Abstract
A tool kit of vectors was designed to manipulate and express genes from a wide range of gram-negative species by using in vivo recombination. Saccharomyces cerevisiae can use its native recombination proteins to combine several amplicons in a single transformation step with high efficiency. We show that this technology is particularly useful for vector design. Shuttle, suicide, and expression vectors useful in a diverse group of bacteria are described and utilized. This report describes the use of these vectors to mutate clpX and clpP of the opportunistic pathogen Pseudomonas aeruginosa and to explore their roles in biofilm formation and surface motility. Complementation of the rhamnolipid biosynthetic gene rhlB is also described. Expression vectors are used for controlled expression of genes in two pseudomonad species. To demonstrate the facility of building complicated constructs with this technique, the recombination of four PCR-generated amplicons in a single step at >80% efficiency into one of these vectors is shown. These tools can be used for genetic studies of pseudomonads and many other gram-negative bacteria.
Cloning using Saccharomyces cerevisiae homologous recombination is a powerful technique in which PCR primers with stretches of homologous DNA are used to target recombination of an amplicon with a vector (9, 21, 25, 27). Cloning of an amplicon with yeast recombination into an intact vector can be done without restriction enzymes, but this requires the use of a selectable or counterselectable marker. The formation of a gap or double-stranded break in the vector by restriction enzyme digestion allows selective cloning of unmarked amplicons (29). An amplicon can seamlessly replace any part of the DNA in a vector with no need for enzyme sites at the junctions of the recombined pieces of DNA, as long as the yeast replication machinery is not replaced and the ends of the amplicons (or other DNA with homologous ends) to be added to a vector are on either side of the digest site (gap) in the vector. Therefore, multiple unmarked pieces of DNA can efficiently be assembled together in a single step using this method. In addition to fusions, directed mutations, restriction sites, and other changes can be designed into the primers. This technique is particularly useful for building vectors and complex constructs.
The basic concept of cloning with gap repair is as follows. A vector able to replicate in yeast with a selective marker, such as URA3, is digested (gapped) with a restriction enzyme. Primers that amplify the region of DNA that one wants to insert into the vector are designed, containing sequences of homology to the yeast-replicating plasmid (30 to 40 bp in length). One or more targeted amplicons and the gapped vector are simultaneously introduced into yeast cells. A subset of these cells recombines the amplicon(s) and vector via Saccharomyces cerevisiae native recombination enzymes, generating stable circularized plasmids. Linearized or gapped vectors are not stable in yeast, while vectors that circularize through homologous recombination are stable. Cells containing circularized plasmids can be selected for by being plated on medium selective for the vector backbone (e.g., deficient in uracil).
We present a tool kit of vectors designed to manipulate and express genes from a wide range of gram-negative species by using S. cerevisiae in vivo recombination.
MATERIALS AND METHODS
Strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strain DH5α was used as a host strain for plasmid construction and propagation, while S17-1 was used for conjugations with Pseudomonas species (37). Pseudomonas aeruginosa strain PA14 and Pseudomonas fluorescens strain WCS365 were used for phenotypic experiments (13, 31). S. cerevisiae strains DC49-7.1c and InvSc1 (Invitrogen Corporation) were used interchangeably for the in vivo homologous recombination used to modify plasmids (1). E. coli and Pseudomonas strains were grown with LB medium, S. cerevisiae was grown with yeast extract-peptone-dextrose (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose), and selections were performed with synthetic defined agar-uracil (Qbiogene 4813-065). E. coli was transformed by electroporation, whereas Pseudomonas was transformed by conjugation or electroporation (5). Ampicillin at 75 μg/ml and 150 μg/ml was used for plasmid selection and propagation, respectively, for E. coli or 500 to 1,000 μg/ml for P. aeruginosa. Gentamicin was used at 10 μg/ml for E. coli and 50 to 100 μg/ml for Pseudomonas strains. Kanamycin was used at 50 μg/ml for E. coli and 150 to 250 μg/ml for P. fluorescens. Tetracycline was used at 15 μg/ml for E. coli and 75 to 150 μg/ml for Pseudomonas species. Nalidixic acid was used at 20 μg/ml for selection against E. coli following E. coli-Pseudomonas sp. conjugations.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169(φ80dlacZΔM15) hsdR17 thi-1 relA1 recA1 | Life Technologies |
| S17-1 | thi pro hsdR hsdM+ ΔrecA RP4-2::TcMu-Km::Tn | 37 |
| P. aeruginosa | ||
| PA14 | Wild type, clinical isolate | 31 |
| RSS255 | PA14 clpX::pRMQS84 | This study |
| EP1 | PA14 clpP::GFPuvr | This study |
| SMC2758 | PA14 rhlB::kanMX4 | 4 |
| P. fluorescens | ||
| WCS365 | Wild type, environmental isolate | 13 |
| SMC2836 | WCS365 ΔlapD | 17 |
| S. cerevisiae | ||
| DC49-7.1c | MATaleu2-3,112 trp1-289 ura3-52 arg4-Δ57,RV | 1 |
| InvSc1 | MATa/MATα leu2/leu2 trp1-289/trp1-289 ura3-52/ura3-52 his3-Δ1/his3-Δ1 | Invitrogen |
| Plasmids | ||
| pBAD18 | E. coli expression vector with PBAD and araC | 15 |
| Mini-CTX | CTX phage neutral site integration vector | 19 |
| pEPSA5 | p15a containing PT5X expression vector | 11 |
| pEX18-Gm | Gram-negative allelic replacement vector | 18 |
| pGFPmut3 | E. coli vector with mutated GFP allele | 6 |
| pJB785TTKm1 | RK2 broad-host-range reporter construct | 34 |
| pRS415 | S. cerevisiae-E. coli shuttle vector | 36 |
| pRS416 | S. cerevisiae-E. coli shuttle vector | 36 |
| pRS426 | S. cerevisiae-E. coli shuttle vector | 36 |
| pUCP20 | Pseudomonas-E. coli shuttle vector | 39 |
| pUG6 | loxP-kanMX-loxP construct | 14 |
| pYC2-CT | S. cerevisiae-E. coli shuttle vector | Invitrogen |
Plasmid construction.
