Abstract
A DNA microarray platform for the characterization of bacterial communities in freshwater sediments based on a heterogeneous set of 70 16S rRNA-targeted oligonucleotide probes and directly labeled environmental RNA was developed and evaluated. Application of a simple protocol for the efficient background blocking of aminosilane-coated slides resulted in an improved signal-to-noise ratio and a detection limit of 10 ng for particular 16S rRNA targets. An initial specificity test of the system using RNA from pure cultures of different phylogenetic lineages showed a fraction of false-positive signals of ∼5% after protocol optimization and a marginal loss of correct positive signals. Subsequent microarray analysis of sediment-related community RNA from four different German river sites suggested low diversity for the groups targeted but indicated distinct differences in community composition. The results were supported by parallel fluorescence in situ hybridization in combination with sensitive catalyzed reporter deposition (CARD-FISH). In comparisons of the data of different sampling sites, specific detection of populations with relative cellular abundances down to 2% as well as a correlation of microarray signal intensities and population size is suggested. Our results demonstrate that DNA microarray technology allows for the fast and efficient precharacterization of complex bacterial communities by the use of standard single-cell hybridization probes and the direct detection of environmental rRNA, also in methodological challenging habitats such as heterogeneous lotic freshwater sediments.
DNA microarrays represent a high-throughput format for the highly parallel application of multiple nucleic acid probes by reverse hybridization and, therefore, one of the most powerful tools in molecular biology. They are now routinely applied in the pharmaceutical industry, clinical diagnostics, and various fields of research such as functional genomics and genetic analysis (18). In recent years, DNA microarrays have also entered the field of microbial ecology. Here, the technology potentially allows for the nearly complete qualitative description of even complex microbial communities within a single experiment by applying large sets of probes targeting various sequence signatures of a phylogenetic marker gene (6).
The first proof of principle for the parallel detection of bacteria using a 16S rRNA-based DNA microarray was given by Guschin et al. in 1997 (12), followed by various studies focusing on selected aspects of 16S rRNA-based arrays (7, 20, 44) and also showing the general applicability of microarray analysis for studying the composition of even complex environmental microbial communities (5, 21, 46). These studies were exclusively based on the initial amplification of the target molecules by PCR. However, despite the potential of this technology, there is thus far no broad application of DNA microarrays in microbial ecology and applied biotechnology. Apparently, this is mainly due to methodological challenges such as providing an appropriate level of hybridization specificity, limited detection sensitivity, and laborious protocols for preparation of the target molecules.
Recently, extracted and directly labeled bacterial community RNAs from the environment were successfully analyzed by microarray hybridization (9, 31). This promises a less distorted view of true community composition, since PCR-based methods have been shown to fail to correctly reflect microbial communities (36, 43, 47). On the other hand, RNA extraction yields from environmental samples are often low and limit the sensitivity of the analysis. While cells within a water column can easily be enriched for the extraction of sufficient amounts of RNA by, e.g., filtration of larger volumes of water (31), a simple enrichment of prokaryotic cells is not possible from soils and sediments for practical reasons. Moreover, the extraction of intact cellular RNA is often challenging due to the inhibitory effects caused by complex organic molecules, mainly humic substances, or nucleases released from eukaryotic cells, which often occur in high numbers within sediment samples (1). However, since microorganisms play an important role in the biogeochemical cycles and mineralization processes of organic and inorganic compounds in marine and freshwater sediments, a PCR-free DNA microarray system for the specific and highly parallel detection of the corresponding populations is of major interest. The goal for the application of such systems is the fast and efficient characterization of microbial communities without the loss of the basic advantages of molecular techniques compared to “classical” cultivation- or “black-box”-based approaches, enabling, e.g., the extensive comparison of samples from different sites or time points at a high level of phylogenetic resolution.
In this study, a microarray platform for the direct detection of fluorescently labeled RNA from freshwater sediments based on 70 16S rRNA targeting already published and well-characterized oligonucleotide probes was developed and characterized. Specificity was initially evaluated with labeled RNAs from pure cultures, and hybridization results from environmental samples were validated by parallel fluorescence in situ hybridization combined with catalyzed reporter deposition (CARD-FISH) to maximize sensitivity of the analysis.
MATERIALS AND METHODS
Bacterial strains and sediment samples.
For initial evaluation of the probe set and hybridization conditions, microarray experiments were conducted with RNA from the four bacterial reference strains Lactococcus lactis (Firmicutes), Rhodopirellula baltica (Planctomycetales), Magnetospirillum gryphiswaldense (Alphaproteobacteria), and Escherichia coli (Gammaproteobacteria). L. lactis was purchased from the DSMZ (Braunschweig, Germany) (DSM 4366) and grown at 30°C in LB medium. R. baltica SH1T (35) and M. gryphiswaldense strain MSR-1 (34) were grown according to Schlesner et al. (35) and Schübbe et al. (38), respectively. For analysis of E. coli RNA, commercially available purified rRNA from E. coli MRE 600 (Boehringer, Ingelheim, Germany) was used.
For parallel microarray and FISH analysis of environmental bacterial communities, surface samples of the uppermost 2 to 5 cm of recent sediments from different German river catchment areas (Ehrenbreitstein, river Rhein, 591 km; Hohenwutzen, river Odra, 655 km; Fahlberg-List, river Elbe, 319.4 km; Dömitz, Müritz-Elde waterway, 1 km) were collected using a grab sampling device and transferred to the laboratory on ice. There, samples were stored at −80°C until further processing.
RNA extraction, purification, and labeling.
Total cellular RNA was extracted from L. lactis and R. baltica cultures according to Peplies et al. (31) and from M. gryphiswaldense according to Oelmüller et al. (29). Total RNA from sediment samples was extracted from approximately 15 g of sediment using a large-scale extraction protocol by MacGregor (25, 26, 42) with the following modifications: bead-beating was done three times for 40 s each time, and final nucleic acid precipitation was done with 1 volume of absolute ethanol and 2 volumes of ammonium acetate. The latter was necessary to prevent contamination of the extracts, which would interfere with the subsequent labeling reaction. For additional purification of the RNA extracts, the MultiScreenTM 96-well Separation System (Millipore, Billerica, MA) packed with Sephadex G-50 superfine gel filtration resin (Sigma-Aldrich, St. Louis, Mo.) was used according to the manufacturer's protocol. The quality and quantity of rRNA within the total RNA extracts were checked by standard agarose gel electrophoresis with a dilution series of an E. coli rRNA standard from Boehringer (Ingelheim, Germany) as well as by capillary gel electrophoresis using a 2100 Bioanalyzer from Agilent Technologies (Palo Alto, Calif.).
Total RNA from pure cultures and sediment samples was directly chemically labeled with Alexa Fluor 488 using a ULYSIS labeling kit from Molecular Probes (Eugene, Oreg.) as previously described for total RNA from bacterioplankton samples (31). An amount of 1 μg of rRNA was added to the labeling reaction. Labeled RNA was eluted in 75 μl of diethyl pyrocarbonate-treated water after the final purification step. Finally, the quantity of labeled rRNA was determined as described above. Labeling efficiency was determined by UV spectrometry according to the manufacturer's protocol using an ND-1000 Spectrophotometer (NanoDrop Technologies, Delaware) allowing for the analysis of a sample volume of 1 μl. Labeled RNA was stored at −18°C.
