in 50-ml polypropylene tubes, and filters were wrapped in aluminum foil. Samples were stored on board at −70°C, transported on dry ice, and then stored at −80°C until use. To extract DNA from sulfide chimney samples, the Bio 101 FastDNA SPIN kit for soil was used according to the manufacturer's instructions. DNA was extracted from basalt and filters (in 50-ml polypropylene tubes) by the addition of sucrose-lysis buffer (10 ml), lysozyme, proteinase K, sodium dodecyl sulfate, and phenol-chloroform according to the method of Gordon and Giovannoni (11) and then ethanol precipitated (20) and resuspended in water.
A number of hydrothermal features in Yellowstone National Park were also screened for Korarchaeota. Samples were collected from 19 sites in summer 2003 and 17 sites in summer 2004 as part of a microbial inventory being conducted of Yellowstone thermal features. Sediment, sinter, mud, and microbial mats were mixed 1:1 in sucrose-lysis buffer, and hydrothermal fluid was mixed 2:1 with RNAlater (Ambion) on site and then stored at −80°C as soon as possible. DNA was extracted from sediment, sinter, and mud by using a Bio 101 FastDNA SPIN kit for soil. For extracting microbial mat DNA, 50 μl of thawed sample was mixed with 100 μl of CTAB buffer (1% cetyltrimethylammonium bromide, 0.75 M NaCl, 50 mM Tris [pH 8], 10 mM EDTA) and then processed like the deep-sea hydrothermal vent basalt and filters. For fluid samples, 2-ml portions were centrifuged 10 min at 16,000 × g, all but 50 μl of supernatant was removed, and then the samples were processed as described for microbial mat samples.
Since Korarchaeota-specific primers had not previously been used to screen natural samples, a range of readily available, primarily nonhydrothermal environmental samples were also collected and tested for Korarchaeota 16S rRNA genes. These included compost (with temperatures measured up to 60°C), plant leaves, flower petals, tree bark, human saliva, biofilms on computer mice, a mosquito, soil (5- to 10-cm-deep Harvard Forest, Petersham, MA, and surface soil of Cambridge, MA), sand (Harvard University campus), river water and sediment (Charles River, Cambridge, MA), lake water (oxic-anoxic interface of Lake Mishawum, Woburn, MA), seawater (coast of Nahant, MA), 32°C spring water (Chena, AK), 57°C tap water (Cambridge, MA), and 60 and 90°C steam exhaust biofilms (campus of Massachusetts Institute of Technology, Cambridge, MA). Except the DNA of sediment, soil, and sand that was extracted by using a Bio 101 FastDNA SPIN kit for soil, DNA was extracted as for Yellowstone microbial mat samples.
For primer design, fifteen sequences of variable size identified as belonging to the Korarchaeota (3, 12, 15, 17, 19, 22, 23, 31, 32) were manually aligned in BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) with a number of Euryarchaeota, Crenarchaeota, and Nanoarchaeota sequences. Two “Korarchaeota” sequences (FZ2bA214 and pOWA133 [22, 31]) that shared very little identity with the other 13 were excluded for the design of the Korarchaeota-specific primers Kor236F (GCT GAG GCC CCA GGR TGG GAC CG) and Kor1236R (CAT CCC GCT GTC CCG CCC ATT GC; numbering corresponds to Escherichia coli positions). Kor236F had a one-half mismatch (degenerate position) with the nine Korarchaeota sequences containing those regions, while containing at least five and a half mismatches with all of the other sequences in GenBank (determined by using Ribosomal Database Project-II release 8.1 program Probe Match [6]). Kor1236R had no mismatches with the 11 Korarchaeota sequences containing those regions while containing at least three mismatches with all other sequences in GenBank. For every sample that was screened by PCR, Kor236F and Kor1236R were used together and individually with Univ1492R (Archaea/Bacteria universal primer [10]) or Arc26F (Archaea universal primer, TCC GGT TGA TCC TGC CGG A), respectively. Univ1492R has no mismatches to the sequenced ends of Korarchaeota 16S rRNA genes (unpublished data), and Arc26F has unknown identity to Korarchaeota 16S rRNA genes, since the 5′ end of a Korarchaeota 16S rRNA gene has not yet been sequenced.