All vectors, with the exception of pMQ87, pMQ89, pMQ57, pMQ65, and pMQ90, were built using yeast gap repair as follows. A yeast-replicating vector to be modified was cut with a restriction enzyme(s). Primers were made to amplify DNA to be joined with the cut vector, including ∼40-bp tails that are homologous to the cut vector on each side of the restriction enzyme cut site (up to 2,000 bp from the cut site). A small sample of the amplicon was run out on a gel to ensure that the desired band was in molar excess to any other bands, and if it was not, the desired band was gel purified. Overnight cultures of S. cerevisiae were cotransformed by the addition of 20 to 200 ng of the cut vector (directly from the restriction enzyme mix) and 50 to 500 ng of the amplicon. These modified vectors all contained a URA3 gene capable of complementing the ura3-52 mutation of the strains used. Transformed cells were either tested individually for the desired construct with colony PCR or pooled, and a preparation of plasmid DNA was made and moved into E. coli cells for screening.
Construction of pMQ30.
Plasmid pMQ30 is derived from pEX18-Gm, a vector used for two-step gene replacement experiments with Pseudomonas species (18), onto which were added elements needed for replication and selection in S. cerevisiae. pMQ30 was made by homologous recombination in yeast (see below for protocol); a 1,633-bp PCR amplicon containing the selectable and counterselectable yeast gene URA3 and the CEN6 and ARSH4 elements was amplified from pYC2-CT (Invitrogen) and recombined with pEX18-Gm that had been linearized with AatII. Both primers had regions homologous to pYC2-CT and ∼40-bp tails, identical to regions of DNA flanking the AatII site of pEX18-Gm. See Table S1 in the supplemental material for primer sequences (primers Ex18-Up and Ex18-Dn).
S. cerevisiae strain DC49-7.1c (ura3-52) was cotransformed with both linear pieces of DNA, and recombinants were selected on uracil dropout medium. Because the CEN6-URA3 amplicon is linear and without telomeres, it is not stably maintained in the yeast cell, but cells in which the linear CEN6-URA3 amplicon and the digested pEX18-Gm had recombined to form a circular stable replicon were able to grow on selective medium. Plasmids were liberated from uracil-prototrophic yeast and electroporated into E. coli, and gentamicin-resistant transformants were selected. Candidates were isolated and screened by restriction analysis and phenotype (uracil prototroph in yeast, gentamicin resistance, and sucrose sensitivity [data not shown]). All of the candidates were verified as correct by restriction analysis, PCR, and phenotypic assays.
Construction of pMQ87 and pMQ89.
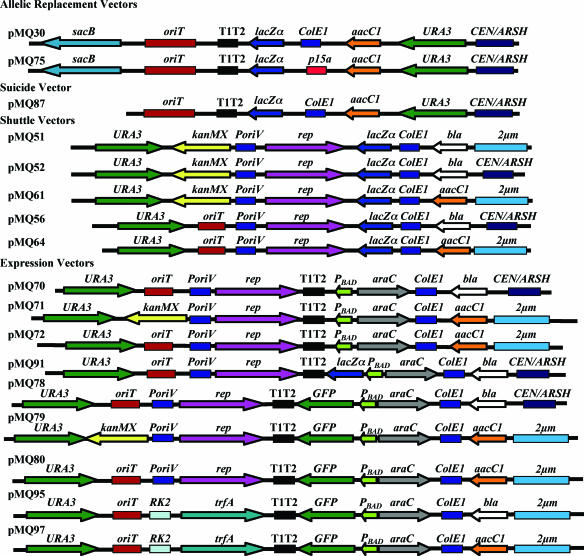
pMQ30 was chosen as a backbone for suicide vectors. The sacB gene was removed to enhance stability of single-crossover constructs as follows. pMQ30 was gapped via digestion with MslI and MscI, whose restriction sites are both in the sacB open reading frame (ORF). We then amplified an internal fragment from the clpX gene of P. aeruginosa strain PA14 by using primers that target the amplicon to recombine with pMQ30 on either side of sacB and replacing sacB and some dispensable vector sequence flanking sacB. The primers each contained an XhoI site so that the clpX sequence could be removed completely following digestion with XhoI and subsequent unimolecular ligation. We also included a StuI site proximal to the yeast replication machinery so that it could be removed by digestion with StuI to generate a very small suicide vector not capable of replication in yeast. Thus, we made pRMQS84, which is pMQ30 with sacB replaced by a P. aeruginosa clpX internal sequence; pMQ87, where the entire clpX sequence had been removed through digestion with XhoI; and pMQ89, which had been digested with StuI to remove the yeast replicon and selectable marker (Fig. 1).
FIG. 1.
Tool kit for use with genes from gram-negative bacteria. Shown are maps of vectors for manipulating bacterial genes by using S. cerevisiae-based homologous recombination. Vector maps are shown as linearized at the first base pair and are not shown to scale. Constructs are clustered based upon suggested utility. sacB, Bacillus subtilis levansucrase gene for counterselection; oriT, origin of conjugal transfer; T1T2, E. coli rrnB transcriptional terminators; lacZα, lacZα with multicloning site driven by the lactose promoter; ColE1, high-copy-number variants of the narrow-host-range ColE1 origin of replication; p15a, narrow-host-range, low-copy origin of replication; aacC1, gentamicin resistance determinant from Tn1696; URA3, orotidine-5′-phosphate decarboxylase gene from S. cerevisiae; CEN/ARSH, low-copy yeast replication and segregation machinery; kanMX, kanamycin/G418 resistance cassette from pUG6; PoriV-rep, pRO1600 broad-host-range replicon; bla, ampicillin and carbenicillin resistance determinant; 2μm, yeast episomal plasmid replicon; PBAD-araC, arabinose-inducible promoter system from E. coli; RK2-trfA, minimal replicon from the extremely-broad-host-range RK2 plasmid; GFP, bright and stable GFPmut3 variant. Plasmid request information can be found at http://www.dartmouth.edu/∼gotoole/vectors.html.
Construction of shuttle vectors pMQ51, pMQ52, and pMQ61.