Oligonucleotide probe set.
A set of 70 redundant and hierarchically structured 16S rRNA-targeted oligonucleotide probes, approximately 20 nucleotides in length and originally designed for fluorescence in situ or membrane hybridization, was chosen from the literature with the focus on covering the phylogenetic groups that are expected in river sediments, based on published cultivation and molecular studies; additional probes targeting selected indicator organisms of the drinking water ordinance and bacteria known for degrading pollutants were also chosen. The probes and their characteristics are listed in Table 1. For the general probes UNIV1392, EUB338, and ALF968, control oligonucleotides comprising single central mismatches (not listed in Table 1) were also applied to assess the specificity of hybridization at a high level of resolution. As an additional general negative control probe, NON338 (reverse complementary to probe EUB338; not listed in Table 1) was used. The current specificity of the probes and the number of mismatches to reference strains were evaluated using the PROBE_MATCH and ARB_EDIT tools of the software package ARB (24) with the current small-subunit rRNA data set of the technical University of Munich (released January 2004; available at http://www.arb-home.de) which contains nearly 40,000 almost full-length sequences (longer than 1,449 nucleotides). In addition, probe specificity was checked against the RDP-II data set (release 9) using the Probe Match function at http://rdp.cme.msu.edu/index.jsp.
TABLE 1.
Oligonucleotide probe set applied for microarray analysis
| Probe no. | Probe name | Target organism(s) | Sequence (5′-3′) | Referencea |
|---|---|---|---|---|
| 1 | UNIV1392 (UNIV1390) | All organisms | ACGGGCGGTGTGTAC | pB |
| 2 | EUK1195 | Most Eukarya | GGGCATCACAGACCTG | pB |
| 3 | Arch915 | Most Archaea | GTGCTCCCCCGCCAATTCCT | pB |
| 4 | EURY514 | Most Euryarchaeota | GCGGCGGCTGGCACC | pB |
| 5 | EUB338 | Most bacteria | GCTGCCTCCCGTAGGAGT | pB |
| 6 | EUB338II | Planctomycetales | GCAGCCACCCGTAGGTGT | pB |
| 7 | EUB338III | Verrucomicrobiales | GCTGCCACCCGTAGGTGT | pB |
| 8 | ACA652 (ACA23A) | Acinetobacter | ATCCTCTCCCATACTCTA | pB |
| 9 | HGC236 | Actinobacteria | AAC AAG CTG ATA GGC CGC | 10 |
| 10 | AC840a (ACI-840-1) | acl subgroup of Actinobacteria | TCGCACAAACCGTGGAAG | 45 |
| 11 | AC840b (ACI-840-2) | acl subgroup of Actinobacteria | TCGCAGAAACCGTGGAAG | 45 |
| 12 | AC1219b | acl subgroup of Actinobacteria | TAGCGTGTTTGCAGCCCT | 45 |
| 13 | AERO1244 | Aeromonas | GCTTGCAGCCCTCTGTACGCG | 4 |
| 14 | ALBO577 | Alcaligenes, Bordetella, and close relatives | CCGAACCGCCTGCGCAC | pB |
| 15 | ALF968 | Alphaproteobacteria, except of Rickettsiales | GGTAAGGTTCTGCGCGTT | pB |
| 16 | Amar839 | Amaricoccus | CTGCGACACCGAACGGCAAGCC | pB |
| 17 | MYBM1171 | Methylobacterium | ATCCACACCTTCCTCGCGGC | 15 |
| 18 | PAR651 | Paracoccus | ACCTCTCTCGAACTCCAG | pB |
| 19 | AQUA841 | Aquabacteria | GCTTCGTTACTGAACAGCAAG | 15 |
| 20 | AT1458 | Azoarcus-Thauera cluster | GAATCTCACCGTGGTAAGCGC | pB |
| 21 | Azo1251 | Azoarcus tolulyticus | CGCGCTTTGGCAGCCCT | pB |
| 22 | BLS1295 | Bacillus-Lactobacillus-Streptococcus cluster | GCAGCCTACAATCCGAACTGAGA | pB |
| 23 | LGC353b | Bacillus | GCGGAAGATTCCCTACTGC | pB |
| 24 | BONE23a | beta1 subgroup of Betaproteobacteria | GAATTCCATCCCCCTCT | pB |
| 25 | BTWO23a | beta2 subgroup of Betaproteobacteria | GAATTCCACCCCCCTCT | pB |
| 26 | LDI23a (LDI) | Leptothrix discophora | CTCTGCCGCACTCCAGCT | pB |
| 27 | SNA23a (SNA) | Sphaerotilus natans | CATCCCCCTCTACCGTAC | pB |
| 28 | Clost I | Subgroup of Clostridia cluster I and II | TTCTTCCTAATCTCTACGCA | pB |
| 29 | III1421 | Cluster III of Clostridiaceae | CTACGGACTTCGGGTGTTCCCG | pB |
| 30 | CLOBU1022 | Clostridia | CCTGCCACCGAAGTGGCT | 4 |
| 31 | CF319a | Flavobacteria-Cytophaga group | TGGTCCGTGTCTCAGTAC | pB |
| 32 | CF319b | Flavobacteria-Cytophaga group | TGGTCCGTATCTCAGTAC | pB |
| 33 | BAC303 | Prevotella and Bacteroides within Bacteroidetes | CCAATGTGGGGGACCTT | pB |
| 34 | CFB286 | Mainly freshwater isolates within CFB cluster | TCCTCTCAGAACCCCTAC | pB |
| 35 | CYA361 | Most Cyanobacteria | CCCATTGCGGAAAATTCC | pB |
| 36 | SRB385Db | Some sulfate-reducing bacteria of the Deltaproteobacteria | CGGCGTTGCTGCGTCAGG | pB |
| 37 | SRB385 | Some sulfate-reducing bacteria of the Deltaproteobacteria | CGGCGTCGCTGCGTCAGG | pB |
| 38 | DSS658 | Desulfosarcina, Desulfofaba, Desulfococcus, Desulfofrigus | TCCACTTCCCTCTCCCAT | pB |
| 39 | DSV1292 | Desulfovibrio desulfuricans | CAATCCGGACTGGGACGC | pB |
| 40 | DSMA488 | Desulfomonile | GCCGGTGCTTCCTTTGGCGG | pB |
| 41 | DBM221 | Desulfobacterium | TGCGCGGACTCATCTTCAAA | pB |
| 42 | DSV687 | Geobacter, Desulfuromusa, Desulfovibrio, Desulfomicrobium | TACGGATTTCACTCCT | pB |
| 43 | ENT183 | Enterobacteriaceae | CTCTTTGGTCTTGCGACG | pB |
| 44 | Enterbact D | Some members of the Enterobacteriaceae | TGCTCTCGCGAGGTCGCTTCTCTT | pB |
| 45 | EURY499 | Methanosarcina, Methanosaeta, Methanomicrobiales groups | CGGTCTTGCCCGGCCCT | pB |
| 46 | EURY496 | Methanomicrobiales group | GTCTTGCCCGGCCCTTTC | pB |
| 47 | LGC | Firmicutes | TCACGCGGCGTTGCTC | pB |
| 48 | LGC354A | Firmicutes | TGGAAGATTCCCTACTGC | pB |
| 49 | LGC354B | Firmicutes | CGGAAGATTCCCTACTGC | pB |
| 50 | LGC354C | Firmicutes | CCGAAGATTCCCTACTGC | pB |
