Abstract
Two Arthrobacter nicotinovorans molybdenum enzymes hydroxylate the pyridine ring of nicotine. Molybdopterin cytosine dinucleotide (MCD) was determined to be a cofactor of these enzymes. A mobA gene responsible for the formation of MCD could be identified and its function shown to be required for assembly of the heterotrimeric molybdenum enzymes.
The highly toxic alkaloid nicotine present in tobacco waste is removed from the environment by mineralization through soil bacteria. The best-characterized pathway of nicotine degradation is the one encoded by the megaplasmid pAO1 of Arthrobacter nicotinovorans (15). It includes two hydroxylations of the pyridine ring of nicotine, at C-6 and C-2, and an oxidation at C-2′ of the pyrrolidine ring, which prepare the molecule for its degradation (4). The enzymes which perform the hydroxylation reactions belong to a group of related heterotrimeric bacterial molybdenum oxidoreductases (14) composed of a middle-sized subunit (about 30 kDa) carrying a flavin adenine dinucleotide (FAD) molecule, a small subunit (about 17 kDa) with two [2Fe-2S] clusters, and a large subunit (about 85 kDa) carrying a molybdenum cofactor (19). Nicotine dehydrogenase (NDH; also called nicotine:acceptor oxidoreductase, hydroxylating) of A. nicotinovorans has been characterized before (9). The genes of its subunits form an operon (12, 15). The genes of a second, related enzyme which is active at pyridine C-2, 6-hydroxy-pseudooxynicotine:acceptor oxidoreductase, hydroxylating (known as “ketone” dehydrogenase [KDH]), showed an unexpected arrangement and were proposed to be carried by a discontinuous gene cluster (1). The large molybdenum-containing subunit (KdhL) gene was tentatively located on pAO1 more than 4,000 bp apart and transcribed divergently from the genes of the middle (KdhM) and small (KdhS) subunits (15). Functional proof of the identity of the KDH subunit genes was missing.
In bacteria, the molybdenum cofactor may take the form of a molybdenum dinucleotide produced by the addition of a nucleoside monophosphate to molybdopterin, a reaction catalyzed by the MobA protein. In Escherichia coli, the cofactor is a molybdopterin guanosine dinucleotide, and the GTP:molybdopterin guanylyl transferase MobA was studied recently in great detail (13). Other bacteria make use of a molybdopterin cytosine dinucleotide (MCD) cofactor (3, 8, 11). A mobA gene encoding the MobA protein with cytidylyl transferase activity has not yet been described. The nature of the molybdenum cofactor of the nicotine hydroxylating enzymes was not known.
Here we demonstrate the identity of the kdh genes by the assembly of the functional holoenzyme from its three subunits and show that the molybdenum cofactor is MCD. We identify the mobA gene required for the synthesis of MCD and show that the assembly of the KDH and NDH subunits depends on a functional mobA gene and thus on the dinucleotide form of the molybdenum cofactor.
Expression of kdhL and ndhL genes from pART2 transformed into A. nicotinovorans.
The E. coli-Arthrobacter shuttle vector pART2 allows nicotine-induced expression of cloned genes in Arthrobacter species from the 6hdno promoter (21) in such a way that the synthesized proteins exhibit a C-terminal His8 tag. The kdhL gene was amplified from A. nicotinovorans whole cells by PCR with the primer pair listed in Table 1, digested with DraI and XbaI, and inserted by ligation into the multiple cloning sites of pART2. E. coli XL-1 Blue transformed with the ligated DNA was selected on Luria-Bertani (LB) plates with 50 μg/ml kanamycin. Clones carrying pART2kdhL recombinant DNA were identified by restriction endonuclease digestion of plasmid DNAs isolated from individual colonies. Cloning of kdhL into this vector resulted, besides the His tag, in an N-terminal MDPTSSTLM amino acid sequence extension of recombinant KdhL (the underlined “M” represents the start methionine of native KdhL). Recombinant KdhL was isolated by Ni-chelating chromatography from extracts of pART2kdhL-transformed A. nicotinovorans grown at 30°C on citrate medium supplemented with vitamins, trace elements, and 3 mM l-nicotine (5), as described previously (6). Analysis of the proteins eluted from the column revealed that besides the large subunit of the enzyme, the native middle and small subunits were coeluted (Fig. 1A), in agreement with the KDH activity of the eluted protein (not shown). When ndhL, amplified by PCR with the primer pair indicated in Table 1, was introduced on pART2 into A. nicotinovorans, all three subunits of enzymatically active NDH were recovered by Ni-chelating chromatography, similar to what was found with kdhL (not shown). For unknown reasons, the yield of recombinant NDH was only 0.25 mg/liter, compared to 2.1 mg/liter for recombinant KDH.