Homologous recombination was used to add the pRO1600 replicon to pRS416 and pRS426, resulting in plasmids pV204 and pV204-2μm, respectively. We were unable to transform Pseudomonas aeruginosa using these plasmids, although we were able to do so with a positive-control pUCP20 with the same replication machinery (39). The URA3 gene was proximal to the Pseudomonas origin, and we hypothesized that transcriptional read-through from this gene in Pseudomonas could inhibit replication. Therefore, we added a kanamycin resistance cassette between the URA3 gene and the pRO1600 origin by using yeast homologous recombination. The kanamycin cassette from pUG6 was chosen rather than a transcriptional terminator because the addition of a selectable marker broadened the usefulness of these plasmids as well as aided in recovery of the correct construct (14). The resulting plasmids, pMQ51 (2μm) and pMQ52 (CEN6), were able to replicate in both P. aeruginosa and P. fluorescens (Fig. 1). The kanamycin resistance cassette from pUG6 is useful for conferring antibiotic resistance in E. coli and S. cerevisiae, where it confers resistance to kanamycin and G418 sulfate, respectively, and P. fluorescens, but not in P. aeruginosa, because of P. aeruginosa's intrinsic high level of kanamycin resistance.
To generate pMQ61, pMQ51 was digested in the ampicillin resistance determinant with BcgI and a fragment 644 bp upstream and 234 bp downstream of the bla gene was replaced with a gentamicin resistance-conferring gene (aacC1) from pEX18-Gm (Fig. 1).
Construction of conjugal shuttle vectors pMQ56, pMQ57, pMQ64, and pMQ65.
pMQ51 and pMQ61 were digested with MluI and modified with an oriT-containing amplicon with 40-bp tails that target replacement of the kanMX cassette by using yeast homologous recombination. The primers used to make the oriT amplicon also contained a PmlI site that was subsequently used to remove the yeast replication machinery. From these recombination events, we made pMQ56 (CEN, ampicillin resistance, oriT) and pMQ64 (2μm, gentamicin resistance, oriT). The yeast machinery of pMQ56 was removed by digestion with PmlI; one site was engineered between oriT and URA3, and the other is within the CEN6 region. The resulting fragment was recircularized with T4 DNA ligase, generating pMQ57. A similar strategy was used with pMQ64, except that we used XmnI, which cuts in the 2μm origin, and PmlI, both of which leave blunt ends. The ligated product lacks the ability to replicate in yeast and was dubbed pMQ65 (Fig. 1).
Primer design.
DNA sequences to be recombined were placed together in silico by using Gene Construction Kit software (Textco, Inc.). Additional features, such as synthetic ribosome binding sites, were added, and the junctions were selected and incorporated as part of the primer sequence.
In vivo cloning procedures (gap repair) and yeast minipreps.
Yeast was grown overnight in yeast extract-peptone-dextrose at 30°C and transformed using a modified “lazy bones” transformation protocol (3, 10). Plasmids were recovered from yeast using the “smash and grab” method (3, 20).
Phenotypic assays.
Rhamnolipid preparation and analysis, surface motility assays, and immunoblotting were performed using published protocols (4, 17, 38). Antihemagglutinin (anti-HA) antibodies were from QED Bioscience, Inc. (18850-01), and anti-green fluorescent protein (GFP) antibodies were from U.S. Biological, Inc. (G8965-10C).
Phase-contrast and epifluorescent microscopy were performed with a model DM IRBE inverted microscope (Leica Microsystems, Wetzlar, Germany), and photomicrographs were taken with a charge-coupled-device camera (model Orca C4742-5; Hamamatsu, Hamamatsu City, Japan) and analyzed with Open Lab 4.0.2 software (Improvision, Coventry, England). Flow cell experiments were performed as previously described (8).
Plasmid maps, sequences, and request information can be found at http://www.dartmouth.edu/∼gotoole/vectors.html.
Nucleotide sequence accession numbers.
Nucleotide sequences for vectors have been deposited in GenBank under accession numbers DQ230317 to DQ230324 and DQ642033 to DQ642043.
RESULTS
Modeling of gram-negative bacterial vectors using S. cerevisiae. (i) Vectors for allelic replacement in gram-negative organisms unable to support replication of the ColE1 origin.
An existing gene replacement vector (pEX18-Gm) (18) was modified such that it was able to replicate in S. cerevisiae, resulting in plasmid pMQ30 (Fig. 1). pEX18-Gm has many fine features, including the counterselectable marker sacB, a high-copy ColE1 variant origin of replication, an origin for conjugal transfer, and a multicloning site in lacZα for blue-white screening (18). Sequences required for stable replication in S. cerevisiae and a selectable/counterselectable marker, URA3, were added to pEX18-Gm by using homologous recombination in yeast. For more details on the construction of pMQ30 and all subsequent vectors, see Materials and Methods; also see Table S1 in the supplemental material for primers, Table S2 in the supplemental material for a key to vector construction, and Table S3 in the supplemental material for host ranges of the replicons used in this study.
pMQ30 was subsequently modified using yeast homologous recombination. The low-copy p15a origin of replication (also referred to as the pACYC origin) for E. coli was used to replace the high-copy ColE1-based origin to generate pMQ75 (Fig. 1). We hypothesized that the lower copy number afforded by the p15a origin would circumvent many toxicity problems associated with excess gene copy number.
pMQ30 was used to mutate the clpP gene of P. aeruginosa (Fig. 2). In this case, we replaced the clpP open reading frame with the GFP open reading frame. We also made clean deletion constructs by amplifying 500- to 1,000-bp regions upstream and downstream of a gene with primers that have homology on one side with the vector and the other side with each other. The two amplicons and the gapped vector were used to cotransform yeast, and recombinant constructs were selected for on medium deficient in uracil (not shown).
FIG. 2.