| 51 | Mmb1121 | Methylomonas album | CATCACGTGTTGGCAACTAA | pB |
| 52 | LEG705 | Legionella | CTGGTGTTCCTTCCGATC | pB |
| 53 | Am445 | Methylocystis, Methylosinus | CTTATCCAGGTACCGTCATTATCGTCCC | pB |
| 54 | Am976 | Methylocystis, Methylosinus | GTCAAAAGCTGGTAAGGTTC | pB |
| 55 | NSO1225 | Betaproteobacterial ammonia-oxidizing bacteria | CGCCATTGTATTACGTGTGA | pB |
| 56 | Nso190 | Betaproteobacterial ammonia-oxidizing bacteria | CGATCCCCTGCTTTTCTCC | pB |
| 57 | NIT3 | Nitrobacter | CCTGTGCTCCATGCTCCG | pB |
| 58 | Ntspa662 | Nitrospira | GGAATTCCGCGCTCCTCT | pB |
| 59 | Nsm156 | Nitrosomonas | TATTAGCACATCTTTCGAT | pB |
| 60 | NSR826 | Freshwater Nitrospira spp. | GTAACCCGCCGACACTTA | pB |
| 61 | Amx820 | Anaerobic ammonium-oxidizing bacteria | AAAACCCCTCTACTTAGTGCCC | 37 |
| 62 | NSR1156 | Nitrospira | CCCGTTCTCCTGGGCAGT | pB |
| 63 | NEU23a (NEU) | Nitrosomonas sp. | CCCCTCTGCTGCACTCTA | pB |
| 64 | PLA46 | Planctomycetes | GACTTGCATGCCTAATCC | pB |
| 65 | Pae997 | Pseudomonas | TCTGGAAAGTTCTCAGCA | pB |
| 66 | Ppu56a | Pseudomonas putida, P. mendocina | GCTGGCCTAACCTTC | pB |
| 67 | Ppu646 | Pseudomonas putida | CTACCGTACTCTAGCTTG | 15 |
| 68 | Strc493 | Most Streptococcus spp. and some Lactococcus spp. | GTTAGCCGTCCCTTTCTGG | pB |
| 69 | Str | Streptococcus | CACTCTCCCCTTCTGCAC | pB |
| 70 | Flavo1004 | Flavobacterium isolate | GGTCTGTTTCCAAACCGG | 4 |
pB, details on oligonucleotide probes are available at probeBase (www.microbial-ecology.net/probebase) (22).
Microarray matrix, probe configuration, and spotting.
5′-amino-modified capture oligonucleotides (Metabion, Martinsried, Germany) were spotted in 10 replicates onto GAPS II aminosilane-coated glass slides from Corning (Schiphol, The Netherlands) using a SpotArray24 spotting device with TeleChem Stealth pins (Packard Biochip Technologies, Billerica, MA).
For reduction of surface-mediated steric hindrance during hybridization, probes were used with polyadenosine triphosphate spacers six nucleotides in length, located at the 5′ end of the capture probes (31), except for probe UNIV1392, which was used with a 18-mer spacer. In addition, a control oligonucleotide with an artificial sequence (5′-GACTGACTGACTGA-3′) was spotted, targeting a 5′ Cy3-labeled reverse complementary oligonucleotide (Metabion, Martinsried, Germany), and added to the hybridization buffer with a final concentration of 15 nM.
The concentration of the capture oligonucleotides in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) spotting buffer with 2% glycerol was 20 μM. Normal spot diameter was approximately 100 μm under the spotting conditions applied. Postprocessing of the spotted slides including covalent immobilization of the capture probe was done according to the GAPS II slide manufacturer's protocol.
Pretreatment and hybridization of DNA microarrays.
Before hybridization, spotted slides were blocked in 50 ml of blocking solution (250 mM NaCl, 5 mM Tris-HCl, pH 8.0, 50% formamide, 0.5× SSC, 0.05% bovine serum albumin, and 1.0% blocking reagent) at 48°C for 1 h and air dried without additional washing as recently described (39). Blocking reagent (10%) was made as follows: 150 mM NaCl, 100 mM maleic acid, pH 7.5, and 100 mg/ml blocking reagent from Roche (Mannheim, Germany).
According to previous results (30, 31), microarrays were hybridized and washed based on a standard FISH protocol (33). Briefly, 60 μl of hybridization solution was made of 2.5 to 250 ng of Alexa Fluor 488-labeled rRNA for pure cultures and 500 ng for the sediment samples, respectively, and 1 pmol of the Cy3-labeled artificial standard oligonucleotide (see above) in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 8.0, 0.01% sodium dodecyl sulfate, and 1% blocking reagent), denatured at 90°C for 2 min, and cooled on ice. The hybridization solution was applied to a pretreated microarray under a 22- by 40-mm LifterSlip (Erie Scientific, Portsmouth, NH). Hybridization was conducted in a Corning hybridization chamber for 12 h at either 46, 54, or 58°C without formamide. After being washed in standard wash buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 8.0, 0.01% sodium dodecyl sulfate) at 48°C for 15 min, slides were dried by centrifugation at 700 rpm for 3 min.
Signal detection and data analysis.
All slides were imaged at a resolution of 10 μm using a ScanArray Express microarray scanner (Packard Biochip Technologies, Billerica, MA) at a sensitivity setting of the photomultiplier of 60% and a laser power of 90%.
For spot detection and quantification of spot signals (mean pixel intensity), the microarray analysis software Quantarray 3.0 (Packard Biochip Technologies, Billerica, MA) was used, and for raw data postprocessing the microarray data analysis software tool MADA (www.mpi-bremen.de/mada) was used. Briefly, each spot signal was tested for statistical significance, and series of probe replicates were tested for outliers using the t test and the outlier test implemented in MADA, respectively. Spot signals were processed further only if at least 6 of the 10 replicates spotted for each probe were considered positive by MADA. Each data point shown represents the arithmetic mean of the non-background-corrected mean pixel intensity of the positive replicates for the corresponding probe, normalized to the median global background signal of the array.
CARD-FISH validation.