TABLE 1.
Oligonucleotides used in this study
| Primer | Orientation | Sequence (5′-3′) | Length of DNA (bp) | Fragment |
|---|---|---|---|---|
| RT-PCR primers | ||||
| 1 | Forward | GGCAACAGACAGGGATGGATAGG | 341 | I |
| 2 | Reverse | GGATCAACAGCAAGTACAGCAACC | ||
| 3 | Forward | TTGGCGCGCTGACACGTCATGCG | 396 | II |
| 4 | Reverse | AGGGTAACGATGGACGCAGCGC | ||
| 5 | Forward | GCGGACTTCCTACGAGACGACCTG | 398 | III |
| 6 | Reverse | GTGATCTCTGCAATGGAAGGCAT | ||
| 7 | Forward | CATGTCTGAGTAGGAAACGC | 325 | IV |
| 8 | Reverse | CCTATCCATGCCTGTCTGTT | ||
| 9 | Forward | CGTACATACTGACCGTTTGC | 300 | V |
| 10 | Reverse | CGCCAGAGTGAGTGCCAAT | ||
| 11 | Forward | GGAGATTGGCACTCACTCTG | 340 | VI |
| 12 | Reverse | GTCAACACTGAGGTCATAGAC | ||
| 13 | Forward | CGGAGCATGAGGACAACTG | 340 | VII |
| 14 | Reverse | CTGTGCCGAAGCCTGAGAG | ||
| 15 | Forward | CAGCTGCAGCAGGGACAC | 410 | VIII |
| 16 | Reverse | CGCGAGCATCCTTCGCA | ||
| 17 | Forward | GCAGAATCCGAAGTACTGG | 350 | IX |
| 18 | Reverse | CAGAGTTCAGGCAGGACG | ||
| 19 | Forward | CACTGAACGAACTGCGAAG | 365 | X |
| 20 | Reverse | GACAGCCTTGAGCCGCGC | ||
| 21 | Forward | GCGTGGATGAGAAGCTAGCGCAGC | 360 | XI |
| 22 | Reverse | CGCGACGCTGAAGATCCAAAACGC | ||
| 23 | Forward | GGCTCATCGTGTTATCTGGGC | 390 | XII |
| 24 | Reverse | CGTCATCGTCGCTATATC | ||
| 25 | Forward | GCCTAGCAATGTCCAACCTTA | 410 | XIII |
| 26 | Reverse | GCAAATCCGTGTCACCCACCG | ||
| Cloning primers | ||||
| 27 | Forward | CATGATGGCAAAGGCTAAAGCG | 2,384 | kdhL |
| 28 | Reverse | CTGGCGGATTGCACTCTAGACCGTTCTGCTTTGTTC | ||
| 29 | Forward | AATGGATAACCCACACTATTACTC | 2,450 | ndhL |
| 30 | Reverse | CAGGAGCAATTCTAGACTCCCTGTCC | ||
| 31 | Forward | GCATCGTCCTGCAGGAGGCGAT | 614 | mobA |
| 32 | Reverse | GAACATCCAGTCTAGACGGCCCGATG | ||
| 33 | Forward | GAACATCCAGTCTAGACGGCCCGATG | 2,998 | kdhL mobA |
| 34 | Reverse | CGGATTGCACTCTAGACCGTTCTGCTTTG | ||
| 35 | Forward | CCTAGCAGATCGGGTCGACCATGCGTGACGTTA | 2,120 | mobA xdhC |
| 36 | Reverse | GCATCGTCCTGCAGGAGGCGATAC |
FIG. 1.
