Abstract
Like several other phytopathogenic fungi, the ascomycete Botrytis cinerea is known to produce the plant hormone abscisic acid (ABA) in axenic culture. Recently, bcaba1, the first fungal gene involved in ABA biosynthesis, was identified. Neighborhood analysis of bcaba1 revealed three further candidate genes of this pathway: a putative P450 monooxygenase-encoding gene (bcaba2), an open reading frame without significant similarities (bcaba3), and a gene probably coding for a short-chain dehydrogenase/reductase (bcaba4). Targeted inactivation of the genes proved the involvement of BcABA2 and BcABA3 in ABA biosynthesis and suggested a contribution of BcABA4. The close linkage of at least three ABA biosynthetic genes is strong evidence for the presence of an abscisic acid gene cluster in B. cinerea.
Phytohormones are synthesized not only by plants but also by microorganisms. Production of ethylene, gibberellins, auxins, and cytokinins has been demonstrated in different bacterial species (for reviews, see references 9, 16, and 28). The best-known example of plant hormones produced by fungi are the gibberellins, named after the ascomycete Gibberella fujikuroi, from which they were first isolated; also, biosynthesis of auxins, ethylene, and cytokinins was reported for a variety of fungal species (for a review, see reference 50). Abscisic acid (ABA) has been shown to be produced by members of different divisions of filamentous fungi, such as the basidiomycete Rhizoctonia solani, the ascomycete Ceratocystis fimbriata, and the zygomycete Rhizopus nigricans (10, 12).
Many of the above-described species are involved in symbiotic or pathogenic interactions with plants. Therefore, the ability of microorganisms to produce plant hormones is thought to play a role during the establishment of these interactions. It was also previously assumed that this capacity could have been inherited from plants via horizontal gene transfer (8). However, in several cases investigated so far, the biosynthetic pathways seem to differ in fungi and higher plants. A good example is the biosynthesis of ethylene, which proceeds from methionine via 1-aminocyclopropanecarboxylic acid in plants (56), whereas in addition to the aminocyclopropanecarboxylic acid pathway, two different pathways were found in fungi: one involves α-keto-γ-methylthiobutyric acid and has been described for Botrytis cinerea, and the other one, present in Penicillium digitatum and Fusarium oxysporum, starts from 2-oxoglutarate (6, 15, 21). On the other hand, gibberellins are produced in fungi and plants along more or less the same biosynthetic routes, but the enzymes involved differ considerably (18).
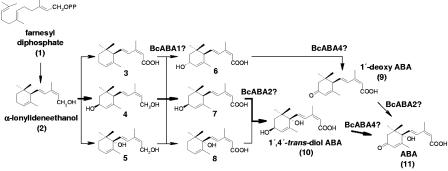
Also, the fungal ABA biosynthetic pathway seems to be different from that in plants. In vascular plants, ABA is derived from cleavage products of carotenoids (for reviews, see references 31 and 36). The ABA pathway in fungi has been biochemically studied in the phytopathogenic ascomycetes Botrytis cinerea and several Cercospora species, and a direct route from farnesyl diphosphate has been postulated (36). However, different biosynthetic intermediates were isolated from different species. In Cercospora cruenta, 1′,4′-dihydroxy-γ-ionylideneacetic acid has been found to be the major metabolite (35), while biosynthesis in C. rosicola proceeds via 1′-deoxy-ABA (32, 33). Only in C. pini-densiflorae and B. cinerea was 1′,4′-trans-diol ABA detected as the main ABA precursor (19, 34) (see Fig. 6). Early steps of the fungal pathway have been investigated in B. cinerea and C. cruenta, indicating a route via allofarnesene and ionylideneethane (22, 23). Recently, bcaba1, the first fungal gene involved in ABA biosynthesis, was identified in B. cinerea (43). It encodes a putative P450 monooxygenase. Mutants lacking a functional copy of the gene are completely inhibited in ABA biosynthesis. ΔBcaba1 mutants accumulate a substance, the molecular mass of which corresponds to that of the postulated ABA intermediates α-ionylideneacetic acid and γ-ionylideneacetic acid, respectively. This suggests a block of the pathway at the stage indicated in Fig. 6. In order to further analyze ABA biosynthesis in Botrytis, we are seeking additional genes involved in this pathway.
FIG. 6.
Postulated biosynthetic pathway of ABA in C. pini-densiflorae (late steps, modified from a method described previously by Okamoto et al. [34]). Bold lines indicate the major route. Intermediates identified are α-ionylideneacetic acid (3), 4′-S-OH-α-ionylideneethanol (4), 1′-OH-α-ionylideneethanol (5), 4′-R-OH-α-ionylideneacetic acid (6), 4′-S-OH-α-ionylideneacetic acid (7), and 1′-OH-α-ionylideneacetic acid (8). The enzymatic steps possibly catalyzed by BcABA1, BcABA2, and BcABA4 are indicated.
In filamentous fungi, genes of secondary metabolite pathways are often found to be organized in gene clusters, such as those responsible for the biosynthesis of sterigmatocystins (4), trichothecenes (20), penicillin (11), ergot alkaloids (52), and gibberellins (49). Therefore, neighborhood analysis of bcaba1 was expected to be a promising tool to identify other genes involved in ABA biosynthesis. In this study, we describe a sequence analysis of the upstream and downstream genomic regions of bcaba1 and the characterization of bcaba2, bcaba3, and bcaba4, three genes located adjacent to bcaba1, by gene inactivation.
MATERIALS AND METHODS
Strains and culture conditions.
Strain SAS56 of Botrytis cinerea Pers.:Fr. (Botryotinia fuckeliana [de Bary] Whetz) is an ascospore isolate from a Vitis-derived field isolate (14), B05.10 is a haploid strain obtained after benomyl treatment of a Vitis isolate (39), and ATCC 58025 is a nonsporulating overproducer of ABA (30).
Escherichia coli strain TOP10F′ (Invitrogen, Groningen, The Netherlands) was used for propagation of plasmids. Propagation of lambda clones was performed using strain LE392 (Stratagene, La Jolla, CA).