Parallel FISH analysis combined with CARD was done for all four river sediments with the horseradish peroxidase-labeled probes EUK1195, EURY514, LGC, EURY499, EURY496, EUB338, NON338 (negative control), EUB II, EUB III, CF319a/CF319b, PLA46, BTWO23a, SRB385/SRB385Db, LGC354A/LGC354B/LGC354C, BLS1295, Am445, and DSS658 (purchased from Biomers, Ulm, Germany) at a hybridization temperature of 37°C and with formamide concentrations of 20% for probes 1 and 2, 40% for probes 3 to 5, 55% for probes 6 to 16, and 60% for probe 17. Hybridization stringencies were chosen according to the oligonucleotide probe database probeBase (22), except for probes EUK1195, BLS1295, Am445, and LGC, for which no information was available, and formamide concentrations were optimized in concentrations series.
Briefly, 2 g of sediment was fixed with 10 ml of 2% paraformaldehyde solution for 1 h at room temperature, and subsequently 200 μl of the fixative was mixed with 1 ml of 1× phosphate-buffered saline. For detachment of the sediment matrix from the cells, the solution was sonicated at 60% power for 10 s with sonication probe MS73 (Sonoplus HD70; Bandelin, Berlin, Germany), followed by centrifugation at 700 relative centrifugal force for 2 min and collection of the supernatant. The procedure was repeated two times, and the combined supernatants were filtered on 0.2-μm-pore-size white polycarbonate membrane filters (Millipore, Eschborn, Germany).
Cells on filter sections were embedded in 0.2% low-gelling-point agarose (MetaPhor; FMC Bioproducts) according to Pernthaler et al. (32) and permeabilized in a probe-specific way as described by Sekar et al. (40) and Ishii et al. (13). Hybridization, signal amplification, counterstaining with 4,6-diamidino-2-phenylindole (DAPI) mounting, and microscopical evaluation were done according to the standard protocol of Pernthaler et al. (32), except that tyramide-fluorescein was used as a substrate for the horseradish peroxidase in a 1:1,000 dilution.
RESULTS
Preparation of labeled target RNAs from sediment samples.
Total RNA from river sediments was extracted with yields of up to 0.13 μg of intact prokaryotic rRNA per gram of sediment from the Ehrenbreitstein (EB), Fahlberg-List (FL), and Dömitz (DÖ) sites. From the Hohenwutzen (HW) site, a much higher yield of approximately 0.80 μg of rRNA per gram of sediment was obtained. The total amount of nucleic acids extracted including DNA contaminations ranged between 0.19 μg/g of sediment and 7.80 μg/g of sediment for sites EB and HW, respectively.
Using an unmodified large-scale RNA extraction protocol, all four extracts appeared brownish and nearly nontransparent, especially in case of the DÖ sample. Since the subsequent labeling reaction was inhibited by the presence of these putatively polyaromatic organic compounds, variations in the parameters of the extraction protocol were tested. Ultimately, the nucleic acid precipitation step was done with 1 volume of absolute ethanol and 2 volumes of ammonium acetate instead of 1 volume of isopropanol and 0.5 volumes of ammonium acetate, resulting in a clear reduction of the amount of unwanted compounds for all four samples. Extraction yields were not affected by this modification. Residual coloring was removed by additional gel filtration, except for the DÖ extract, which remained slightly brownish. Alternative purification strategies initially tested, e.g., based on columns or standard gel electrophoresis, led to the nearly complete loss of the target rRNA.
The relative labeling efficiency of the target molecules was similar for all four samples. Spectroscopic data indicated the presence of one label for every 43 nucleotides, on average.
Evaluation and optimization of microarray hybridization with pure bacterial cultures.
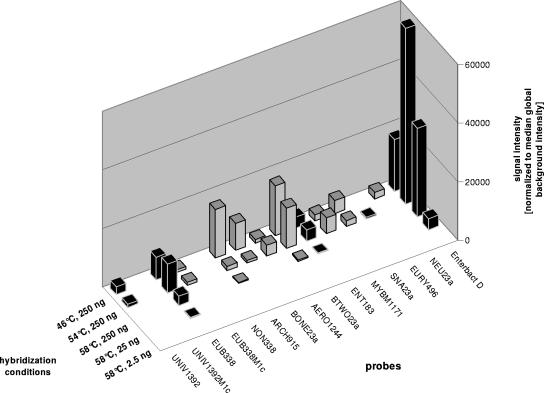
The general suitability of the oligonucleotide probe set listed in Table 1 for microarray analysis of 16S rRNA targets in terms of specificity was evaluated by initial hybridization of 250 ng of labeled rRNA from the E. coli reference strain at 46°C (Fig. 1). Besides the four correct positive signals for probes UNIV1392, EUB338, ENT183, and Enterbact D, eight false-positive signals were found within the data set (10.7% of all potential false-positive events). Five of the false-positive signals possessed five or even more mismatches to the 16S rRNA of E. coli. No hits at all were found for these probes on the 23S rRNA of E. coli. Except for probe ARCH915, probes with five or more mismatches showed comparably low signal intensities. To optimize the specificity of analysis, the hybridization temperature was gradually raised. While the identical number of false-positive signals was found at 54°C, only two weak false-positive signals were observed at 58°C for the probes BTWO23a (one mismatch) and ARCH915 (five mismatches). The correct positive signal of probe UNIV1392 was lost at 58°C. Remarkably, for the correct positive probes EUB338 and Enterbact D, the strongest signals were observed at 54°C. While hybridization of 25 ng (1:10 dilution) of E. coli rRNA at 58°C led to correct positive signals only for the probes EUB338 and Enterbact D, no signals were observed after hybridization of 2.5 ng (Fig. 1). Since the hybridized rRNA pool consisted roughly of 40% of 16S rRNA, as verified by capillary gel electrophoresis, the detection limit of our format was found to be as low as approximately 10 ng of a particular population of 16S rRNA molecules but seemed to be probe specific in response to the different hybridization efficiencies of the probes.
FIG. 1.
Effect of hybridization temperature and target amount on microarray hybridization of pure rRNA from the E. coli reference strain with the complete set of probes listed in Table 1. Correct positive and false-positive signals are indicated by black and gray bars, respectively. For clarity, only probes with positive signals plus the control probes UNIV1392M1c and NON338 are shown. Mean standard deviation for the replicates of all positive probes was 14.0% of normalized signal intensities, with a minimum of 0.8% for probe Enterbact D at 58°C and 250 ng and a maximum of 44.0% for probe ARCH915 at 46°C and 250 ng (error bars are not indicated in the chart).