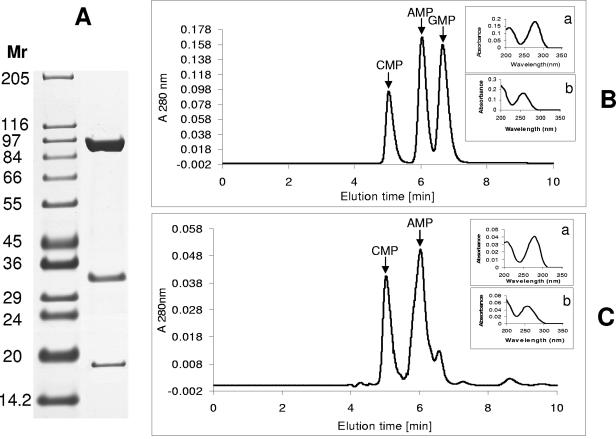
Purification of KDH and determination of CMP as a component of its molybdenum dinucleotide cofactor. (A) His-tagged KdhL was used to isolate the KDH holoenzyme by Ni-Sepharose chelation from extracts of bacteria transformed with pART2kdhL. Shown are the three protein subunits of KDH revealed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (B) HPLC elution profile of CMP, AMP, and GMP standard. (C) KDH was treated as described previously (18), and the released mononucleotides were analyzed by HPLC. CMP and AMP peaks are indicated by arrows. The insets in panels B and C show the UV spectra of CMP, with an absorption peak at 280 nm (a), and of AMP and GMP, with an absorption peak at 260 nm (b).
Characterization of the nucleotide moiety of the molybdenum cofactor of KDH.
The molybdenum contents of two independent KDH preparations were first determined in triplicate in a Perkin-Elmer 4110 ZL atomic absorption spectrometer. A content of 0.80 mol molybdenum per mol of enzyme was found, which is close to the 0.87 mol molybdenum per mol of enzyme reported for NDH (9) and is an indication that KDH was loaded with the molybdenum cofactor. Next, KDH was incubated at 95°C for 10 min in the presence of sulfuric acid (3% [by volume]), which leads to the release of the nucleotide from the molybdenum dinucleotide cofactor and of AMP from FAD, the cofactor associated with KdhM (18). Following centrifugation, the supernatant was analyzed by high-performance liquid chromatography (HPLC) on a reversed-phase column (5 μm by 250 mm by 4.6 mm) (Aqua; Phenomenex, Aschaffenburg, Germany) and eluted isocratically at room temperature with 0.1% trifluoroacetic acid in water at a flow rate of 0.75 ml/min. A rapid scanning detector was used (model 206PHD; Linear Instruments Corp., Sykam, Gilching, Germany) for detection and spectrum collection. Besides AMP, the analysis revealed the presence of CMP but not of GMP (Fig. 1). The small peak present as a shoulder of the AMP peak had a slightly different elution time from that of GMP and did not show the typical purine absorption spectrum. When FAD was treated as described above and analyzed by HPLC, only an AMP peak was revealed (not shown). The identification of CMP in the extract indicated that KDH belongs to the MCD-dependent enzymes. When the same analysis was performed with His-tagged KdhL isolated from a mobA-deficient strain (see below), no nucleotide was detected.
Identification of transcripts of the kdhL-mobA gene cluster in wild-type and ORF310-disrupted strains by reverse transcription-PCR (RT-PCR).