B. cinerea strains were grown on 2% malt extract (Oxoid Ltd., Basingstoke, Hampshire, England) amended with 0.5% glucose, 0.1% casein peptone (Difco Laboratories, Sparks, MD), 0.1% Casamino Acids (Difco), 0.1% yeast extract (Duchefa Biochemie BV, Haarlem, The Netherlands), and 0.02% RNA sodium salt. For ABA production and RNA isolation, fungi were cultivated in 300-ml Erlenmeyer flasks with 100 ml of a defined liquid medium, described previously by Sprecher (46) and modified as described previously by Kettner (27), containing 20 g of lactose/liter instead of glucose (Sprecher medium). Fungi and culture filtrates were harvested after 3 to 7 days of cultivation on a rotary shaker at 150 rpm and 20°C. For DNA isolation, mycelium was grown for 3 to 4 days at 20°C on complex medium agar (37) with a cellophane overlay.
DNA isolation.
Fungal genomic DNA was isolated as described previously by Cenis (5). Lambda DNA was isolated according to a standard method (41). Plasmid DNA was isolated using a plasmid DNA preparation kit (Genomed, Bad Oeynhausen, Germany).
Southern blot analysis.
Genomic DNA was digested with restriction enzymes, size separated on a 1% agarose gel, and blotted onto Hybond-N+ membranes (Amersham Pharmacia, Freiburg, Germany) according to a method described previously by Sambrook et al. (41). Hybridization was carried out in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 0.5% sodium dodecyl sulfate, and 0.01% salmon sperm DNA at 65°C for 16 to 20 h in the presence of a randomly primed [α-32P]dCTP-labeled probe. Membranes were washed in 2× SSPE-0.1% sodium dodecyl sulfate at 65°C before being exposed to an autoradiographic film.
RNA blot analysis.
RNA was isolated from mycelial samples using the RNAgents Total RNA isolation system (Promega, Mannheim, Germany). Samples of 15 to 20 μg of RNA were transferred on to Hybond-N+ filters after electrophoresis on a 1% agarose gel containing formaldehyde according to the method described previously by Sambrook et al. (41). Blot hybridizations were carried out in a solution containing 0.6 M NaCl, 0.16 M Na2HPO4, 0.06 M EDTA, 1% N-lauroylsarcosine (Sigma), 10% dextran sulfate (Eppendorf AG, Hamburg, Germany), and 0.01% salmon sperm DNA, pH 6.2, as described above for Southern blots.
Sequencing.
DNA sequencing of recombinant plasmid clones was performed with a LI-COR 4200 automatic sequencer (MWG Biotech, Munich, Germany) using the Thermo Sequenase Fluorescent Labeled Primer cycle sequencing kit (Amersham Pharmacia). For sequence analysis, the program Seqman (DNA Star) was used.
Construction of replacement vectors.
For construction of the gene replacement vectors pABA2Rep, pABA3Rep, and pABA4Rep, the plasmid pOliHP (40), carrying the Escherichia coli hygromycin phosphotransferase gene hph under the control of the Aspergillus nidulans oliC promoter and trpC terminator, was used as a basis vector. In order to construct pABA2Rep, a 0.47-kb PCR fragment was amplified from the 5′ region of bcaba2 using the following primers (see Fig. 3): ABA2-5′F (5′-CCT TCG GGT ACC TAC CGA ATC-3′ [primer 1]) and ABA2-5′R (5′-GAG TGT CGA CAG ATC TTT GAC-3′ [primer 2]) (an artificial KpnI site is indicated in boldface type). From the 3′ end of bcaba2, a 0.53-kb fragment was amplified using the following primers: ABA2-3′F (5′-GAA ATG AAG CTT ATT ACT TCC CG-3′ [primer 3]) and ABA2-3′R (5′-GAC GGA ATT CAA GTC CTA TAC TG-3′ [primer 4]) (artificial HindIII and EcoRI sites are indicated in boldface type). Both fragments were cloned into pCR2.1-TOPO (Invitrogen), cut with KpnI/SalI and HindIII/EcoRI, respectively, and cloned into the corresponding sites of pOliHP. By cutting with KpnI and EcoRI, the replacement cassette was isolated from the vector prior to transformation. Accordingly, vectors pABA3Rep and pABA4Rep were constructed using primers ABA3-5′F (5′-GGT CGG TTC CAG GCT CTC GAG-3′, containing an artificial XhoI site [in boldface type]), ABA3-5′R (5′-ATT CTG TCG ACG TGA GAG CAC TC-3′, containing an artificial SalI site [in boldface type]), ABA3-3′F (5′-CAG TCT TTG AAG CTT CTG TTA C-3′), ABA3-5′R (5′-CAC CCC AAG AAT TCC GAC TGG C-3′, containing an artificial EcoRI site [in boldface type]), ABA4-5′F (5′-GAA CAA GAT GGT ACC CCA CTG G-3′, containing an artificial KpnI site [in boldface type]), ABA4-5′R (5′-CAC CAG CCA GAT TTC TCG AGC C-3′, containing an artificial XhoI site [in boldface type]), ABA4-3′F (5′-CCA AGC TTA TTC TGT GCT CTG CC-3′, containing an artificial HindIII site [in boldface type]), and ABA4-3′R (5′-CAT GAG GAA CTG CGA ATT CTG GC-3′, containing an artificial EcoRI site [in boldface type]).
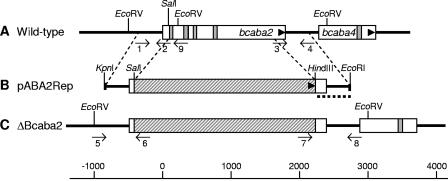
FIG. 3.
Scheme of the gene replacement approach for bcaba2. Physical maps of bcaba2 from wild-type strain ATCC 58025 (A), the gene replacement fragment pABA2Rep (B), and bcaba2 from a ΔBcaba2 replacement mutant (C) showing the organization of exons (white bars), introns (gray bars), the hygromycin resistance cassette (hatched bars), and flanking regions of bcaba2 (bold lines) are shown. Arrowheads indicate orientations of the genes. Binding sites of primers 1 to 9, used for PCR analysis of replacement mutants (see Materials and Methods), as well as the 3′ flank of pABA2Rep (dotted line) used as a probe for Southern analysis, are indicated.