Subsequently, the specificity of the microarray analysis was also tested with three additional reference strains. A comprehensive overview of the signal patterns obtained by hybridization of all four strains at 46 and 58°C is given in Fig. 2. All probes for which hybridization was expected possessed a correct positive signal at 46°C with the exception of probe UNIV1392 for the Lactococcus reference strain. However, a total of 11, 11, and 8 false-positive signals were found for RNAs of Magnetospirillum, Rhodopirellula, and Lactococcus, respectively. Together with the results of the E. coli reference rRNA, approximately 14% of all probes targeting none of the 16S RNAs showed false-positive signals, strongly hampering data interpretation. Except for probe ARCH915, which was not further considered, no correlation of nonspecific hybridization and probe characteristics such as G+C content, length, or presence of long G+C stretches could be observed, and 29% of the false-positive signals were based on more than four mismatches. By increasing the hybridization temperature to 58°C, the number of false-positive signals for RNAs of Magnetospirillum, Rhodopirellula, and Lactococcus could be reduced to three, seven, and two, respectively. Now, approximately 5% of all probes targeting none of the 16S RNAs showed nonspecific hybridization signals (including the results of the E. coli reference rRNA) based on one to three mismatches, except for probes Am445 and DSS658, both with four mismatches to Magnetospirillum. However, probe Am445 represents the longest of all probes applied with 28 nucleotides and has only slight similarity to the probe binding sites of the other three reference strains, and for probe DSS658 three of the four mismatches are at the 5′ end terminal position of the probe binding site of Magnetospirillum. Correct positive signals were lost for only probes UNIV1392 and LGC with the E. coli (Fig. 1) and Lactococcus reference strains, respectively. The optimized signal patterns allowed for improved data interpretation based on the multiple probe approach. Phylogenetic assignment of Magnetospirillum and Rhodopirellula was limited by the lack of additional probes targeting these particular organisms or corresponding higher-level phylogenetic groups since the probe set was not primarily designed for the identification of selected organisms on a high level of resolution; however, E. coli and Lactococcus could be clearly phylogenetically assigned by the redundant and nested probes available.
FIG. 2.
Comprehensive overview of the signal patterns obtained by microarray hybridization of 250 ng of rRNA from all four reference strains at 46 and 58°C. Correct positive and false-positive signals are indicated by black and open rectangles, respectively. The values on top of each column indicate the number of false-positive signals of the corresponding hybridization experiment. For clarity, normalized signal intensities and standard deviations are not shown.
Comparative microarray analysis of sediment samples and CARD-FISH validation.
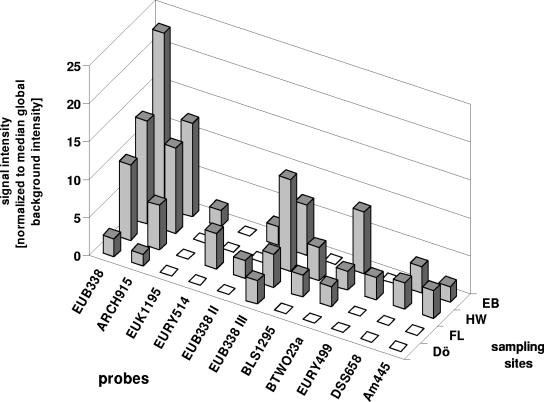
Total community RNAs from sediments of the four river sites were hybridized to the complete set of 70 probes (Table 1) under optimized conditions (58°C) in independent microarray experiments. A total of eight, eight, seven, and three positive signals for the sites EB, HW, FL, and DÖ, respectively (Fig. 3), were found with distinct differences in the qualitative composition of the resulting signal patterns as well as in normalized signal intensities for probes that showed signals with all four samples. While for the two general probes EUB338 and ARCH915, strongest signals were observed at site EB (23.0- and 12.4-fold of median global background intensity, respectively) and weakest signals were observed at site DÖ (2.4- and 1.6-fold, respectively), the strongest signal for probe EUB338 III was found at site HW with 12.1-fold of median global background intensity (6.6-fold at EB, 4.4-fold at FL, and 3.1-fold at DÖ). All other positive probes showed signals at not more than two different sites with a maximum and minimum signal intensity of 8.3-fold for probe BTWO23a and 2.1-fold for probe Am445, respectively, both at site EB. Probes EUK1195, EURY514, and EURY499 showed unique signals at sites EB, FL, and HW with intensities of 2.1-, 4.7-, and 2.9-fold of median global background intensity, respectively. No signals were observed for the general negative control probe NON338 or for the single mismatch control of probe EUB338.
FIG. 3.
Comparative microarray analysis of fluorescently labeled total RNA from sediments of the four different river sites: EB, HW, FL, and DÖ. In each experiment, 500 ng of rRNA was hybridized at 58°C for 18 h to the complete set of 70 oligonucleotide probes listed in Table 1. For clarity, only probes with positive signals are shown. The mean standard deviation for the replicates of all positive probes was 14.8% of normalized signal intensities, with a minimum of 3.6% for probe ARCH915 at site DÖ and a maximum of 44.4% for probe ARCH915 at site HW (error bars are not indicated in the chart).
Parallel CARD-FISH analysis was conducted with sediments of all four sites and 20 probes from the microarray probe set, including all 10 probes that showed positive signals in the corresponding microarray experiments and 10 randomly chosen probes to also validate absent microarray signals. Probe ARCH915 was excluded from the FISH analysis due to common problems with hybridization specificity (unpublished observation). High morphological diversity was found within all samples. Total cell numbers ranged between 6.0 × 107 and 1.9 × 109 cells/g of sediment for sites DÖ and HW, respectively, and are listed in Table 2, as well as the relative cellular abundances of the populations analyzed. With the general bacterial probes EUB338, EUB II, and EUB III, total abundances of 97, 82, 76, and 41% were detected at the sites EB, HW, FL, and DÖ, respectively. A good correlation of the two data sets was found since 89% of the microarray data points validated were in good agreement with the corresponding FISH counts in a way that positive microarray signals matched with relative cellular abundances of 2% or higher and absent microarray signals matched with absent FISH signals or very small populations (clearly below 1%). Moreover, a dependency of microarray signal intensities and the size of particular populations are also indicated by our data. For example, for probe EUB338 a highly significant linear correlation of the normalized microarray signals and relative cellular abundances detected at the four river sites was found (n = 4; r = 0.989). False-positive microarray signals were suggested for only probes EUK1195 and Am445, both with RNA from site EB, since no corresponding FISH counts were observed. These two signals represent the lowest of all intensities within the four microarray data sets. Probe EURY496 showed no microarray signal at all, although a corresponding relative abundance of 2% was found at site HW. For probe EURY514, abundances of 1% were found at sites EB, HW, and FL, but a corresponding microarray signal was only detected for site FL. A similar situation was observed for probes EUB II, BLS1295, DSS 658, and Am445, which showed relative cellular abundances of 3, 2, 3, and 1%, respectively, in combination with absent microarray signals, but here positive microarray signals were observed for higher abundances at different sites (6% for probe EUBII, 4% for BLS1295, 4% DSS658, and 3% for Am445).
TABLE 2.