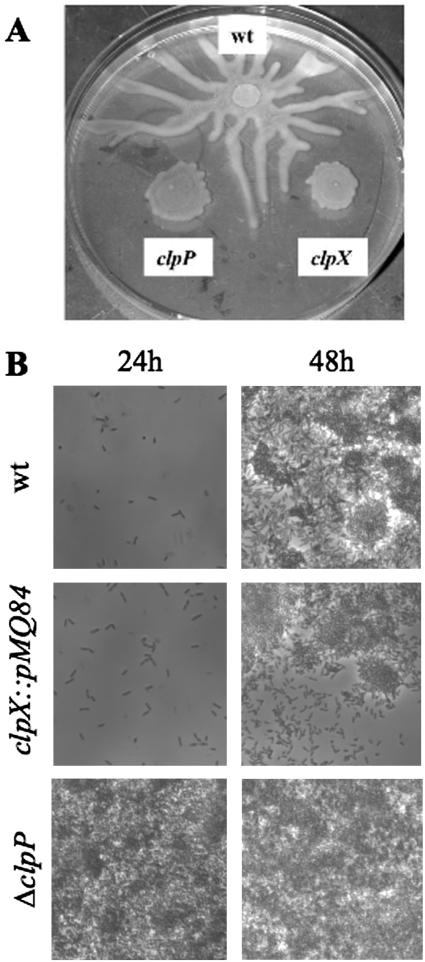
P. aeruginosa clpP deletion and clpX disruption mutants have altered surface-associated phenotypes. (A) Overnight cultures of P. aeruginosa strain PA14 (wild type [wt]) and isogenic ΔclpP and clpX::pRMQS84 strains were spotted on swarming agar plates and incubated at 37°C for 24 h (4, 38). Swarming in the wt strain is indicated by the tendril-like structures originating from the point of inoculation and spreading across the agar surface. The absence of ClpP and ClpX correlated with a reduction in surface motility. (B) ΔclpP mutants exhibit increased biofilm formation under flow conditions in M63 medium supplemented with arginine. Shown here are photomicrographs of wt, ΔclpP, and clpX::pMQ84 biofilms 24 and 48 h after inoculation.
(ii) E. coli-S. cerevisiae shuttle vectors usable as suicide vectors in gram-negative organisms unable to support replication of the ColE1 origin.
Vectors were designed specifically to disrupt genes via single-crossover events or to complement genes in their native context. The use of gene disruption by single-crossover events resulting in internal duplication is very quick and easy compared to allelic replacement and is also useful to rapidly test the importance of a gene to a biological phenomenon. This approach is especially useful when genomic or proteomic data implicate multiple genes in a biological process that would otherwise require considerable time to mutate through traditional allelic replacement.
We chose pMQ30 as a backbone for a suicide vector. The sacB gene was removed via homologous recombination to increase the stability of single-crossover constructs in the bacterial chromosome, generating pMQ87 (Fig. 1).
The yeast replication machinery was also removed by using a yeast recombination-introduced StuI restriction site, yielding pMQ89 (not shown).
(iii) E. coli-S. cerevisiae-Pseudomonas shuttle vectors.
Shuttle vectors that replicate in a range of gram-negative organisms as well as S. cerevisiae were made for complementation and other purposes. To make yeast-E. coli-Pseudomonas shuttle vectors, a Pseudomonas stability-conferring sequence from pUCP20 (originally from the cryptic P. aeruginosa plasmid pRO1600) was added to two yeast-E. coli shuttle vectors of the pRS400 series (35, 36). The pRO1600 replicon is sufficient for replication in many Pseudomonas species as well as Klebsiella species and has been shown to be stable even in the lack of selection (see Table S3 in the supplemental material) (28, 39). The pRS400 backbones we chose were pRS416 and pRS426, which differ by both copy number in the yeast cell and size. pRS416 is a centromeric (CEN) plasmid and therefore is maintained at 1 to 2 copies per cell, while pRS426 is a 2μm plasmid, maintained at about 25 copies per cell (36). Both plasmids have the ColE1-derived pBluescript high-copy origin of replication, a lacZα-associated multicloning site, an ampicillin resistance gene (bla) for gram-negative bacteria, and a URA3 marker for yeast. These plasmids also have T7 and Plac promoters that can be utilized for gene expression. The Plac promoter is useful in pseudomonads as a low-level constitutive promoter.
Homologous recombination was used to add the pRO1600 replicon and a kanamycin resistance cassette to pRS416 and pRS426, resulting in plasmids pMQ51 (2μm) and pMQ52 (CEN6), which were able to replicate in both P. aeruginosa and P. fluorescens (Fig. 1 and data not shown).
pMQ51 was further modified with addition of a gentamicin resistance gene by use of yeast homologous recombination to generate pMQ61 (Fig. 1).
(iv) Shuttle vectors containing oriT.
Homologous recombination was used to add the RP4 oriT to pMQ51 and pMQ61 shuttle vectors. The resulting plasmids, pMQ56 (CEN, ampicillin resistance, oriT) and pMQ64 (2μm, gentamicin resistance, oriT), were made with primers that contained a PmlI site that was subsequently used to remove the yeast replication machinery from these plasmids. The yeast machinery of pMQ56 and pMQ61 was removed, generating vectors pMQ57 and pMQ65, respectively (Fig. 1 and data not shown).
These oriT-containing plasmids are capable of replication in and conjugation into P. aeruginosa (data not shown). Conjugations were done using E. coli strain S17-1, which bears transfer elements from RP4 on its chromosome (37).
(v) Expression vectors.
All of the preceding vectors contain the Plac promoter driving the lacZα gene. These vectors could be used for expression of genes in Pseudomonas simply by recombining or ligating an open reading frame and ribosome binding site of interest downstream from the Plac promoter; however, Pseudomonas spp. do not have the lac repressor, and therefore expression from Plac is constitutive. The PBAD promoter and the araC repressor are known for tight control of gene expression in E. coli and other species, so we chose this regulatory system for use with pseudomonads (15, 26). Newman and Fuqua have previously described the utility of the PBAD promoter-bearing vectors for pseudomonads and other gram-negative organisms (26). PBAD and araC and transcriptional terminators were amplified from pBAD18 and recombined onto several of the shuttle vectors described above by completely replacing the lacZα gene and Plac promoter, generating pMQ70 from pMQ56, pMQ71 from pMQ61, and pMQ72 from pMQ64 (Fig. 1). The PmlI-PmlI yeast replication fragment was removed from pMQ70 to yield pMQ90. The lacZα gene was recombined into pMQ70 under the control of the PBAD promoter to enable arabinose-dependent blue-white screening in appropriate E. coli strains (pMQ91) (Fig. 1). E. coli strain DH5α bearing pMQ91 forms blue colonies on selective medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plus l-arabinose but not on the same plates without l-arabinose (data not shown).
(vi) Increased host range.
The very-broad-host-range replication machinery of the RK2 plasmid was used to extend the usefulness of this tool kit. RK2-based vectors are commonly used in the study of diverse gram-negative genera (see Table S2 in the supplemental material). We replaced the pRO1600 replication machinery with the minimal RK2 replicon by homologous recombination. This replicon has been shown to be remarkably stable without selection in P. aeruginosa and E. coli but requires selection in Azotobacter vinelandii (2). The genetic-information swap was performed with pMQ78 and pMQ80 to generate pMQ95 and pMQ97, respectively (Fig. 1). PA14 was transformed with these vectors, and GFP expression was found to be similar to that exhibited by PA14 with pMQ78 and pMQ80 (data not shown).