Transformation of B. cinerea.
Protoplasts were generated as described previously (43). Protoplasts (107) were added to 10 μg of the replacement cassette and transformed according to a method described previously by ten Have et al. (48). Hygromycin B-resistant colonies were transferred onto agar plates containing Gamborg B5 medium (Duchefa Biochemie BV, Haarlem, The Netherlands) and 2% glucose complemented with 70 μg/ml of hygromycin B.
Homologous integration events and pure transformants were identified by PCR using the following primers (see Fig. 3): ABA2a (5′-CCC TCC CGT CTA GAC CCT CCC G-3′ [primer 5]), pLOF-oliP (5′-GGT ACT GCC CCA CTT AGT GGC AGC TCG CG-3′ [primer 6]), pAN-T (5′-CCC AGA ATG CAC AGG TAC AC-3′ [primer 7]), ABA2b (5′-TAT CAT GAT ATA GCG AGA TGA GG-3′ [primer 8]), ABA2c (5′-CCA GTT GCA ATA ACC CAC GAG CCG-3′ [primer 9]), ABA3a (5′-GAT AGC CAA CAA CTC TGA CTG GTG G-3′), ABA3b (5′-TTG GTA CTG TCG GTG GCT AGG CC-3′), ABA3c (5′-CTA CTA CAT GCG TAA CAA GCG-3′), ABA4-1 (5′-CGC TGC TTC GAG AAC GCC GA-3′), ABA4-WT (5′-GCT GCC GTT CTT GTG AGC CC-3′), and ABA4-2 (5′-GGA GGT GCC TGG TCT GCA A-3′).
EIA for cis-(+)-abscisic acid.
An enzyme immunoassay (EIA) for the detection of ABA in culture filtrates was performed on 96-well microplates (flat bottom; Sarstedt Inc., Newton, NC) in triplicate measurements as described previously by Weiler (55). A rabbit polyclonal antibody raised against mouse immunoglobulin G, an anti-ABA mouse monoclonal antibody, and an ABA-labeled alkaline phosphatase (tracer) were kindly provided by E. W. Weiler, Ruhr University, Bochum, Germany. p-Nitrophenyl phosphate (Biomol Feinchemikalien GmbH, Hamburg, Germany) was used as a phosphatase substrate. ABA standards ranged from 0.05 pmol/ml to 50 pmol/ml. For each sample, ABA was assayed in at least two dilutions. The optical density at 405 nm was measured by a microplate reader (model 550; Bio-Rad Laboratories GmbH, Munich, Germany).
LC-MS analyses.
Fungi were grown on Czapek yeast (autolysate) extract agar (44), which was previously shown to be suitable for ABA production, for 7 days in the dark. Culture extracts were prepared by a plug extraction method modified from that described previously by Smedsgaard (45). Three 6-mm agar plugs were cut from each culture and transferred into a 1.5-ml disposable autosampler screw-cap vial. The plugs were extracted twice by sonication for 60 min using 500 μl ethyl acetate with 0.5% formic acid for the first extraction. The ethyl acetate extract was transferred to a clean vial and evaporated to dryness. The plugs were reextracted with 500 μl 2-propanol. The 2-propanol extract was transferred to the vial with the dry residues from the first extraction and again evaporated to dryness. The combined residues were redissolved by sonication for 10 min in 400 μl methanol and filtered before analysis. Liquid chromatography-mass spectrometry (LC-MS) analyses were done as negative electrospray mass spectrometry on an Agilent 1100 system connected to a Micromass LCT (Waters, United Kingdom) time-of-flight mass spectrometer with a z-spray source and a LockSpray probe. The separation was done on a 50-mm by 2-mm-inside-diameter Luna C18(II) column (Phenomenex) at a flow rate of 0.3 ml/min with a linear gradient of H2O (containing 20 mM formic acid) and CH3CN; the gradient was changed from 10% CH3CN to 100% over 20 min and then maintained at 100% CH3CN for 10 min before returning to starting conditions in 5 min and maintaining the starting conditions for another 5 min. Leucine-enkephaline dissolved in 50% acetonitrile with 0.2% formic acid was used as lockmass (M − H− at 554.2615 Da) at a flow rate of approximately 5 μl/min. The instrument was tuned to maximum sensitivity in negative electrospray mode at a resolution better than a 5,500 full-width half maximum and calibrated on a mixture of sugars. A sample cone voltage of 20 V was used. The mass accuracy was verified to be better than 5 ppm by injection of a known test sample. Mass spectra were collected from m/z 100 to 900 at 0.5 scans/s; every third scan was taken from the reference spray.
Nucleotide sequence accession numbers.
Sequences have been deposited in the GenBank database under accession numbers AJ851088 for bcaba2, AM237449 for bcaba3, and AM237450 for bcaba4.
RESULTS
Characterization of the genomic region of bcaba1.
In order to analyze the DNA region adjacent to the ABA biosynthesis gene bcaba1, a cDNA fragment of the gene was used to screen a genomic EMBL3 library of strain SAS56. Two positive phages were purified and subcloned after restriction with HindIII, SacI, and EcoRI, respectively. In total, about 17 kb of the SAS56 genomic DNA was sequenced, encompassing 6 kb upstream and 9.4 kb downstream of bcaba1 (Fig. 1).
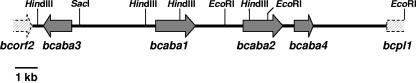
FIG. 1.
Scheme of the putative ABA gene cluster of B. cinerea. EcoRI, SacI, and HindIII restriction sites are indicated.