Control reactions were performed to determine the optimal annealing temperatures of the Korarchaeota 16S rRNA gene screening PCR. These were the lowest temperatures at which marine group I Crenarchaeota (TFA4) and marine group II Euryarchaeota (TFA3) 16S rRNA gene negative controls did not amplify enough DNA to be visualized on an ethidium bromide-stained agarose gel. Clones TFA3 and TFA4 were created from PCR products that were Arc26F-Univ1492R PCR amplified from DNA of the deep-sea hydrothermal vent water described above.
When we screened for Korarchaeota 16S rRNA genes, positive control reactions (45°C annealing temperature) with universal Archaea (Arc26F-Univ1492R) and/or Bacteria (Bac27F [26]-Univ1492R) primers yielded at least one correctly sized band for all samples, suggesting that samples that did not yield products in Korarchaeota-specific PCR did not fail due to inhibition or lack of DNA. As additional positive controls, KorY38 plasmid (described below) was added to, and DNA was successfully amplified from, replicates of the Kor236F-Kor1236R reaction of nonhydrothermal samples and a replicate of each Korarchaeota-specific PCR for Yellowstone 2004 samples.
PCR mixtures contained 0.625 U of Taq, 1× buffer (QIAGEN), 2 mM deoxynucleoside triphosphates, 4 mM MgCl2, 200 nM concentrations of each primer, and 0.04% bovine serum albumin. After an initial denaturation of 94°C for 2 min, there were 35 cycles of 94°C for 45 s, 55°C (Kor236F-Univ1492R) or 62°C (Arc26F-Kor1236R and Kor236F-Kor1236R) for 45 s, and 72°C for 2 min, followed by 72°C for 2 min. A portion of each Korarchaeota-specific PCR-amplified sample was visualized on a 1% agarose gel. When a band of the appropriate size was observed, DNA in the remaining reaction volumes was purified by using a PCR purification kit (QIAGEN), cloned into pCR2.1 (Invitrogen) or pDrive (QIAGEN) and then transformed into TOP10 E. coli (20). The resulting colonies were screened by PCR with the primers M13F and M13R under the same conditions as for the Kor236F-Univ1492R reactions to identify clones that contained inserts of the appropriate size. At least one clone per reaction was sequenced by using the primer M13F with an ABI BigDye terminator v3.1 cycle sequencing kit on an ABI Prism 3100 genetic analyzer. Initial classification of partial sequences (typically >500 nt) was performed by using BLAST (1). If a Korarchaeota match was indicated, the Arc26F-Univ1492R amplicon from that sample was also cloned, and at least one representative was sequenced in an effort to obtain longer Korarchaeota 16S rRNA gene sequences. Full sequencing of the most complete Korarchaeota 16S rRNA gene clone from every site was accomplished with M13R. Only Korarchaeota partial sequences ≤99% identical to others found at a site were also fully sequenced, since we used a large number of PCR cycles to detect low numbers of Korarchaeota, at the expense of a low mutation rate that is optimal for studying microdiversity (13).
To screen for additional Korarchaeota genes in Korarchaeota-positive samples and for undetected Korarchaeota genes in negative samples with correctly sized amplicons, additional screening was conducted. First, another round of PCR and cloning was performed on the original sample; however, the PCR for this round was not as successful and generally produced fewer clones. When fewer or greater than 10 clones were generated, they were screened by sequencing or colony hybridizations, respectively. For the latter, PCR with the primers M13F and M13R was performed on four random clones to confirm that they contained inserts. All colonies were then lysed, and their DNA was fixed to Hybond N+ membranes (http://www.fhcrc.org/labs/roberts/Protocols/ColonyHybridization.html). The DIG High-Prime DNA labeling and detection kit (Roche) was used as directed, with one 70°C and two 80°C washes; labeled probe was created from DNA Kor236F-Kor1236R PCR amplified from the KorY42A plasmid (described below). The Korarchaeota clones Kor9NEPR, KorY03, and KorY38 (described below) were used as positive controls, and Euryarchaeota (TFA3) and Crenarchaeota (TFA4) 16S rRNA gene clones were used as negative controls. Although Korarchaeota could not be proven definitively absent from samples in which their 16S rRNA genes were not amplified, the positive controls, multiple primer combinations, high number of PCR cycles, and second round of screening decreased the possibility that Korarchaeota were undiscovered due to technical problems.