Inactivation of clpP but not clpX stimulates biofilm formation in P. aeruginosa.
The ClpP protease has been shown to be conditionally required for Pseudomonas fluorescens biofilm formation (30). With Staphylococcus aureus, clpP mutants have been shown to exhibit elevated levels of biofilm production, while mutants of the ClpP-binding partner, clpX, produce diminished biofilms (12). We used some of the tools described in this work to examine the role of these two important proteins in P. aeruginosa surface-associated behaviors.
The clpP ORF was replaced directly with GFPuvr by using a pMQ30-based construct (data not shown). A clpP::GFPuvr allelic replacement construct was generated in a single recombination step. Briefly, primers were used to amplify clpP with flanking regions homologous to pMQ30. A second pair of primers was used to amplify a GFPuvr with flanking regions homologous to clpP. First, the clpP amplicon was digested in the open reading frame with AatII. Then, both of these amplicons were added to a yeast transformation reaction along with pMQ30 that had been digested with SmaI. Uracil prototrophs were pooled, and plasmid DNA was acquired and transformed into E. coli. Gentamicin-resistant colonies that were also white on X-Gal were collected and analyzed by restriction digestion and sequenced. The proximity of the homologous targeting DNA to the restriction site does not seem to be important for the efficacy of these reactions. The clpP amplifying primers were also used to add the wild-type locus to pMQ30 for future allelic replacement or complementation scenarios (pRMQS44).
The clpP deletion strain grew slowly and exhibited reduced surface swarming motility (Fig. 2A and data not shown). Although clpP mutants grew slowly, they formed more robust biofilms than the wild-type strain in M63 medium supplemented with arginine under flow conditions (Fig. 2B).
We used pRMQS84, described above, to disrupt clpX. The plasmid was introduced into P. aeruginosa by conjugation, and all gentamicin-resistant candidates were found to have pRMQS84 disrupting clpX (data not shown). We found that clpX was not essential for Pseudomonas aeruginosa, unlike for some other bacteria, such as Caulobacter crescentus (22). clpX::pRMQS84 mutants exhibited slightly reduced growth rates and altered surface motility (Fig. 2A and data not shown). The effect of clpX disruption upon biofilm formation under sheer conditions was determined with M63 medium using arginine as a sole carbon source. Under the conditions utilized, clpX mutants formed biofilms in a manner similar to that of the wild type (Fig. 2B).
pMQ87 was subsequently used to mutate a number of genes implicated in biofilm formation by simply transforming P. aeruginosa with the pMQ87-based construct via conjugation or electroporation followed by selection for gentamicin-resistant colonies (Daniel Kadouri, Daniel MacEachran, and G. A. O'Toole, unpublished observations).
Use of inducible expression vectors in pseudomonads.
GFPmut3 from pGFPmut3 was introduced into vectors pMQ70 to pMQ72 by homologous recombination for use as a reporter. GFPmut3 is a stable and bright variant of GFP (6). The primers used to amplify GFPmut3 contained an engineered ribosome binding site 8 bp upstream of the start codon of GFP as well as 40-bp tails that target recombination downstream of the PBAD promoter on pMQ70 to pMQ72. Uracil-auxotrophic yeast (InvSc1) cells were transformed by the addition of both the GFP-containing amplicon and pMQ70 (or pMQ71 or pMQ72 in separate reactions) that had been digested with SmaI. Yeast transformants were pooled, and plasmid DNA was prepared and electroporated into E. coli which was subsequently plated on selective medium containing 100 mM l-arabinose. Antibiotic-resistant colonies were screened for GFP fluorescence by use of epifluorescent microscopy. Constructs verified to contain GFP were dubbed pMQ78, pMQ79, and pMQ80, respectively (Fig. 1). Fluorescence in E. coli was found to be dependent upon the presence of l-arabinose, and there was little leakiness, based on the lack of observable GFP fluorescence, even though these plasmids have very high copy numbers in E. coli (data not shown). The plasmids were conjugated into P. aeruginosa strain PA14 and tested for fluorescence during growth in a dilution series of l-arabinose. We found that at 100 mM l-arabinose the majority of cells fluoresced brightly, while PA14 bearing a no-GFP vector control (pMQ72) did not fluoresce (Fig. 3A and data not shown). Cells grown with 1 mM arabinose were clearly less bright, cells grown with 0.1 mM arabinose had detectable but very low levels of fluorescence, and those grown without arabinose exhibited no fluorescence at the exposure used (Fig. 3A).
FIG. 3.
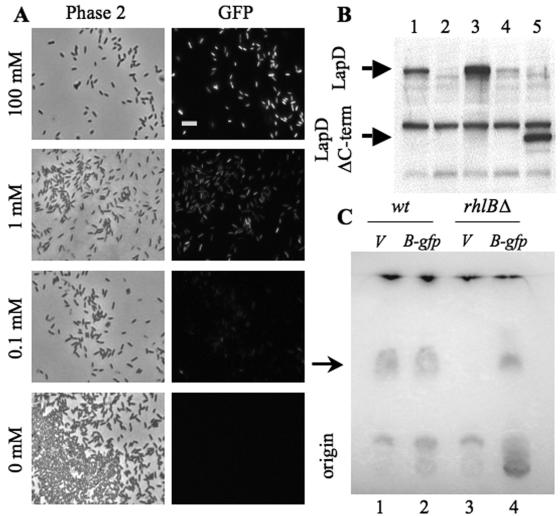
PBAD-driven expression in Pseudomonas species. (A) Aliquots of P. aeruginosa cultures bearing pMQ78 grown overnight in LB with ampicillin and various concentrations of l-arabinose were viewed with either phase-contrast microscopy (left) or epifluorescent microscopy (right). Micrographs of GFP fluorescence were taken with identical exposure times. No fluorescence was detected in the absence of l-arabinose or in cells containing an identical vector without GFP (pMQ72 [not shown]). Bar = 10 μm. (B) LapD immunoblot of membrane fractions from P. fluorescens. (Lane 1) Wild-type samples exhibit the predicted 71-kDa LapD protein. (Lane 2) Anti-LapD antibodies do not react with samples from a ΔlapD strain. (Lane 3) P. fluorescens bearing pMQ72 plus lapD exhibits relatively high levels of LapD in the presence of 0.2% l-arabinose (grown in M63 plus 0.2% glucose plus 0.5% Casamino Acids) and (lane 4) very little LapD in the absence of l-arabinose. (Lane 5) Fractions with pMQ72+lapDΔC-term exhibited a predicted 45-kDa protein in the presence of 0.2% l-arabinose. (C) Thin-layer chromatographic analysis of P. aeruginosa. The wild type and the rhlB::kanMX mutant were grown in PPGAS with 100 mM l-arabinose for 24 h at 30°C as reported previously (4). The arrow indicates dirhamnolipids. (Lane 1) Wild type (wt) grown with vector alone (V) (pMQ78). (Lane 2) Wild type grown with a plasmid-borne rhlB-GFPmut3 fusion construct (B-gfp) under the control of the PBAD promoter (pRMQS96). (Lane 3) rhlB::kanMX mutant bearing the vector alone. (Lane 4) rhlB::kanMX mutant bearing the PBAD-rhlB-GFPmut3 plasmid. These data show that the rhlB::kanMX mutant is deficient in formation of dirhamnolipids and that this mutation is complemented by an RhlB-GFPmut3 fusion protein.