Quantitative FISH analysis and total cell counts of the four river sedimentsa
| Probe | EB
|
HW
|
FL
|
DÖ
|
||||
|---|---|---|---|---|---|---|---|---|
| Normalized microarray signal (±SD)b | % Hybridized cells of all DAPI-stained objects | Normalized microarray signal (±SD)b | % Hybridized cells of all DAPI-stained objects | Normalized microarray signal (±SD)b | % Hybridized cells of all DAPI-stained objects | Normalized microarray signal (±SD)b | % Hybridized cells of all DAPI-stained objects | |
| EUB 338 | 23.0 (±2.6) | 71 | 13.6 (±5.1) | 57 | 10.0 (±0.4) | 53 | 2.3 (±0.2) | 35 |
| EUB II | 2.5 (±0.1) | 6 | ND | 3 | 2.3 (±0.1) | 6 | ND | <1 |
| EUB III | 6.6 (±0.4) | 20 | 12.1 (±3.0) | 22 | 4.4 (±0.7) | 17 | 3.1 (±1.2) | 6 |
| EURY514 | ND | 1 | ND | 1 | 4.7 (±0.3) | 1 | ND | <1 |
| EURY499 | ND | <1 | 2.9 (±0.6) | 2 | ND | <1 | ND | ND |
| EURY496 | ND | <1 | ND | 2 | ND | <1 | ND | ND |
| EUK1195 | 2.1 (±0.1) | ND | ND | ND | ND | ND | ND | ND |
| BLS1295 | ND | 2 | 4.4 (±0.9) | 4 | 2.9 (±0.3) | 5 | ND | <1 |
| BTWO23a | 8.1 (±0.5) | 8 | 2.5 (±0.5) | 4 | 2.7 (±0.2) | 4 | ND | <1 |
| DSS658 | 3.4 (±0.6) | 4 | 3.5 (±0.8) | 6 | ND | 3 | ND | ND |
| Am445 | 2.1 (±0.2) | <1 | 3.7 (±0.9) | 3 | ND | 1 | ND | ND |
| CF319a+b | ND | ND | ND | ND | ND | ND | ND | ND |
| PLA46 | ND | ND | ND | ND | ND | ND | ND | ND |
| SRB385, SRB385Db | ND | ND | ND | <1 | ND | <1 | ND | ND |
| LGC | ND | ND | ND | <1 | ND | <1 | ND | ND |
| LGC354A-C | ND | ND | ND | ND | ND | ND | ND | ND |
Total cell counts were as follows: for EB, 4.0 × 108; for HW, 1.9 × 109; for FL, 1.3 × 109; DÖ, 6.0 × 107. Values in boldface are discussed in the text. Underlined values represent relative cellular abundances of 1% or higher in combination with absent microarray signals (false negatives). Since for the same probes, microarray signals were found for higher abundances, the absent signals are assumed to reflect a probe-specific detection limit of the format. All other values (not in boldface or underlined) represent matches of microarray signals and relative cellular abundances of 2% or higher as well as matches of absent microarray signals and relative cellular abundances well below 1% (indicated by <1 or ND, respectively).
According to the results shown in Fig. 3.
DISCUSSION
Since microarray technology has been introduced to environmental microbiology for high-throughput identification purposes, various studies demonstrated the applicability of the format, but only few have focused on the direct hybridization of rRNA extracted from complex prokaryotic communities (9, 31), even though it enables a less distorted view than PCR-based protocols of true community composition. The main challenge of the microarray-mediated RNA detection is to provide sufficient amounts of target molecules as well as to ensure adequate quality of the extracts for the subsequent labeling and hybridization reaction, especially in case of soil and sediment samples (41). While degradation of the rRNA extracts was not observed and no indications for a substantial extraction bias were suggested by the results of parallel FISH analysis as discussed below, low extraction yields had to be compensated by large amounts of starting material (15 g) compared to the study of El Fantroussi et al. (9), in which total RNA was extracted and hybridized from 0.5 g of soil. However, even for sediments the quality of RNA extracts can vary widely with the sampling site and time and do not represent a constraint of the microarray technology itself.
Applicability of the well-characterized probe set for the microarray-mediated direct detection of cellular rRNA in terms of specificity was initially tested with total RNA extracted from four phylogenetically distinct bacterial strains using a recently developed protocol (31). This was done to screen on a broad basis for probes that tend to give false-positive signals under the conditions applied. In contrast to our earlier study, where highly specific signal patterns could be obtained, in the current data set a considerable number of false-positive signals (∼14% of all probes) was found under identical conditions, and 29% of the nonspecific hybridization events were based on more than four mismatches. It must be pointed out that discrimination of single mismatches could be reproduced for the three probes applied in both studies (UNIV1392, EUB338, and ALF968). Therefore, we assume that the overall decrease in specificity reflects the hybridization characteristics of particular probes within the extended and heterogeneous probe set (32 to 87% G+C content; mean, 59% ± 10%). A conspicuous example is probe ARCH915, which even under optimized conditions showed strong signals with all four reference targets comprising up to five mismatches. Inherent nonspecific hybridization of this probe has also been found for other formats such as FISH (unpublished observation) and is presumably due to the long G/C stretch of 8 nucleotides within its sequence pattern that is not present in any other probe of the set. The comparable low intensities of the major part of the false-positive signals also suggest that the improved sensitivity of the protocol reported in this study contributed to the increased number of false-positive signals observed. However, by increasing the hybridization stringency, their number could be decreased to an adequate level without the loss of a notable number of correct positive signals caused by melting effects. A further increase of hybridization stringency led to an accumulated number of false-negative results (data not shown). Nevertheless, our results clearly support the outcome of recent studies (21, 23) which pointed out the requirement of nested probe sets to compensate for remaining false-positive and false-negative results since they cannot completely be avoided in a monostringent hybridization of multiple probes. However, in silico preselection of oligonucleotide probes according to probe characteristics such as the free energy ΔG of a given probe-target hybrid, as suggested by Loy et al. (23), should help to minimize the number of false data points in the future.
For the validation of the microarray data obtained from the environmental samples, parallel FISH in combination with highly sensitive CARD (32), also known as tyramide signal amplification, was used. Whole-cell hybridization techniques have routinely been applied for the molecular characterization of complex microbial communities for more than a decade, and they also provide quantitative data on the cellular level in terms of relative or even absolute abundances of selected populations (3). Recently, FISH analysis was also adapted to the characterization of microbial communities in lotic freshwater sediments (15, 17). Moreover, identical RNA-targeting probes can be used in both formats, enabling high comparability of the two data sets. Compared to microarray analysis, all probes are hybridized under individually optimized conditions in FISH analysis. So far, validation of microarray data in the field of environmental microbiology has mainly been based on nonquantitative methods, such as cloning and sequencing of the genetic markers (5, 21), or used a low level of resolution by focusing only on selected microarray results (23) or by applying very limited probe sets (16), respectively.
FISH validation provided evidence that highly specific signal patterns were obtained by microarray analysis of the four river sediments. Within the 22 positive microarray signals (positive signals for probe ARCH915 are not included), only two data points were not supported by the parallel FISH counts. This corresponds to a fraction of 2.5% of false-positive signals for the 80 microarray hybridization events considered (20 different probes with four samples minus 20 signals assigned as correct positive) and is in good agreement with the data from initial pure culture analysis. Remarkably, the two signals assigned as false positive (probes EUK1195 and Am445 with sample EB) were the weakest within the four microarray data sets (both 2.1-fold of the median global background). Presumably, they represent noise that could not be removed during data processing. This assumption is also supported by the absolute global background signal of the four hybridizations, which was the highest for sample EB (1.4-fold of sample FL and 1.9-fold of samples HW and DÖ). Optionally, the parameters for the test of the statistical significance of each spot signal were intensified, but then additional weak correct positive microarray signals, as indicated by FISH analysis, were lost. This led to an increased number of false-negative data points (data not shown).