In the region directly upstream of bcaba1, no significant similarities to sequences present in the public databases were found. However, by comparison with an expressed sequence tag library of strain ATCC 58025 established from mycelia grown under ABA biosynthesis conditions (43), an open reading frame (bcaba3) of 1,254 bp was identified; it contains no intron and thus encodes a polypeptide of 417 amino acids. bcaba3 is located 3.7 kb upstream of bcaba1 and is divergently transcribed (Table 1 and Fig. 1). Upstream of bcaba3, another putative open reading frame (bcorf2) shows similarity to a ferulic acid esterase-encoding gene of Neurospora crassa. Downstream of bcaba1 (2.2 kb), a putative second P450 monooxygenase-encoding gene (bcaba2) was detected. Due to the presence of two putative translation initiation codons that are closely linked, the exact length of the coding region cannot be determined. The open reading frame comprises either 1,771 or 1,810 bp interrupted by four introns of 50, 66, 50, and 60 bp, resulting in an encoded polypeptide of 514 or 527 amino acids. The derived sequence shows similarity to fungal P450 monooxygenases, especially trichothecene C-15 hydroxylases of some Fusarium species. bcaba1 as well as bcaba2 are transcribed in the same direction as a third gene located 0.5 kb downstream of bcaba2. The open reading frame (bcaba4) of 842 bp, interrupted by an intron of 65 bp, encodes a protein with significant homology to a group of short-chain dehydrogenases/reductases (SDRs) from bacteria and fungi. The right border of the sequenced DNA region is defined by the 0.8-kb 5′ end of a gene showing similarity to fungal pectin lyase genes (bcpl1) (Fig. 1). All intergenic regions were analyzed with the exon-trapping software program Xpound (http://bioweb.pasteur.fr/-seqanal/interfaces/-xpound-simple.html), revealing that the probability of finding more genes in the investigated sequence is low (data not shown).
TABLE 1.
Characteristics of genes located adjacent to bcaba1
| Gene | Length (bp) | Intron(s) (bp) | Encoded polypeptide (aa) | Function (putative) |
|---|---|---|---|---|
| bcorf2 | Ferulic acid esterase | |||
| bcaba3 | 1,254 | 417 | ||
| bcaba1 | 1,769 | 48, 49, 83, 59 | 509 | P450 monooxygenase |
| bcaba2 | 1,771/1,810 | 50, 66, 50, 60 | 514/527 | P450 monooxygenase |
| bcaba4 | 842 | 65 | 258 | Short-chain dehydrogenase/ reductase |
| bcpl1 | Fungal pectin lyase |
Because the genomic DNA library used in these studies was derived from strain SAS56, which probably does not produce abscisic acid in axenic culture, we wanted to test whether the organization of the sequenced genes is the same in the nonsporulating ABA overproducer ATCC 58025 and the highly pathogenic strain B05.10 used in most molecular studies. Therefore, a PCR approach was performed using primers binding to the 5′ and 3′ ends of the respective genes in order to amplify the intergenic regions. The resulting PCR products were sequenced in order to exclude unspecific binding of primers, and their identity with the corresponding sequences of strain SAS56 was confirmed (data not shown). Thus, it can be concluded that the organization of the genes from bcorf2 to bcpl1 is conserved in strains SAS56, B05.10, and ATCC 58025.
Expression analysis of neighboring genes.
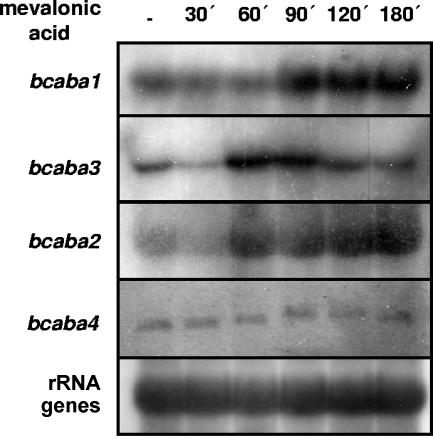
Genes in fungal metabolic pathway gene clusters are mostly coregulated (reviewed in reference 26). Hence, expression analysis of neighboring genes could provide an indication of a possible involvement in the same pathway. Expression of bcaba1 shows a persistent induction 90 min after the addition of the ABA precursor mevalonic acid (MVA) to the medium (43). In accordance with these investigations, mycelium of ABA-producing ATCC 58025 was harvested at different time points after MVA feeding and hybridized in a Northern blot analysis to internal cDNA or genomic DNA fragments of bcaba3, bcaba2, and bcaba4, respectively. As for bcaba1, a persistent induction could also be detected for bcaba2, starting, in contrast to bcaba1, after 60 min (Fig. 2). Expression of bcaba3 was likewise enhanced at 60 and 90 min after the addition of MVA but declined again after 120 min, whereas bcaba4 was constitutively expressed at a low level (Fig. 2). Thus, all three genes are transcribed in mycelia grown under ABA production conditions, but a similar expression pattern as that for bcaba1 is evident only for bcaba2; i.e., the four putative ABA genes are not strictly coregulated.
FIG. 2.
Expression analysis of bcaba1-4. Total RNA was extracted from strain ATCC 58025, which was grown for 3 days in Sprecher medium. Mycelia were harvested 30, 60, 90, 120, and 180 min after the addition of 3.8 mM mevalonic acid lactone. cDNA fragments of the respective genes were used for probing. Loading of lanes with RNA was checked by probing with rRNA genes.
Target inactivation of bcaba3, bcaba2, and bcaba4.