The efficacy of these expression vectors was also assessed in P. fluorescens. The lapD gene of P. fluorescens is adjacent to the lapA gene and is implicated in biofilm formation (16, 17). The open reading frame of lapD, including 34 bp upstream and 25 bp downstream, was recombined into pMQ72 via yeast homologous recombination (pMQ72+lapD). Additionally, a truncated version of the gene lacking the C-terminal domain was cloned into pMQ72 (pMQ72+lapDΔC-term). These plasmids were electroporated into P. fluorescens and grown in broth culture supplemented with either 0% or 0.2% l-arabinose. Membrane fractions were prepared, run on polyacrylamide gels, transferred to nylon membrane, and immunoblotted with polyclonal anti-LapD antibodies (17). We observed relatively high levels of LapD in cultures grown with 0.2% arabinose and very little LapD from those grown without arabinose (Fig. 3B, lanes 3 and 4). We also observed stable expression of the LapDΔC-term polypeptide (Fig. 3B, lane 5). LapD antibodies showed predicted patterns with control wild-type and lapD deletion strains (Fig. 3B, lanes 1 and 2).
Complementation of ΔrhlB with rhlB-GFP.
pMQ30 has previously been utilized to mutate the rhlB (rhamnolipid biosynthetic pathway) gene in P. aeruginosa (4). Rhamnolipids are biosurfactants and are hypothesized to modulate surface motility and biofilm architecture (4, 8). A biochemical approach was taken to directly determine the formation of rhamnolipids in the wild-type and rhlB::kanMX strains. Rhamnolipids were extracted from culture medium, separated on thin-layer chromatography plates, and detected by exposure to an anthrone-sulfuric acid mixture (Fig. 3C, lanes 1 and 3). RhlB is a rhamnosyltransferase that acts to add rhamnose moieties to rhamnolipid precursors, generating monorhamnolipids. Monorhamnolipids are subsequently converted to dirhamnolipids by RhlC. In the absence of RhlB, no rhamnolipids should be present, and that is what we observed from rhamnolipid preparations from rhlB::kanMX cultures, as was previously reported (4) (Fig. 3C, lane 3).
One advantage of using yeast homologous recombination is that seamless translational fusions can be made without multiple, possibly mutagenic, rounds of PCR associated with classical “sewing” techniques. We designed primers to fuse the rhlB ORF to the N terminus of GFP in pMQ78 and pMQ80. These primers direct a translational fusion of the last codon of rhlB to the first codon of GFP. pMQ78 and pMQ80 were digested with SmaI between the PBAD promoter and GFPmut3 and added to transformation reactions along with the targeted rhlB amplicon. The resulting plasmids, pRMQS96 and pRMQS98, were introduced into PA14 and the rhlB::kanMX strain. Rhamnolipid production was assessed as noted above for strains incubated in the presence of 100 mM l-arabinose. We observed that pRMQS98 was able to restore rhamnolipid production to the rhlB::kanMX strain (Fig. 3C, compare lanes 3 and 4). The GFP fusion can subsequently be used for microscopic visualization of the protein or as a convenient epitope tag.
This protocol is especially useful in making artificial operons for the study of prokaryotic genes. For example, in a single step we have cloned a two-gene operon while fusing a gene for cyan fluorescent protein or yellow fluorescent protein to the 3′ end of the first gene in the operon (C. M. Toutain and G. A. O'Toole, unpublished results). Each of these synthetic operons can then be expressed under the control of its own promoter or under regulation of PBAD or other promoters of choice or moved to pMQ30 (Fig. 1) to replace the chromosomal locus.
High-efficiency recombination of multiple pieces of DNA.
The fusion of several pieces of DNA can easily be performed using yeast. This is especially useful for building complicated vectors and making gene fusions and site-directed mutations. As an example, we performed a complex reaction where we added four pieces of DNA to a vector in a single step. With pMQ87 (Fig. 4), in one step we (i) added the T5X inducible promoter-repressor system (11), (ii) added GFP, (iii) added F64L and S65T mutations to GFP, and (iv) fused a C-terminal 3HA epitope tag to GFP. The resulting plasmid was named pRMQS106 (Fig. 4A and B). At the same time, we removed the entire lacZα gene and Plac promoter of pMQ87.
FIG. 4.