All new and previously identified putative Korarchaeota 16S rRNA gene sequences were analyzed with the RDPII v8.1 program Chimera Check (6). Two recently published Korarchaeota sequences, WB3D011 and OPPE037 (24), were found to be chimeras and excluded. Phylogenetic analyses included 31 possible Korarchaeota sequences. Ten of these were identified in the present study. Nineteen were previously published as Korarchaeota, including the fifteen available when the primers were designed. Two sequences, pUWA9 and 20a-1, were identified in GenBank by using BLAST. pUWA9 was published as a Thermoplasmata 16S rRNA gene (31), and 20a-1 is an unpublished sequence obtained from Aegean Sea sediment (accession no. AJ299148). Putative Korarchaeota sequences were compiled and aligned in ARB (14), with 43 16S rRNA gene sequences from representatives of major archaeal groups included to test the monophyly of Korarchaeota 16S rRNA genes. Escherichia coli (Bacteria) and Homo sapiens (Eukarya) rRNA genes were used as outgroups. After further refinement of the alignment in BioEdit considering the predicted secondary structure of small subunit rRNA (5), alignments of various lengths (651 nt, 119 to 770, n = 20; 738 nt, 237 to 975, n = 22; 946 nt, 29 to 975, n = 17; 1,048 nt, 342 to 1,390, n = 15; and 1,361 nt, 29 to 1,390, n = 12) were exported to PAUP (27). Trees were constructed and bootstrap resampled (500 replicates) with maximum parsimony by 500 heuristic random-addition sequence searches using the tree bisection reconnection branch-swapping option and treating gaps as new states.
To infer the relationships among all sequences within the Korarchaeota, the phylogram from the analysis of the 946-nt alignment (for which the most complete and phylogenetically representative Korarchaeota sequences were available) was imported into ARB, where individual partial sequences were added to the tree by the parsimony add option, without changing the tree topology. The ARB-determined topological placement and length of added partial sequence branches agreed with those in individual trees generated from alignments specific to each partial sequence.
Ten putative Korarchaeota 16S rRNA genes were amplified from 9 of the 63 diverse samples screened, all from sites at temperatures of ≥55°C, with a single sequence identified from DNA of the active sulfide chimney (Kor9NEPR) and nine sequences identified from 8 of the 41 Yellowstone samples examined (Fig. 1). No chimeras were identified among these sequences. Indels and nucleotide changes are nonconsecutive in all sequences, except in a six-base loop region of KorY41B's Helix 44 (ARB helix numbering) that contains two insertions and three changes compared to its most closely related sequences. KorY38 (amplified with Arc26F and Univ1492R) also contains two and one additional mismatches with primers Kor236F and Kor1236R, respectively, and 20a-1 has one additional mismatch with Kor236F. Therefore, additional Korarchaeota-specific primers may be warranted in the future.
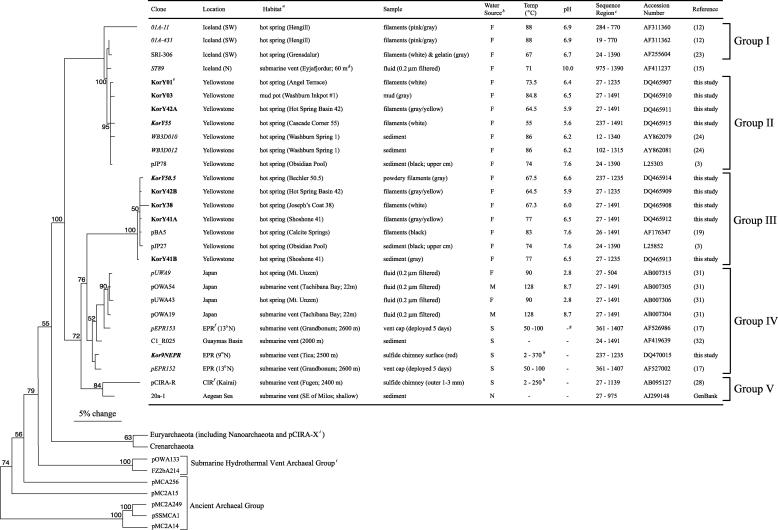
FIG. 1.