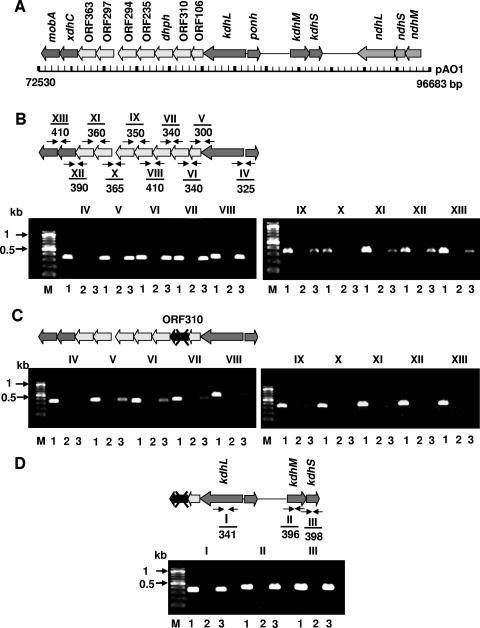
The three subunit genes of ndh are arranged in the order M-S-L (Fig. 2A) and form a transcriptional unit (12). The kdh genes are discontinuous and separated by 4.3 kb (Fig. 2A). The assumed kdhL gene is the first of a gene cluster. It is followed by the ORF106 and ORF310 genes, with unknown functions, by the gene for 2,6-dihydroxypyridine hydroxylase, and by the ORF235, ORF294, ORF297, and ORF363 genes, with unknown functions. The last two genes of this cluster are a gene similar to xdhC (17) and one similar to mobA (Fig. 2B). The ORF310 gene was disrupted with a chloramphenicol resistance cassette (cmx) (10) by homologous recombination. To this end, cmx was inserted into the PmlI site of the ORF310 gene carried on pH6EX3 (2), which is unable to replicate in A. nicotinovorans. The pH6EX3 construct carrying the cmx-disrupted ORF310 gene was transformed into A. nicotinovorans by electroporation and selected on chloramphenicol (22 μg/ml) plates, and colonies carrying a disrupted ORF310 gene were identified by PCR (7). Attempts to disrupt the genes similar to xdhC and mobA were unsuccessful.
FIG. 2.
Transcriptional analysis of the kdh and mobA gene clusters. (A) Schematic representation of the pAO1 region addressed in this work. (B) RT-PCR with primer pairs (Table 1) derived from the end of one gene and the start of the next one. No RT-PCR product was obtained with primer pair X. (C) Same RT-PCR analysis as that in panel B, but with cDNAs prepared from RNAs extracted from the cmx-disrupted ORF310 strain. No transcripts of genes downstream of the cmx-disrupted ORF310 gene were produced. (D) RT-PCR with cDNAs prepared from the strain with cmx-disrupted ORF310 and with primer pairs derived from the coding regions of the kdh genes, showing that kdh transcripts are present in this strain. The sizes in bp of the PCR-amplified cDNA fragments are indicated below the primer pair numbers. Lanes 1, 2, and 3 indicate PCRs performed with pAO1 as a positive control template, RNA as a negative control template, and cDNAs as experimental templates, respectively.
RT-PCR was performed with nicotine-grown A. nicotinovorans bacteria as described previously (6, 20), using primer pairs derived from the end of one gene and from the start of the next gene (Table 1). The genes from kdhL to the ORF294 gene were apparently transcribed into one RNA, and those from the ORF297 gene to mobA were transcribed into a second RNA molecule (Fig. 2B), since no amplification product was obtained between the ORF294 and ORF297 genes (Fig. 2B, lanes X). The same analysis performed with RNA prepared from the ORF310-disrupted strain revealed that transcripts of genes downstream of the inserted chloramphenicol resistance cassette were no longer detectable, including those from the ORF297 gene to mobA (Fig. 2C). Transcripts of kdhL as well as kdhMS were present (Fig. 2D). These results suggest that insertion of cmx into the ORF310 gene leads to the inactivation of downstream genes, including those similar to xdhC and mobA.