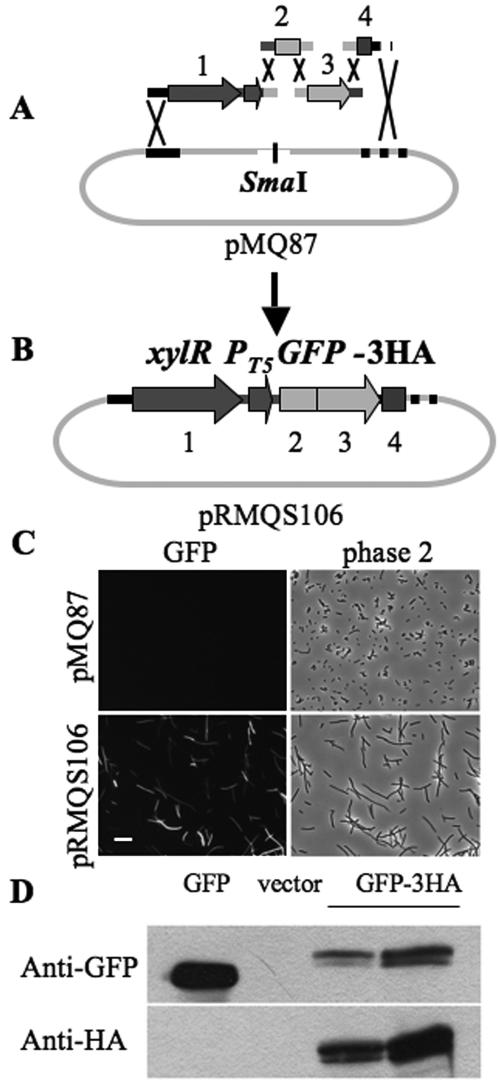
Yeast recombination facilitates the construction of multiple pieces of DNA. (A and B) Schematics depicting the recombination of four PCR-based amplicons into a gapped vector in a single step. (A) pMQ87 that has been digested with SmaI (gapped) and the four PCR-based amplicons: 1, xylR repressor-T5X promoter; 2, a 5′ region of GFPuvr with two mutations added to the primer; 3, a 3′ region of GFPuvr with two mutations added to the primer; 4, a 3HA tag. (B) Schematic diagram representing the recombinant product which has a modified GFP-3HA allele under the control of a xylose-regulated promoter (no. 1 to 4) all upon a yeast-E. coli shuttle vector. (C) Phase-contrast and epifluorescent micrographs of E. coli strains bearing either pMQ87 (no GFP) or pRMQS106 (GFP-3HA). Images were taken with identical exposure times. Magnification, ×400. Bar = 10 μm. (D) Immunoblots of lysates of E. coli cells bearing pMQ80 (PBAD-GFP), pMQ87 (vector alone), or pRMQS106 (PT5X-GFP-3HA). Blots were probed with either anti-GFP or anti-HA antiserum. Cells were grown overnight in the presence of gentamicin and 0.4% glucose and then pelleted and diluted at 1:20 in LB supplemented with gentamicin and either 40 mM l-arabinose for the pMQ80 culture (GFP) or 0.4% (vol/vol) d-xylose for pMQ87 (vector) and pRMQS106 (GFP-3HA). Diluted cultures were grown for 3 h at 37°C before aliquots were taken for analysis. Lysates of cells bearing two independent pRMQS106 constructs are shown.
The four amplicons are (i) a xylose repressor-promoter with primers to target recombination with pMQ87 and GFP including an artificial ribosome binding site, (ii) a portion of the GFPuv ORF, corresponding to the first 63 amino acids, with primers that target recombination with the promoter sequence including an artificial ribosome binding site on one side and GFPuv targeting sequence with F64L and S65T mutations on the other side, (iii) a portion of the GFPuv ORF, corresponding to the rest of the gene, including targeting sequences to the 5′ portion of GFPuv including the F64L and S65T mutations and with HA epitope targeting sequence on the 3′ end, and (iv) a 3HA epitope tag with targeting sequences to GFP and pMQ87 (Fig. 4A and B). S. cerevisiae was cotransformed by the four amplicons and pMQ87 that had been digested with SmaI.
We observed a >10-fold ratio of transformants from the cut vector plus all four amplicons compared to the number of transformants from the cut vector alone. Hundreds of transformants were pooled, and their plasmids were obtained and used to transform E. coli. Colonies grown on selective medium plus 0.4% d-xylose were observed with fluorescent microscopy, and 11 of the 12 colonies tested exhibited bright green fluorescence (Fig. 4C). Additional candidate colonies were transferred to individual wells of a microtiter dish, each containing LB plus gentamicin plus 0.4% d-xylose, and grown overnight at 37°C. We examined the wells for GFP fluorescence with an inverted microscope and found that 52 out of 60 (86.7%) wells exhibited bright fluorescence. Cells in the microtiter plate were subjected to several rounds of freezing and thawing, and detergent was added to lyse cells. Lysates were then spotted onto a nitrocellulose membrane and immunoblotted using anti-HA antibodies. In control experiments, these commercially available antibodies detected no background bands in E. coli lysates on Western blots (Fig. 4D and data not shown). We found that while a control lysate bearing an empty vector exhibited no cross-reactivity to the HA antibody, 53 out of 60 (88%) candidate lysates reacted with anti-HA antibodies. Together, these results are consistent with >80% of candidate colonies containing constructs that had undergone five-way recombination events producing functional fusion proteins. This frequency is likely an underrepresentation of the actual frequency of recombinants for two reasons. (i) Original yeast transformants contained a mixture of uncut vectors that had not been digested in the original gap-generating digestion, gapped vectors that had closed through nonhomologous end joining, and gapped vectors that had closed through the targeted recombination events. Rather than screening through yeast transformants, we pooled them and electroporated yeast-plasmid preparations into E. coli. (ii) Gap-repaired vectors bearing the targeted recombinant construct may contain PCR-generated or recombination-generated mutations leading to a lack of GFP-3HA expression or a lack of production of a functional protein.
Western blot analysis was performed with cells bearing pRMQS106 (GFP-3HA) to further verify the construct. Cell lysates of E. coli strains bearing pMQ80 (PBAD-GFP), pMQ87 (negative control), or pRMQS106 (PT5X-GFP-3HA) were immunoblotted with anti-GFP and anti-HA antibodies (Fig. 4D). With anti-GFP antibodies, a single band was identified from pMQ80 lysates, corresponding to approximately 25 kDa (GFP has a predicted mass of 26.9 kDa); no band was observed with the pMQ87 lysate, as expected; and a larger band was observed associated with pRMQS106 lysates (GFP-3HA has a predicted mass of ∼31 kDa). Anti-HA antibodies revealed bands only with pRMQS106 lysates, and these corresponded to the same size as those observed with the anti-GFP antibodies (Fig. 4D). The two plasmids used in Fig. 4 were sequenced, and all five recombinant junctions from each construct were as predicted. Likewise, we sequenced the junctions for the majority of recombinant junctions for the vectors described here and all were as predicted, consistent with reports showing the high fidelity of recombination in S. cerevisiae.