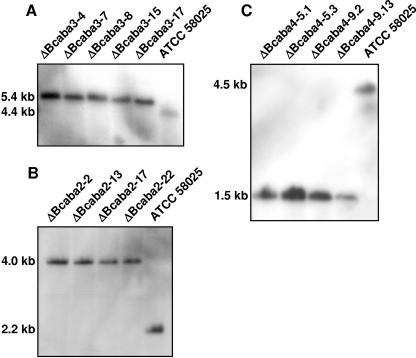
All three genes were functionally tested using a gene replacement approach: fragments pABA3Rep, pABA2Rep, and pABA4Rep (Fig. 3), (see Materials and Methods) were used to transform protoplasts of ABA-producing strain ATCC 58025. As an example, the approach for the deletion of bcaba2 is represented in Fig. 3. Hygromycin-resistant transformants were examined by PCR for the presence of the expected knockout allele using primer pairs 5/6 and 7/8 in the given example of the ΔBcaba2 mutants (Fig. 3). The purity of the transformants was checked by PCR as well by using primers 5 and 9 for the ΔBcaba2 transformants. Five out of 19 ΔBcaba3 transformants and four out of 24 ΔBcaba2 transformants exhibited only the appropriate diagnostic PCR fragments but not the respective wild-type-specific fragments anymore (data not shown). Since all of the 18 ΔBcaba4 mutants obtained still contained the wild-type-specific PCR fragment, two of the mutants were purified by protoplast isolation. Pure transformants were further analyzed by Southern hybridization. Genomic DNA of the ΔBcaba3, ΔBcaba2, and ΔBcaba4 mutants was digested with SacI, EcoRV, and HindIII, respectively, and hybridized to the labeled 3′ flank of the corresponding replacement vector. Transformants ΔBcaba3-4, ΔBcaba3-7, ΔBcaba3-8, ΔBcaba3-15, and ΔBcaba3-17 showed a hybridizing fragment of 5.4 kb, while they lacked the 4.4-kb fragment characteristic of the wild type (Fig. 4A). Accordingly, the ΔBcaba2 and ΔBcaba4 mutants tested showed knockout fragments of 4 kb and 1.5 kb, respectively, whereas the respective wild-type fragments were missing (Fig. 4B and C). Therefore, they can be regarded as homokaryotic deletion mutants. None of the mutants showed a significant alteration of growth compared to that of the wild type on any of the media used in this study.
FIG. 4.
Southern blot analysis of wild-type strain ATCC 58025 and ΔBcaba3 (A), ΔBcaba2 (B), and ΔBcaba4 (C) replacement mutants. DNA was digested with SacI, EcoRV, and HindIII, respectively, blotted, and hybridized with the 3′ flank of the respective replacement vectors.
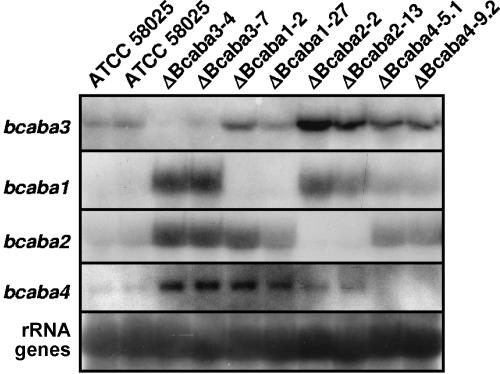
In a first step, transformants were investigated by enzyme immunoassay for the production of cis-(+)-abscisic acid. Culture filtrates of ΔBcaba3 transformants, in contrast to the wild-type strain, no longer contained ABA. However, culture filtrates of ΔBcaba2 and ΔBcaba4 mutants showed an immunoreaction indicating that they might still produce ABA (data not shown). Since cross-reactivity of an ABA precursor possibly accumulating in the culture of the knockout mutants cannot be excluded, the transformants were further studied by negative electrospray LC-MS analyses (Fig. 5) using narrow ion traces (50-ppm window).
FIG. 5.
Expression analysis of the genes bcaba1-4 in wild-type strain ATCC 58025 as well as ΔBcaba3, ΔBcaba1, ΔBcaba2, and ΔBcaba4 mutants. Total RNA was extracted from mycelium grown for 4 days in Sprecher medium. PCR fragments of the respective genes were used for probing. Loading of lanes with RNA was checked by probing with rRNA genes. Because of the strong signals in some of the mutant lanes, exposure time had to be short; therefore, the wild-type lane shows rather weak signals (compare to Fig. 2).
ABA elutes at 6.45 min on the 50-ppm ion trace at m/z 263.128, corresponding to the deprotonated molecular ion from the analysis of an ABA standard (data not shown). Similarly, when looking at narrow ion traces (50 ppm) around m/z 263.128 (deprotonated ABA), the presence of ABA could be confirmed in wild-type strain ATCC 58025 but was not found in either the ΔBcaba3 transformants or the ΔBcaba2 mutants tested (Table 2). Further analyses of ΔBcaba2-2 and ΔBcaba2-13 showed a distinct peak at a retention time of 8.38 min in the ion trace of m/z 247.133 (50 ppm), which was absent in ATCC 58025 (Table 2) as well as in mutants defective in the other cluster genes (data not shown). The deduced mass corresponds to a molecule with the formula C15O3H19, which might represent deprotonated 1′-deoxy-ABA, although unequivocal identification of this compound would require further analysis. In contrast to the ΔBcaba3 and ΔBcaba2 mutants, the ΔBcaba4 strains still contained ABA but apparently at a lower level than that found in the wild type (Table 2). On the other hand, a peak in the ion trace of m/z 265.144 (50 ppm) that eluted after 5.36 min was much more pronounced in these transformants than in ATCC 58025 (Tab. 2). This value corresponds to the mass of deprotonated 1′,4′-trans-diol-ABA, but again, further investigation is necessary to confirm the identity of this intermediate.
TABLE 2.
LC-MS analysis to determine ion counts for selected ionsa
| Strain | Ion count for ions at a mass (Da) of:
|
||
|---|---|---|---|
| 263.128b | 247.133c | 265.144d | |
| ATCC 58025 | 1,330 | 40 | 129 |
| ATCC 58025 | 1,390 | 43 | 69 |
| ΔBcaba3-4 | ND | ND | ND |
| ΔBcaba3-7 | ND | ND | ND |
| ΔBcaba1-2 | ND | ND | ND |
| ΔBcaba1-27 | ND | ND | ND |
| ΔBcaba2-2 | ND | 1,550 | ND |
| ΔBcaba2-13 | ND | 2,060 | ND |
| ΔBcaba4-5.1 | 665 | 135 | 5,420 |
| ΔBcaba4-9.2 | 762 | 271 | 7,850 |
A 50-ppm width. Counts below 40 were regarded as noise. ND, not detected.
Retention time of 6.45 min.
Retention time of 8.38 min.
Retention time of 5.36 min.