Phylogram of Korarchaeota based on 16S rRNA gene sequences. The samples, sites, and chemical and physical characteristics of the environments from which they were PCR amplified, length of the sequences, accession numbers, and references are also listed. Major phylogenetic groupings are bracketed on the right. The tree topology and bootstrap values were generated by parsimony analysis of the 946-nt alignment. Partial sequences were added by using the ARB parsimony tool (italicized clone names) and thus have no bootstrap values. The sequences identified in the present study are shown in boldface. Escherichia coli, Homo sapiens, and 46 diverse Archaea small subunit rRNA genes were used as outgroups (shown with branches shortened and Euryarchaeota and Crenarchaeota branches collapsed). The scale bar = 5% change in nucleotide sequence. Superscript letters: a, hot springs and mudpots are terrestrial; b, F = freshwater, S = seawater, M = mix, N = not reported; c, numbering is based on E. coli; d, depth below seawater surface (where not noted = surface); e, the Korarchaeota clone naming terminology is “Kor” + Y (Yellowstone) + inventory sample number; f, EPR = East Pacific Rise, CIR = Central Indian Ridge; g, a dash means not reported; h, sulfide chimney separated cold ambient seawater and hot hydrothermal fluid; i, previously referred to as Korarchaeota.
Phylogenetic analyses indicated that the proposed Korarchaeota sequences pOWA133 (31), FZ2bA214 (22), and pCIRA-X (28), though first published as such, fall outside the group as originally defined (2). These sequences diverge greatly from others within the two largest regions (9-nt stems) conserved among all Korarchaeota 16S rRNA gene sequences: Helix 6, where only 4 of 9 nt matched, and Helix 11, where each shared ≤1 of 9 nt. The original analyses of pOWA133 and pCIRA-X using neighbor joining had placed these sequences in the Korarchaeota with only weak support, while FZ2bA214 was named a Korarchaeota because it clustered with pOWA133 (22). Indeed, Takai et al. questioned their korarchaeotal identification of pOWA133 and have since considered it a separate group (30).
The remaining 28 sequences formed a coherent clade designated here as Korarchaeota with 100% bootstrap support in all trees. We define these as Korarchaeota because of their strong phylogenetic support as a group (including the sequences originally named Korarchaeota) and conservation of the two signature sequences described above. Although support for the hypothesis of a basal evolutionary position for Korarchaeota remains weak, when bootstrap support was at all present (≥50%), Korarchaeota branched sister to the clade of Crenarchaeota and Euryarchaeota (946-nt tree), with only the pOWA133/FZ2bA214 clade (946-nt tree) or Ancient Archaeal Group sequences (651-, 946-, and 1,361-nt trees) ever branching deeper (Fig. 1).
Analysis of the 28 Korarchaeota 16S rRNA gene sequences indicate five subgroups exist based on various degrees of phylogenetic support and geographic location (Fig. 1). The overall topology of trees constructed using other length sequence alignments agreed with the 946-nt tree, except the 738-nt tree in which groups I and II branched with group III Korarchaeota, and groups IV and V were not resolved.