No transcripts of these genes could be detected in A. nicotinovorans bacteria grown in the absence of nicotine (not shown).
NDH and KDH enzyme activities in the cmx-disrupted strain and complementation of the strain with the pAO1 genes similar to xdhC and mobA.
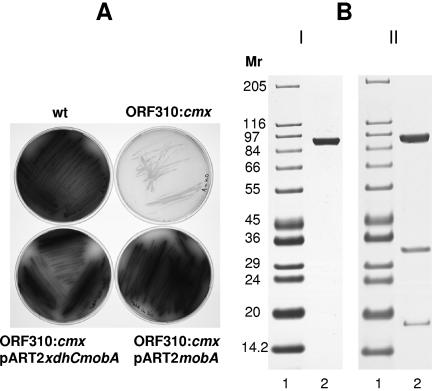
In the cmx-disrupted strain, no NDH or KDH activity could be measured. Transcripts of the kdhL and kdhMS genes (Fig. 2D) or ndh genes (not shown), however, were present. Since no transcripts of the genes similar to xdhC and mobA were seen in the disrupted strain but the XdhC and MobA proteins were implicated in molybdenum cofactor insertion into xanthine dehydrogenase (16, 17) and molybdopterin dinucleotide biosynthesis (13), respectively, we introduced these genes into the ORF310-disrupted strain. The catabolism of nicotine requires active NDH and KDH, and its end product, nicotine blue, is an indicator that the pathway is active. The wild-type strain produced blue pigment on nicotine plates, but the disrupted strain, which showed no NDH or KDH activity, did not (Fig. 3, wt and ORF310::cmx).
FIG. 3.
Complementation of A. nicotinovorans strain ORF310::cmx/pAO1 with mobA carried on pART2. (A) Blue pigment development on plates of citrate-nicotine medium inoculated with the A. nicotinovorans wild-type strain (wt), with the cmx-disrupted ORF310 strain (ORF310::cmx), with the cmx-disrupted ORF310 strain complemented with pART2xdhCmobA, and with the cmx-disrupted ORF310 strain complemented with pART2mobA. (B) His-tagged KdhL subunit derived from the cmx-disrupted ORF310 strain transformed with pART2kdhL (I) and KDH holoenzyme isolated from the cmx-disrupted ORF310 strain transformed with pART2kdhLmobA (II). The results shown are from sodium dodecyl sulfate-polyacrylamide gel electrophoresis of proteins purified by Ni-chelating chromatography.
Both xdhC and mobA or xdhC and mobA individually were amplified from A. nicotinovorans whole cells in PCRs with the primer pairs listed in Table 1, and the restriction enzyme-digested PCR products were inserted into the multiple cloning site of pART2 (21). When the genes similar to xdhC and mobA were introduced on pART2 into the disrupted strain, the transformants regained the ability to produce blue pigment on nicotine plates (Fig. 3A, ORF310:cmx/pART2xdhCmobA). Complementation with the gene similar to mobA only proved sufficient to restore blue pigment formation (Fig. 3A, ORF310::cmx/pART2mobA). The cmx-disrupted strain complemented with pART2xdhC only did not produce blue pigment (not shown) and looked identical to the plate shown for the cmx-disrupted strain without complementation. The function of the gene similar to xdhC in A. nicotinovorans remains to be established.
Recovery of holoenzyme containing His-tagged KdhL from cmx-disrupted ORF310 strain and mobA-complemented A. nicotinovorans.
A. nicotinovorans with cmx-disrupted ORF310 was transformed with pART2kdhL. When a bacterial extract was prepared from this strain and passed over Ni-chelating Sepharose, only recombinant KdhL was recovered (Fig. 3B). However, when the strain was transformed with pART2 carrying mobA in addition to kdhL (pART2mobAkdhL), all three subunits of KDH were isolated, and the holoenzyme showed the same specific activity as the holoenzyme assembled in the wild-type strain transformed with pART2kdhL (Fig. 3B). The same result was obtained when an extract of pART2ndhL-transformed bacteria was used (not shown).