Expression analysis of ABA biosynthesis genes in ABA-deficient mutants.
In order to analyze whether a deletion of one ABA biosynthesis gene has an impact on the transcription level of other cluster genes, a Northern blot analysis was performed. RNA was isolated from mycelium of wild-type strain ATCC 58025 and the ΔBcaba1, ΔBcaba2, ΔBcaba3, and ΔBcaba4 mutants grown under ABA biosynthesis conditions and hybridized to labeled PCR fragments of all four genes.
As shown in Fig. 5, in most cases, an up-regulation of the three remaining cluster genes can be observed if one of the four genes is deleted. For instance, bcaba1, bcaba2, and bcaba4 show a much higher transcription level in the ΔBcaba3 mutants than in wild type strain ATCC 58025. Accordingly, bcaba2 and bcaba4 are up-regulated in the strains ΔBcaba1-2 and ΔBcaba1-27, while only a slightly enhanced transcription is shown for bcaba3 in these mutants. Transcription of bcaba3, bcaba1, and bcaba4 is likewise increased in the ΔBcaba2 strains, although at a lower degree in the case of bcaba4. In the ΔBcaba4 transformants, which still produce some ABA, the enhancement in transcription of the other cluster genes is less pronounced.
DISCUSSION
Genes involved in the biosynthesis of fungal metabolites, especially those that are not necessarily required for growth, are often physically linked (reviewed in reference 26). It has been suggested that this arrangement could play a role in the transcriptional regulation of the genes (20) or favor the transmission of certain properties by horizontal gene transfer (54). In this study, we present evidence for a gene cluster of at least four genes responsible for ABA biosynthesis in B. cinerea. Two genes involved in the production of this plant hormone, the previously characterized gene bcaba1 (43) and bcaba2, are found to be directly linked to each other. Both genes are obviously regulated in a similar (but not identical) manner. They show an enhanced level of transcription after the addition of mevalonic acid. Coregulation is a common feature of fungal metabolic pathway genes. In Aspergillus parasiticus, coordinated regulation of the aflatoxin biosynthesis genes is mediated by the transcription factor AflR together with its putative coactivator, AflJ (3, 7, 13). Two genes, Tri6 and Tri10, are involved in the regulation of the trichothecene cluster genes in Fusarium sporotrichioides (38, 47). Whether such pathway-specific regulatory elements are also participating in ABA biosynthesis in B. cinerea remains to be elucidated.
Regulation of bcaba3 and bcaba4 seems to differ from that of the other two genes. While bcaba3 shows a transiently enhanced transcription level after mevalonic acid donation, bcaba4 exhibits constitutive expression under these conditions. It has been observed that certain genes of fungal metabolic pathway gene clusters do not follow coregulation; e.g., P450-3 is the only gene in the Fusarium fujikuroi gibberellin biosynthesis gene cluster that is not repressed by nitrogen (51).
On the other hand, all four cluster genes appear to be subject to some kind of negative feedback regulation, a feature that has also previously been described for certain genes of trichothecene biosynthesis (1): transcription of bcaba1-4 is enhanced in those mutants that do not produce ABA, the final product of the pathway. However, the addition of ABA to the wild type did not influence the expression of bcaba2 (data not shown), arguing against a simple feedback mechanism; on the other hand, it is possible that externally supplied ABA is not taken up and that ABA cannot be monitored extracellularly.
Deletion of the bcaba2-4 genes finally proved their role in ABA production. Instead of ABA, ΔBcaba2 mutants accumulate a substance that is not present in the wild type, the molecular mass of which corresponds to 1′-deoxy-ABA, indicating that BcABA2 as a P450 monooxygenase would be responsible for the hydroxylation of carbon atom C-1′ (Fig. 6). Also, the fact that a cross-reaction in EIA analysis of the culture filtrates has been detected could indicate a block of the pathway at this late stage. Weiler (55) previously described that the carbonyl function at C-4′ is required for the immunoreactivity of the molecule. 1′-Deoxy-ABA has been described as an ABA precursor in Cercospora spp. such as C. pini-densiflorae (34). The conversion of 1′-deoxy-ABA to ABA has been detected in cell extracts from C. rosicola (2). Hirai et al. (19) previously showed that 1′,4′-trans-diol-ABA is the main precursor of ABA in B. cinerea, and those authors were not able to isolate any 1′-deoxy-ABA from cultures, whereas Neill et al. (33) reported the detection of 1′-deoxy-ABA in B. cinerea culture filtrates also. Thus, this could indicate the presence of a branched pathway similar to that of C. pini-densiflorae. Another possibility is that, assuming the pathway via 1′,4′-trans-diol-ABA is indeed the major one used by B. cinerea, 1′-deoxy-ABA might be a side product of the original pathway resulting from nonspecific enzymatic conversion of 4′-OH-α-ionylideneacetic acid when hydroxylation at C-1′ is blocked. Vaughan and Milborrow (53) also demonstrated a nonenzymatic oxidation at C-4′ of 1′,4′-cis and 1′,4′-trans-diol-ABA under acidic conditions.
Nonenzymatic oxidation at C-4′ would also be a possible explanation for the presence of ABA in the ΔBcaba4 mutants, assuming that BcABA4 is responsible for the respective enzymatic reaction. ΔBcaba4 transformants accumulated a substance that might represent 1′,4′-trans-diol-ABA. The bcaba4 gene encodes a putative SDR. SDRs represent a highly divergent large family of NAD(P)(H)-dependent enzymes with a wide range of substrate specificities (24, 25). In Arabidopsis thaliana, the SDR ABA2 has been identified as the enzyme catalyzing the conversion of xanthoxin to abscisic aldehyde, the penultimate step in plant ABA biosynthesis, which involves the oxidation of the 4′-hydroxyl to a ketone group, desaturation of a 2′,3′ bond, and the opening of the 1′,2′-epoxide ring (17). It has been suggested that these ring modifications might also result from a single enzymatic reaction, the oxidation at the C-4′ carbon atom, followed by a spontaneous epoxid rearrangement (42).