Group I sequences were all identified from Iceland samples. Sequence 01A-11 was placed outside the clade containing other group I and II sequences. However, this sequence is provisionally included in group I because it shares five (nonconsecutive) nucleotides exclusively found in other group I sequences and no nucleotides in common with just group II sequences. KorY01, -03, -42A, and -55, all of which were amplified from Yellowstone National Park samples, are closely related to each other and other group II sequences (99.0 to 99.8% identity). The sequence ST89 was too short (440 nt) to resolve whether it is group I or group II. Group III, also comprised of sequences amplified from Yellowstone National Park samples, included KorY38, -41A, -42B, and -50.5, which are closely related to one other and to two other previously described sequences (98.6 to 99.7% identity). KorY41B, also amplified from a Yellowstone hot spring, was basal in group III. Group IV includes sequences amplified from samples collected in or near the Pacific Ocean. Four sequences, recovered from Japanese hot springs and submarine vents, form a clade with relatively strong support (bootstrap 90; n = 3 for analysis). Support for other sequences clustering as a group with the Japanese sequences was strong when longer sequence lengths were used in analyses (1,048 nt, n = 6, bootstrap 85; 1,361 nt, n = 4, bootstrap 97). Since all of these sequences also contain an AT in Helix 24 that is not present in other Korarchaeota, they are provisionally placed together in group IV. Group V currently contains only two sequences (bootstrap 84), amplified from shallow and deep-sea hydrothermal vent samples from the Mediterranean Sea and Indian Ocean, respectively.
Korarchaeota, currently found only in high-temperature hydrothermal settings, appear to be influenced by geographic isolation, visible even from analysis of their very conserved 16S rRNA gene sequences. Some microbial populations inhabiting distantly separated extreme environments have been shown to exhibit high levels of endemism consistent with geographic isolation (18, 25, 35). However, to discern possible biogeographic patterns among Korarchaeota, additional surveys for their 16S rRNA genes and other markers will be needed to provide higher resolution.
High-temperature hydrothermal settings (50 to 128°C), including sulfide chimney walls and caps, fluid, mud, sediment, and microbial mats, are still the only places Korarchaeota 16S rRNA genes have been detected (Fig. 1). To date, Korarchaeota 16S rRNA genes have not been retrieved in microbial diversity surveys of chemically, physically, and proximally similar but predominantly nonhydrothermal habitats, such as sulfide-rich cold springs, hydrocarbon seeps, anaerobic methane oxidation-produced carbonate chimneys, deep-seawater or sediment, subseafloor samples, or off-axis deep-sea hydrothermal vents (see, for example, references 7, 9, 16, 21, 33, and 34, where the primers should have amplified known Korarchaeota 16S rRNA genes equally well). The apparent thermophilic preference of Korarchaeota is supported by the high G+C content of their rRNA (mean = 65.3% G+C [standard deviation = 1.2]), a trait that has been shown to correlate with high optimal growth temperature (8). In contrast, the sample type, water source (marine and freshwater), and pH (2.8 to 10.0) of environments found to contain Korarchaeota 16S rRNA genes have been variable (Fig. 1). As microbial communities are characterized in ever more detail, including metagenomic studies of as-yet-uncultured Korarchaeota, the environmental parameters which determine Korarchaeota distribution will be elucidated.
Over a decade since their discovery, almost nothing had been revealed about representatives of the Korarchaeota. Insights into their environmental distribution and phylogeny provide a foundation for continuing the hunt for these enigmatic organisms, their optimal conditions for growth in culture, and for future genetic and metabolic studies.
Nucleotide sequence accession numbers.
The sequences reported in the present study have been deposited in GenBank under accession numbers DQ465907-20 and DQ470015.
Acknowledgments
We thank Anna-Louise Reysenbach for conception of the Yellowstone microbial inventory; Kendra Mitchell, Olan Jackson-Weaver, Mike Bobb, Sara Caldwell, George Silva, Chuck Fisher, the crew of the R/V Atlantis, and the Alvin group for assistance in collection of hydrothermal samples; Meredith Fisher, Evan Lau, Tara Harmer, and Chris Luke for selected samples from cool environments; Lief Fenno for collection and analysis of the Chena, Alaska, spring water; and Irene Newton for help with the colony hybridizations.