A scattered arrangement of the subunit genes of an enzyme is unusual. It requires the correlated biosynthesis and stoichiometric assembly of subunits produced from genes transcribed from different promoters. Therefore, experimental proof was required for the assignment of the kdh genes (1). This was provided by the assembly of the recombinant KdhL subunit with the native KdhM and KdhS subunits into the active KDH holoenzyme.
KDH was shown here to be an enzyme with an MCD cofactor. The MobA variant responsible for its synthesis was unknown. Our results strongly suggest that the pAO1 mobA-like gene encodes this molybdenum cofactor cytidylyl transferase. Despite many attempts, our efforts to isolate MobA in soluble form failed. Therefore, we could not test its enzyme specificity in vitro. However, the mobA complementation studies support the functional assignment of MobA as a molybdopterin cytosine dinucleotide biosynthesis protein. The low yield of NDH holoenzyme prevented formal proof of the nature of its cofactor. Since the lack of mobA expression abolished the enzyme activity of KDH as well as NDH, we consider it a reasonable assumption that NDH also contains a molybdopterin cytosine dinucleotide.
Our results show that the synthesis of MCD, and thus that of a functional MCD biosynthesis protein, MobA, is required for the assembly of the heterotrimeric KDH holoenzyme. Inability to synthesize the dinucleotide form of the molybdenum cofactor because of a deficient mobA gene resulted in the failure to assemble the holoenzyme.
Acknowledgments
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to R.B.
REFERENCES
- 1.Baitsch, D., C. Sandu, and R. Brandsch. 2001. A gene cluster on pAO1 of Arthrobacter nicotinovorans involved in the degradation of the plant alkaloid nicotine: cloning, purification and characterization of 2,6-dihydroxypyridine 3-hydroxylase. J. Bacteriol. 183:5262-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthold, H., M. Scanarini, C. C. Abney, B. Frorath, and W. Northemann. 1992. Purification of recombinant antigenic epitopes of the human 68-kDa (U1) ribonucleoprotein antigen using the expression system pH6EX3 followed by metal chelating affinity chromatography. Protein Expr. Purif. 3:50-56. [DOI] [PubMed] [Google Scholar]
- 3.Bononin, I., B. M. Martins, V. Purvanov, S. Fetzner, R. Huber, and H. Dobbek. 2004. Active site geometry and substrate recognition of the molybdenum hydroxylase quinoline 2-oxidoreductase. Structure 12:1425-1435. [DOI] [PubMed] [Google Scholar]
- 4.Brandsch, R. 2006. Microbiology and biochemistry of nicotine degradation. Appl. Microbiol. Biotechnol. 69:493-498. [DOI] [PubMed] [Google Scholar]
- 5.Brühmüller, M., A. Schimz, L. Messmer, and K. Decker. 1975. Covalently bound FAD in d-6-hydroxynicotine oxidase. J. Biol. Chem. 250:7747-7751. [PubMed] [Google Scholar]
- 6.Chiribau, C.-B., C. Sandu, M. Fraaije, E. Schiltz, and R. Brandsch. 2004. A novel γ-N-methylaminobutyrate demethylating oxidase involved in catabolism of the tobacco alkaloid nicotine by Arthrobacter nicotinovorans pAO1. Eur. J. Biochem. 271:4677-4684. [DOI] [PubMed] [Google Scholar]