DISCUSSION
In vivo cloning via gap repair is efficient, easy, and amenable to high-throughput applications. Here we describe a complete set of tools made for modification of bacterial genes by using yeast homologous recombination. While others have previously described the use of gap repair in cloning, a bacterium-centered set of tools has not been made. For example, Raymond and colleagues have previously described using this technique for cloning of Pseudomonas DNA and have described the clever use of oligonucleotide linkers that further broaden the flexibility of yeast recombination-mediated cloning (32, 33).
We used these vectors to manipulate genes from Pseudomonas aeruginosa and P. fluorescens to emphasize the power of a new set of molecular tools that take advantage of the S. cerevisiae native recombination proteins. Although these vectors were made with Pseudomonas spp. in mind, they can be used with many gram-negative organisms and thus are broadly useful. The bacterial origins of replication placed upon one or more of these vectors include ColE1, p15a, pRO1600, and RK2, allowing the use of these vectors to replicate in more than 30 species of bacteria. These tools will allow researchers who study many types of prokaryotic organisms, including nonmodel species, to conveniently modify DNA by using recombination.
Other systems for in vivo cloning have been described previously, and some can be used in conjunction with the vectors and system described here (7, 23). The vectors described here can be used with many existing tools designed for use with yeast, such as a set of constructs that allow epitope tagging of a protein of interest with any of a group of several epitopes with a single set of primers via yeast homologous recombination (24).
Cloning with yeast homologous recombination is an additional tool to facilitate molecular studies of a wide range of gram-negative organisms rather than a replacement for traditional in vitro cloning. However, this tool kit can be readily substituted for many applications of traditional cloning, particularly for large-scale cloning projects and for the construction of plasmids requiring multiple inserts, site-directed mutations, or precise junctions. While multiple amplicons can also be fused via splicing by overlap extension PCR, this method requires serial rounds of amplification, increasing the frequency of PCR-generated mutations, and is more limited with regard to the size of the final amplicon. Also, recombination can be targeted hundreds of base pairs away from the gap site on a vector, providing a large advantage over traditional cloning methods where modifications are confined within existing restriction sites. This allows complete exchange of features on a vector, such as replicons or promoter/regulator systems. Another advantage is that, although yeast-based cloning can take longer, as yeast colonies can take 2 to 4 days to appear on selective plates, these methods often require much less handling time. Amplicons are used directly, and vector digests can be used without any purification or desalination. Additionally, the method of yeast transformation described here requires no lengthy preparation of competent cells. The disadvantages are the increased cost due to longer primers and the increased amount of time for yeast cultures to grow.
Supplementary Material
Acknowledgments
We thank Dan MacEachran, Benedict Kemp, and Stu Powers for critical reading of the manuscript and members of the O'Toole lab for trying many of the described constructs. We thank Herbert Schweizer, Dean Dawson, and Janet Ajioka for the kind gifts of plasmids, strains, and glass beads.
This work was supported by NIH training grants T32 AI07363 and F32 GM66658-01A1 to R.M.Q.S. and NIH R21-AI055774 and AI51360 to G.A.O.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bascom-Slack, C. A., and D. Dawson. 1998. A physical assay for detection of early meiotic recombination intermediates in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:512-520. [DOI] [PubMed] [Google Scholar]
- 2.Blatny, J. M., T. Brautaset, H. C. Winther-Larsen, K. Haugan, and S. Valla. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl. Environ. Microbiol. 63:370-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 4.Caiazza, N. C., R. M. Shanks, and G. A. O'Toole. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187:7351-7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, K. H., A. Kumar, and H. P. Schweizer. 2005. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMarini, D. J., C. L. Creasy, Q. Lu, J. Mao, S. A. Sheardown, G. M. Sathe, and G. P. Livi. 2001. Oligonucleotide-mediated, PCR-independent cloning by homologous recombination. BioTechniques 30:520-523. [DOI] [PubMed] [Google Scholar]
- 10.Elble, R. 1992. A simple and efficient procedure for transformation of yeasts. BioTechniques 13:18-20. [PubMed] [Google Scholar]
- 11.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Zhan-yang, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 12.Frees, D., A. Chastanet, S. Qazi, K. Sorensen, P. Hill, T. Msadek, and H. Ingmer. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 54:1445-1462. [DOI] [PubMed] [Google Scholar]
- 13.Geels, F. P., and B. Schippers. 1983. Reduction of yield depressions in high frequency potato cropping soil after seed tuber treatments with antagonistic fluorescent Pseudomonas spp. Phytopathol. Z. 108:207-214. [Google Scholar]
- 14.Guldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman, L.-M., B. Carson, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 17.Hinsa, S. M., and G. A. O'Toole. 2006. Identification and characterization of the LapD protein: LapD modulates the secretion of LapA. Microbiology 152:1375-1383. [DOI] [PubMed] [Google Scholar]
- 18.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 19.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Jansen, G., C. Wu, B. Schade, D. Y. Thomas, and M. Whiteway. 2005. Drag&Drop cloning in yeast. Gene 344:43-51. [DOI] [PubMed] [Google Scholar]
- 22.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, M. Z., and S. J. Elledge. 2005. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nat. Genet. 37:311-319. [DOI] [PubMed] [Google Scholar]
- 24.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 25.Mallet, L., and M. Jacquet. 1996. Intergenic Flip Flop, a method for systematic gene disruption and cloning in yeast. Yeast 12:1351-1357. [DOI] [PubMed] [Google Scholar]
- 26.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 27.Oldenburg, K. R., K. T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen, R. H., G. DeBusscher, and W. R. McCombie. 1982. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J. Bacteriol. 150:60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orr-Weaver, T. L., and J. W. Szostak. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 80:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 31.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 32.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Karul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raymond, C. K., E. H. Sims, and M. V. Olson. 2002. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 12:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos, P. M., I. D. Bartolo, J. M. Blatny, E. Zennaro, and V. Svein. 2001. New broad-host-range promoter probe vectors based on the plasmid RK2 replicon. FEMS Microbiol. Lett. 195:91-96. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski, R. S. 1993. Extrachromosomal cloning vectors of Saccharomyces cerevisiae, p. 225-241. In K. G. Hardy (ed.), Plasmids, a practical approach, 2nd ed. IRL Press, Oxford, United Kingdom.
- 36.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. BioTechnology 1:784-790. [Google Scholar]
- 38.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.