The enzymatic steps proposed to be catalyzed by BcABA1, BcABA2, and BcABA4 are indicated in Fig. 6. For BcABA3, no such proposal can be made, since the respective gene did not show any homology to known sequences, and ΔBcaba3 mutants did not accumulate any substance that could be correlated to the proposed intermediates of the B. cinerea ABA pathway. A role of BcABA3 as a positive pathway regulator can be excluded, since in the ΔBcaba3 mutants, the other cluster genes are still transcribed (Fig. 6).
However, in order to support these conclusions and to study the enzymatic function of BcABA1-4 in detail, further biochemical analyses together with feeding experiments using possible pathway intermediates are essential.
Although further gene products are likely to be necessary for catalyzing the remaining biosynthetic steps in ABA biosynthesis (Fig. 6), bcaba3 and bcaba4 probably represent the borders of this cluster. Sequencing up- and downstream revealed a gene with similarity to a ferulic acid esterase-encoding gene from Neurospora crassa and a putative pectin lyase-encoding gene, respectively, both of them very likely not involved in ABA production.
One problem that still remains to be solved is the question of why, in contrast to ATCC 58025, strains B05.10 and SAS56 do not produce ABA in axenic culture. At least B05.10 shows (low) expression of bcaba1 and bcaba2 under the same conditions in which ATCC 58025 produces large amounts of ABA. By PCR analysis, we could exclude one possible explanation, the absence of certain genes of the described gene cluster, as the organization of bcaba1-4 is the same in all three strains tested. However, the presence and expression of the respective genes do not mean that they are necessarily functional. Malonek et al. (29) previously investigated gibberellin production in a producing and a nonproducing mating population of Gibberella fujikuroi. They showed that the loss of the ability to produce gibberellins resulted from several point mutations in the coding sequence and promoter regions of certain gibberellin biosynthetic genes. Whether the gene products of the B. cinerea ABA cluster in B05.10 and SAS56 are still functional has to be elucidated by further investigations, especially with respect to the impact of ABA biosynthesis on the virulence of these highly pathogenic isolates. The impact of the genes (and therefore the impact of ABA biosynthesis) on pathogenicity is currently under investigation.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Tu 50/9).
We thank Anke Bölting for excellent technical assistance and E. Weiler (Bochum) for providing ABA-specific antisera and for help in the establishment of the ABA EIA.
REFERENCES
- 1.Alexander, N. J., S. P. McCormick, T. M. Larson, and J. E. Jurgenson. 2004. Expression of Tri15 in Fusarium sporotrichioides. Curr. Genet. 45:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Al-Nimri, L. F., and R. C. Coolbaugh. 1991. Conversion of 1′-deoxy-2H-ABA to 2H-ABA in cell-free extracts from Cercospora rosicola. J. Plant Growth Regul. 10:63-66. [Google Scholar]
- 3.Bhatnagar, D., K. C. Ehrlich, and T. E. Cleveland. 2003. Molecular genetic analysis and regulation of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 61:83-93. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. W., J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams, and T. J. Leonard. 1996. Twenty-five co-regulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 93:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cenis, J. L. 1992. Rapid extraction of fungal DNA for PCR amplification. Nucleic Acids Res. 20:2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chagué, V., Y. Elad, R. Barakat, P. Tudzynski, and A. Sharon. 2002. Ethylene biosynthesis in Botrytis cinerea. FEMS Microbiol. Ecol. 40:143-149. [DOI] [PubMed] [Google Scholar]
- 7.Chang, P. K. 2003. The Aspergillus parasiticus protein AFLJ interacts with the aflatoxin pathway-specific regulator AFLR. Mol. Genet. Genomics 268:711-719. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, D. J., and M. A. Regan. 1980. Evolution of a biochemical pathway: evidence from comparative biochemistry. Annu. Rev. Plant Physiol. 31:639-645. [Google Scholar]
- 9.Costacurta, A., and J. Vanderleyden. 1995. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 21:1-18. [DOI] [PubMed] [Google Scholar]
- 10.Crocoll, C., J. Kettner, and K. Dörffling. 1991. Abscisic acid in saprophytic and parasitic species of fungi. Phytochemistry 30:1059-1060. [Google Scholar]
- 11.Diez, B., S. Gutierrez, J. L. Barredo, P. van Solingen, L. H. M. van der Voort, and J. F. Martin. 1990. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J. Biol. Chem. 265:16358-16365. [PubMed] [Google Scholar]
- 12.Dörffling, K., W. Petersen, E. Sprecher, I. Urbasch, and H.-P. Hanssen. 1984. Abscisic acid in phytopathogenic fungi of the genera Botrytis, Ceratocystis, Fusarium, and Rhizoctonia. Z. Naturforsch. 39:683-684. [Google Scholar]
- 13.Ehrlich, K. C., B. G. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230:249-257. [DOI] [PubMed] [Google Scholar]
- 14.Faretra, F., E. Antonacci, and S. Pollastro. 1988. Sexual behavior and mating system of Botryotinia fuckeliana, teleomorph of Botrytis cinerea. J. Gen. Microbiol. 134:2543-2550. [Google Scholar]
- 15.Fukuda, H., T. Fujii, and T. Ogawa. 1986. Preparation of a cell-free ethylene-forming system from Penicillium digitatum. Agric. Biol. Chem. 50:977-981. [Google Scholar]
- 16.Fukuda, H., T. Ogawa, and S. Tanase. 1993. Ethylene production by microorganisms. Adv. Microbiol. Physiol. 35:275-306. [DOI] [PubMed] [Google Scholar]
- 17.González-Guzmán, M., N. Apostolova, J. M. Bellés, J. M. Barrero, P. Piqueras, M. R. Ponce, J. L. Micol, R. Serrano, and P. L. Rodríguez. 2002. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14:1833-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedden, P., A. L. Phillips, M. C. Rojas, E. Carrera, and B. Tudzynski. 2002. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20:319-331. [DOI] [PubMed] [Google Scholar]
- 19.Hirai, N., M. Okamoto, and K. Koshimizu. 1986. The 1′,4′-trans-diol of abscisic acid, a possible precursor of abscisic acid in Botrytis cinerea. Phytochemistry 25:1865-1868. [Google Scholar]
- 20.Hohn, T. M., S. P. McCormick, and A. E. Desjardins. 1993. Evidence for a gene cluster involving trichothecene pathway biosynthetic genes in Fusarium sporotrichioides. Curr. Genet. 24:291-295. [DOI] [PubMed] [Google Scholar]
- 21.Hottiger, T., and T. Boller. 1991. Ethylene biosynthesis in Fusarium oxysporum f. sp. tulipae proceeds from glutamate/2-oxoglutarate and requires oxygen and ferrous ions in vivo. Arch. Microbiol. 157:18-22. [Google Scholar]
- 22.Inomata, M., N. Hirai, R. Yoshida, and H. Ohigashi. 2004. The biosynthetic pathway to abscisic acid via ionylideneethane in the fungus Botrytis cinerea. Phytochemistry 65:2667-2678. [DOI] [PubMed] [Google Scholar]
- 23.Inomata, M., N. Hirai, R. Yoshida, and H. Ohigashi. 2004. Biosynthesis of abscisic acid by the direct pathway via ionylideneethane in a fungus, Cercospora cruenta. Biosci. Biotechnol. Biochem. 68:2571-2580. [DOI] [PubMed] [Google Scholar]
- 24.Jörnvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzáles-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 25.Jörnvall, H., J.-O. Höög, and B. Persson. 1999. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445:261-264. [DOI] [PubMed] [Google Scholar]
- 26.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 27.Kettner, J. 1991. Untersuchungen zur Biosynthese von Abscisinsäure bei der Interaktion des pflanzenpathogenen Pilzes Botrytis cinerea Pers. mit der Kulturtomate Lycopersicon esculentum Mill. Dissertation. Universität Hamburg, Hamburg, Germany.