- 7.Chiribau, C.-B., C. Sandu, G. L. Igloi, and R. Brandsch. 2005. Characterization of PmfR, the transcriptional activator of the pAO1-borne purU-mabO-folD operon of Arthrobacter nicotinovorans. J. Bacteriol. 187:3062-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobbek H., L. Gremer, R. Kiefersauer, R. Huber, and O. Meyer. 2002. Catalysis at a dinuclear [CuSMo(=O)OH] cluster in a CO dehydrogenase resolved at 1.1-A resolution. Proc. Natl. Acad. Sci. USA 99:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freudenberg, W., K. König, and J. R. Andreesen. 1988. Nicotine dehydrogenase from Arthrobacter oxidans: a molybdenum-containing hydroxylase. FEMS Microbiol. Lett. 52:13-18. [Google Scholar]
- 10.Gartemann, K. H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J. Bacteriol. 183:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gremer, L., and O. Meyer. 1996. Characterization of xanthine dehydrogenase from the anaerobic bacterium Veillonella atypica and identification of a molybdopterin-cytosine-dinucleotide-containing molybdenum cofactor. Eur. J. Biochem. 236:862-866. [DOI] [PubMed] [Google Scholar]
- 12.Grether-Beck, S., G. L. Igloi, S. Pust, E. Schiltz, K. Decker, and R. Brandsch. 1994. Structural analysis and molybdenum-dependent expression of the pAO1-encoded nicotine dehydrogenase genes of Arthrobacter nicotinovorans. Mol. Microbiol. 13:929-936. [DOI] [PubMed] [Google Scholar]
- 13.Guse, A., C. E. M. Stevenson, J. Kuper, G. Buchanan, G. Schwarz, G. Giordano, A. Magalon, R. R. Mendel, D. M. Lawson, and T. Palmer. 2003. Biochemical and structural analysis of the molybdenum cofactor biosynthesis protein MobA. J. Biol. Chem. 278:25302-25307. [DOI] [PubMed] [Google Scholar]
- 14.Hille, R. 1996. The mononuclear molybdenum enzymes. Chem. Rev. 96:2757-2816. [DOI] [PubMed] [Google Scholar]
- 15.Igloi, G. L., and R. Brandsch. 2003. Sequence of the 165-kilobase catabolic plasmid pAO1 from Arthrobacter nicotinovorans and identification of a pAO1-dependent nicotine uptake system. J. Bacteriol. 185:1976-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov, N. V., F. Hubálek, M. Trani, and D. E. Edmondson. 2003. Factors involved in the assembly of a functional molybdopyranopterin center in recombinant Comamonas acidovorans xanthine dehydrogenase. Eur. J. Biochem. 270:4744-4754. [DOI] [PubMed] [Google Scholar]
- 17.Neumann, M., M. Schulte, N. Jünemann, W. Stöcklein, and S. Leimkühler. 5 April 2006, posting date. Rhodobacter capsulatus XDHC is involved in molybdenum cofactor binding and insertion into xanthine dehydrogenase. J. Biol. Chem. [Online.] doi: 10.1074/jbc.M601617200. [DOI] [PubMed]
- 18.Parschat, K., B. Hauer, R. Kappl, R. Kraft, J. Hüttermann, and S. Fetzner. 2003. Gene cluster of Arthrobacter ilicis Rü61a involved in the degradation of quinaldine to anthranilate. J. Biol. Chem. 278:27483-27494. [DOI] [PubMed] [Google Scholar]
- 19.Rajagopalan, K. V. 1997. Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans. 25:757-761. [DOI] [PubMed] [Google Scholar]
- 20.Sandu C., C.-B. Chiribau, and R. Brandsch. 2003. Characterization of HdnoR, the transcriptional repressor of the 6-hydroxy-d-nicotine oxidase gene of Arthrobacter nicotinovorans pAO1, and its DNA-binding activity in response to l- and d-nicotine derivatives. J. Biol. Chem. 278:51307-51315. [DOI] [PubMed] [Google Scholar]
- 21.Sandu, C., C.-B. Chiribau, P. Sachelaru, and R. Brandsch. 2005. Plasmids for nicotine-dependent and -independent gene expression in Arthrobacter nicotinovorans and other Arthrobacter species. Appl. Environ. Microbiol. 71:8920-8924. [DOI] [PMC free article] [PubMed] [Google Scholar]