Another good example for the requirement of multiple probes with redundant group coverage are the CARD-FISH results obtained for probes EURY499 and EURY496. While both probes consistently suggested a relative cellular abundance of 2% for the group of Methanomicrobiales and relatives for sample HW, a corresponding microarray signal could be detected only for probe EURY499. The occurrence of false-negative results is a common phenomenon in microarray analysis (21, 30) and is based on parameters that affect hybridization efficiency in a probe-specific way such as variation in the accessibility of probe binding sites caused by secondary structures of the target molecules (30). Interestingly, even small variations in the probe sequence appear potentially to result in clear differences in the hybridization efficiency. The 16S rRNA binding site of probe EURY496 is shifted by only two nucleotides as well as elongated by one nucleotide compared to probe EURY499. However, this finding corresponds to the data of Mir and Southern (28), who reported variations in hybridization signal intensities of up to a 33-fold decrease when the probe binding sites of 12-mer oligonucleotides were shifted by a single position.
In general, microarray and FISH data both suggest the presence of only part of the phylogenetic groups targeted by the probe set (approximately 20%) as well as minor differences in community composition at the four river sites investigated. For the general probe EUB338, cellular abundances were found that were in the range reported by Kloep et al. (15) for lotic freshwater sediments at different sites. While the low cellular abundances of the Planctomycetales found for various river sediments by Kloep et al. correspond well to the results of this study, probe CF319a yielded high cellular abundances in all sediments investigated by Kloep et al. (4 to 17%) but showed no positive signals in this study. However, Kloep et al. reported clear differences in the presence and abundance of particular phylogenetic groups, depending on seasonal shifts and the impact of the physicochemical parameters that characterize the sampling sites, habitats, and microenvironments. Also, the influence of specific surface features of bacteria affiliated with the Cytophaga-Bacteroidetes phylum on the RNA yield cannot be sufficiently judged at the moment. Finally, the probe set includes not only oligonucleotides targeting sequence signatures of microorganisms that are assumed to be ecologically relevant but also diagnostic probes for the evaluation of water quality, targeting, e.g., members of the Enterobacteriaceae.
An important aspect in DNA microarray analysis of environmental microbial communities is the sensitivity of the protocol applied, especially if an initial PCR amplification step in target molecule preparation is to be avoided. In this context, the 16S rRNA represents the phylogenetic marker of choice due its high copy number of up to 105 per cell (3). In addition, by far the largest number of sequence variants is deposited in the public databases for this marker, allowing for high-quality in silico evaluation of probe specificity. In the present study, four absent microarray signals were classified as false negative according to the parallel CARD-FISH analysis. For probes EUB II, BLS1295, DSS658, and Am445, no microarray signals were detected at particular sites, but corresponding relative cellular abundances of 3, 2, 3, and 1%, respectively, were observed. Remarkably, all four probes showed positive microarray signals at other sites in combination with an increased abundance ranging from 3 to 6% (next lowest abundance with a positive microarray signal). This finding suggests a probe-specific detection limit and is in good agreement with the commonly observed variation in hybridization efficiency of different capture probes (23, 30). Considering the complete data set, the detection limit of the format in terms of relative cellular abundance was found to be in the range of 2 to 5% after hybridization of 500 ng of environmental rRNA. In comparison, Loy et al. (23) have hybridized 400 ng of PCR products of the 16S rRNA gene and reported the detection of members of the betaproteobacterial order Rhodocyclales with a relative cellular abundance of less than 1% only after applying primer pairs that were highly specific for this particular phylogenetic group. Using more conserved primers targeting the pmoA gene of methanotrophic bacteria, Bodrossy et al. (5) were able to detect populations down to 5% of the community, as indicated by cloning and sequencing of the PCR products. El Fantroussi et al. (9) also detected bacterial populations by direct hybridization of 2 μg of total RNA extracted from sediment samples, but no additional information on quantitative community composition was provided. Our data show that the detection limit of the microarray-mediated direct profiling of environmental ribosomal RNAs is in the range of corresponding PCR-based approaches. This finding is clearly connected to the initial blocking procedure for aminosilane-coated slides adapted from Schübbe et al. (39), which strongly reduced adsorption of the labeled target molecules to the slide surface during hybridization compared to our previous protocol (31). The general applicability of signal amplification techniques to microarray analysis has already been demonstrated (8, 14) and will help to further increase the sensitivity of environmental studies in the future.
In the context of quantification of microbial populations, previous studies have shown that population sizes cannot directly be deduced from comparing the signal intensities of different probes within a single data set (23, 30), again due to the variations in probe hybridization efficiency. Nevertheless, comprehensive quantitative data can theoretically be achieved by empirical calibration of each capture probe applied since a linear correlation of signal intensities and the absolute amount of hybridized nucleic acids targeted by particular probes was repeatedly reported (11, 48). In the present study, we provide evidence that differences in signal intensities measured for particular probes at different sites reflect variations in relative cellular abundances of the corresponding populations. Examples are probe BTWO23a, targeting the beta-2 subgroup of the Betaproteobacteria (2), which showed the strongest microarray signal together with the highest cellular in situ abundance at site EB, and probe EUB338, which possesses a distribution of microarray signal intensities at the four sites that matched well with the distribution of the corresponding cellular abundances. In consequence, DNA microarrays not only allow for the qualitative detection of major populations but also provide a means to follow the dynamics of selected populations, e.g., by time series. However, it must be stressed that an inevitable limitation for quantification of microbial populations by reverse hybridization is represented by nucleic acid extraction biases (27, 41) and variations in the cellular RNA content (3).
In conclusion, we demonstrated the suitability of the DNA microarray format for the specific and sensitive detection of prokaryotic ribosomal RNAs extracted with low yields from complex environmental communities without further PCR amplification and based on common 16S rRNA-targeted oligonucleotide probes. Our results clearly suggest that microarrays can be used in a screening procedure for the reliable detection of predominant populations with cellular abundances down to 2% as revealed by parallel, highly sensitive CARD-FISH analysis. However, additional probes have to be applied in the future to cover more of the diversity expected for such habitats. Also the use of additional redundant and nested probes is an important aspect to further optimize data interpretation. Besides the specific detection of dominant populations, DNA microarrays will also allow the generation of complex fingerprints of microbial communities using modified protocols for the detection of additional, less abundant populations. Combined with, e.g., nonmetric multidimensional scaling, these data will help to quickly compare microbial communities on different levels of resolution, ranging from different river sites to different habitats and microenvironments. Fully integrated flowthrough hybridization and detection systems (19) will further increase the quality of microarray data and significantly speed up the procedure in the future. This is important to enable the high-throughput molecular characterization of environmental microbial communities also in terms of a highly parallel sample processing.