This study was supported by an NIH Genetics Training Grant graduate fellowship to T.A.A., grants from NASA (NAG5-10906) and NOAA National Undersea Research Center for the West Coast and Polar Regions (03-0092) to C.M.C., and NSF Biotic Surveys and Inventories (0206773) to C.D.T.-V.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barns, S. M., C. F. Delwiche, J. D. Palmer, and N. R. Pace. 1996. Perspectives on archaeal diversity, thermophily, and monophyly from environmental rRNA sequences. Proc. Natl. Acad. Sci. USA 93:9188-9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brochier, C., S. Gribaldo, Y. Zivanovic, F. Confalonieri, and P. Forterre. 2005. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 6:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowen, J. P., S. J. Giovannoni, F. Kenig, H. P. Johnson, D. Butterfield, M. S. Rappe, M. Hutnak, and P. Lam. 2003. Fluids from aging ocean crust that support microbial life. Science 299:120-123. [DOI] [PubMed] [Google Scholar]
- 8.Dalgaard, J. Z., and R. A. Garrett. 1993. Archaeal hyperthermophile genes. In M. Kates, D. J. Kushner, and A. T. Matheson (ed.), The biochemistry of Archaea (Archaebacteria). Elsevier, Amsterdam, The Netherlands.
- 9.Elshahed, M. S., F. Z. Najar, B. A. Roe, A. Oren, T. A. Dewers, and L. R. Krumholz. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl. Environ. Microbiol. 70:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for Campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, D. A., and S. J. Giovannoni. 1996. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl. Environ. Microbiol. 62:1171-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjorleifsdottir, S., S. Skirnisdottir, G. O. Hreggvidsson, O. Holst, and J. K. Kristjansson. 2001. Species composition of cultivated and noncultivated bacteria from short filaments in an Icelandic hot spring at 88°C. Microb. Ecol. 42:117-125. [DOI] [PubMed] [Google Scholar]
- 13.Klepac-Ceraj, V., M. Bahr, B. C. Crump, A. P. Teske, J. E. Hobbie, and M. F. Polz. 2004. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6:686-698. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marteinsson, V. T., S. Hauksdottir, C. F. V. Hobel, H. Kristmannsdottir, G. O. Hreggvidsson, and J. K. Kristjansson. 2001. Phylogenetic diversity analysis of subterranean hot springs in Iceland. Appl. Environ. Microbiol. 67:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 17.Nercessian, O., A. L. Reysenbach, D. Prieur, and C. Jeanthon. 2003. Archaeal diversity associated with in situ samplers deployed on hydrothermal vents on the East Pacific Rise (13°N). Environ. Microbiol. 5:492-502. [DOI] [PubMed] [Google Scholar]
- 18.Papke, R. T., N. B. Ramsing, M. M. Bateson, and D. M. Ward. 2003. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 5:650-659. [DOI] [PubMed] [Google Scholar]
- 19.Reysenbach, A. L., M. Ehringer, and K. Hershberger. 2000. Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4:61-67. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Schrenk, M. O., D. S. Kelley, S. A. Bolton, and J. A. Baross. 2004. Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ. Microbiol. 6:1086-1095. [DOI] [PubMed] [Google Scholar]
- 22.Schrenk, M. O., D. S. Kelley, J. R. Delaney, and J. A. Baross. 2003. Incidence and diversity of microorganisms within the walls of an active deep-sea sulfide chimney. Appl. Environ. Microbiol. 69:3580-3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjorleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki, M., M. S. Rappe, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 28.Takai, K., T. Gamo, U. Tsunogai, H. Hirayama, K. H. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 29.Takai, K., and K. Horikoshi. 1999. Genetic diversity of Archaea in deep-sea hydrothermal environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takai, K., D. P. Moser, M. DeFlaun, T. C. Onstott, and J. K. Fredrickson. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67:5750-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takai, K., and Y. Sako. 1999. A molecular view of archaeal diversity in marine and terrestrial hot water environments. FEMS Microbiol. Ecol. 28:177-188. [Google Scholar]
- 32.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tourova, T. P., T. V. Kolganova, B. B. Kuznetsov, and N. V. Pimenov. 2002. Phylogenetic diversity of the archaeal component in microbial mats on coral-like structures associated with methane seeps in the Black Sea. Microbiology 71:196-201. [PubMed] [Google Scholar]
- 34.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitaker, R. J., D. W. Grogan, and J. W. Taylor. 2003. Geographic barriers isolate endemic populations of hyperthermophilic Archaea. Science 301:976-978. [DOI] [PubMed] [Google Scholar]