Acknowledgments
We thank A. Ellrott for setting up and maintaining technical equipment as well as for support with the microarray data analysis software tool MADA, B. MacGregor for help with the RNA extraction, and R. Amann and C. Würdemann for fruitful discussions. P. Ohlf is gratefully acknowledged for excellent technical assistance.
This work was supported in the scope of interdepartmental research by the German Federal Ministry of Transport, Building and Housing, the German Federal Ministry for the Environment, Nature Conservation and Nuclear Safety, and the Max Planck Society.
REFERENCES
- 1.Alm, E. W., and D. A. Stahl. 2000. Critical factors influencing the recovery and integrity of rRNA extracted from environmental samples: use of an optimized protocol to measure depth-related biomass distribution in freshwater sediments. J. Microbiol. Methods 40:153-162. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K.-H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böckelmann, U., W. Manz, T. R. Neu, and U. Szewzyk. 2000. Characterization of the microbial community of lotic organic aggregates (“river snow”) in the Elbe River of Germany by cultivation and molecular methods. FEMS Microbiol. Ecol. 33:157-170. [DOI] [PubMed] [Google Scholar]
- 5.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 6.Bodrossy, L., and A. Sessitsch. 2004. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 7:245-254. [DOI] [PubMed] [Google Scholar]
- 7.Chandler, D. P., G. J. Newton, J. A. Small, and D. S. Daly. 2003. Sequence versus structure for the direct detection of 16S rRNA on planar oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2950-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denef, V. J., J. Park, J. L. M. Rodrigues, T. V. Tsoi, S. A. Hashsham, and J. M. Tiedje. 2003. Validation of a more sensitive method for using spotted oligonucleotide DNA microarrays for functional genomics studies on bacterial communities. Environ. Microbiol. 5:933-943. [DOI] [PubMed] [Google Scholar]
- 9.El Fantroussi, S., H. Urakawa, A. E. Bernhard, J. J. Kelly, P. A. Noble, H. Smidt, G. M. Yershov, and D. A. Stahl. 2003. Direct profiling of environmental microbial populations by thermal dissociation analysis of native rRNAs hybridized to oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2377-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erhart, R., D. Bradford, R. J. Seviour, R. Amann, and L. L. Blackall. 1997. Development and use of fluorescent in situ hybridization probes for the detection and identification of Microthrix parvicella in activated sludge. Syst. Appl. Microbiol. 20:310-318. [Google Scholar]
- 11.Guo, Z., R. A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii, K., M. Mussmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 14.Karsten, S. L., V. M. D. Van Deerlin, C. Sabatti, L. H. Gill, and D. H. Geschwind. 15 January 2002. An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res. 30:e4. [Online.] http://nar.oxfordjournals.org/cgi/content/full/30/2/e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloep, F., W. Manz, and I. Röske. 2006. Multivariate analysis of microbial communities in the River Elbe (Germany) on different phylogenetic and spatial levels of resolution. FEMS Microbiol. Ecol. 56:79-94. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachmund, C., B. Köcher, W. Manz, and P. Heininger. 2003. Chemical and microbiological in situ characterization of benthic communities in sediments with different contamination levels. J. Soils Sediments 3:188-196. [Google Scholar]
- 18.Lander, E. S. 1999. Array of hope. Nat. Genet. 21:3-4. [DOI] [PubMed] [Google Scholar]
- 19.Lehr, H.-P., M. Reimann, A. Brandenburg, G. Sulz, and H. Klapproth. 2003. Real-time detection of nucleic acid interactions by total internal reflection fluorescence. Anal. Chem. 75:2414-2420. [DOI] [PubMed] [Google Scholar]
- 20.Liu, W.-T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 21.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loy, A., C. Schulz, S. Lucker, A. Schopfer-Wendels, K. Stoecker, C. Baranyi, A. Lehner, and M. Wagner. 2005. 16S rRNA gene-based oligonucleotide microarray for environmental monitoring of the betaproteobacterial order “Rhodocyclales”. Appl. Environ. Microbiol. 71:1373-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacGregor, B., D. Moser, E. Alm, K. Nealson, and D. A. Stahl. 1997. Crenarchaeota in Lake Michigan sediment. Appl. Environ. Microbiol. 63:1178-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacGregor, B., V. Bruchert, S. Fleischer, and R. Amann. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451-464. [DOI] [PubMed] [Google Scholar]
- 27.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mir, K., and E. M. Southern. 1999. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 17:788-792. [DOI] [PubMed] [Google Scholar]
- 29.Oelmüller, U., N. Krüger, A. Steinbüchel, and G. Cornelius. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73-84. [Google Scholar]
- 30.Peplies, J., F. O. Glöckner, and R. Amann. 2003. Optimization strategies for the DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peplies, J., S. C. K. Lau, J. Pernthaler, R. Amann, and F. O. Glöckner. 2004. Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ. Microbiol. 6:638-645. [DOI] [PubMed] [Google Scholar]
- 32.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes, p. 207-226. In John H. Paul (ed.), Methods in microbiology, vol. 30. Marine microbiology. Academic Press, San Diego, Calif. [Google Scholar]
- 34.Schleifer, K. H., D. Schüler, S. Spring, M. Weizenegger, R. Amann, W. Ludwig, and M. Koehler. 1991. The genus Magnetospirillum new-genus, description of Magnetospirillum gryphiswaldense new-species, and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum new-combination. Syst. Appl. Microbiol. 14:379-385. [Google Scholar]
- 35.Schlesner, H., C. Rensmann, B. J. Tindall, D. Gade, R. Rabus, S. Pfeiffer, and P. Hirsch. 2004. Taxonomic heterogeneity within the Planctomycetales as derived by DNA-DNA hybridization, description of Rhodopirellula baltica gen. nov., sp nov., transfer of Pirellula marina to the genus Blastopirellula gen. nov. as Blastopirellula marina comb. nov. and emended description of the genus Pirellula. Int. J. Syst. Evol. Microbiol. 54:1567-1580. [DOI] [PubMed] [Google Scholar]
- 36.Schmalenberger, A., F. Schwieger, and C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 38.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schübbe, S., C. Würdemann, J. Peplies, U. Hayen, C. Wawer, F. O. Glöckner, and D. Schüler. Submitted for publication.
- 40.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sessitsch, A., S. Gyamfi, N. Stralis-Pavese, A. Weilharter, and U. Pfeifer. 2002. RNA isolation from soil for bacterial community and functional analysis: evaluation of different extraction and soil conservation protocols. J. Microbiol. Methods 51:171-179.12133609 [Google Scholar]
- 42.Stahl, D. A., B. Flesher, H. R. Mansfield, and L. Montgomery. 1988. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl. Environ. Microbiol. 54:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urakawa, H., S. El Fantroussi, H. Smidt, J. C. Smoot, E. H. Tribou, J. J. Kelly, P. A. Noble, and D. A. Stahl. 2003. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warnecke, F., R. Sommaruga, R. Sekar, J. S. Hofer, and J. Pernthaler. 2005. Abundances, identity, and growth state of Actinobacteria in mountain lakes of different UV transparency. Appl. Environ. Microbiol. 71:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wintzingerode, F. V., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 48.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]