- 28.MacMillan, J. 2001. Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20:387-442. [DOI] [PubMed] [Google Scholar]
- 29.Malonek, S., M. C. Rojas, P. Hedden, P. Gaskin, P. Hopkins, and B. Tudzynski. 2005. Functional characterization of two cytochrome P450 monooxygenase genes, P450-1 and P450-4, of the gibberellic acid gene cluster in Fusarium proliferatum (Gibberella fujikuroi MP-D). Appl. Environ. Microbiol. 71:1462-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marumo, S., M. Katayama, E. Komori, Y. Ozaki, M. Natsume, and S. Kondo. 1982. Microbial production of abscisic acid by Botrytis cinerea. Agric. Biol. Chem. 46:1967-1968. [Google Scholar]
- 31.Milborrow, B. V. 2001. The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J. Exp. Bot. 52:1145-1164. [PubMed] [Google Scholar]
- 32.Neill, S. J., R. Horgan, D. C. Walton, and T. S. Lee. 1982. The biosynthesis of abscisic acid in Cercospora rosicola. Phytochemistry 21:61-65. [Google Scholar]
- 33.Neill, S. J., R. Horgan, D. C. Walton, and C. A. M. Mercer. 1987. The metabolism of α-ionylidene compounds by Cercospora rosicola. Phytochemistry 26:2515-2519. [Google Scholar]
- 34.Okamoto, M., N. Hirai, and K. Koshimizu. 1988. Biosynthesis of abscisic acid from α-ionylideneethanol in Cercospora pini-densiflorae. Phytochemistry 27:3465-3469. [Google Scholar]
- 35.Oritani, T., M. Ichimura, and K. Yamashita. 1984. A novel abscisic acid analog, (+)-(2Z,4E)-5-(1′,4′-dihydroxy-6′,6′-dimethyl-2′-methylene-cyclohexyl)-3-methyl-2,4-pentadienoic acid, from Cercospora cruenta. Agric. Biol. Chem. 48:1677-1678. [Google Scholar]
- 36.Oritani, T., and H. Kiyota. 2003. Biosynthesis and metabolism of abscisic acid and related compounds. Nat. Prod. Rep. 20:414-425. [DOI] [PubMed] [Google Scholar]
- 37.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 38.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quidde, T., A. E. Osbourn, and P. Tudzynski. 1998. Detoxification of α-tomatine by Botrytis cinerea. Physiol. Mol. Plant Pathol. 52:151-165. [Google Scholar]
- 40.Rolke, Y., S. Liu, T. Quidde, B. Williamson, A. Schouten, K.-M. Weltring, V. Siewers, K. B. Tenberge, B. Tudzynski, and P. Tudzynski. 2004. Functional analysis of H2O2-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 5:17-23. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schwartz, S. H., K. M. Léon-Kloosterziel, M. Koornneef, and J. A. D. Zeevaart. 1997. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siewers, V., J. Smedsgaard, and P. Tudzynski. 2004. The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microbiol. 70:3868-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samson, R. A., E. S. Hoekstra, J. C. Frisvad, and O. Filtenborg (ed.). 2002. Introduction to food- and airborne fungi, 6th ed. Ponson & Looyen, Wageningen, The Netherlands.
- 45.Smedsgaard, J. 1996. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. 760:264-270. [DOI] [PubMed] [Google Scholar]
- 46.Sprecher, E. 1959. Über die Guttation bei Pilzen. Planta 53:565-574. [Google Scholar]
- 47.Tag, A. G., G. F. Garifullina, A. W. Peplow, C. Ake, T. D. Phillips, T. M. Hohn, and M. N. Beremand. 2001. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67:5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ten Have, A., W. Mulder, J. Visser, and J. A. L. van Kan. 1998. The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol. Plant-Microbe Interact. 11:1009-1016. [DOI] [PubMed] [Google Scholar]
- 49.Tudzynski, B., and K. Hölter. 1998. The gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 50.Tudzynski, B., and A. Sharon. 2002. Biosynthesis, biological role and application of fungal hormones, p. 183-211. In H. D. Osiewacz (ed.), The mycota X: industrial applications. Springer-Verlag, Berlin, Germany.
- 51.Tudzynski, B., M. Mihlan, M. C. Rojas, P. Linnemannstöns, P. Gaskin, and P. Hedden. 2003. Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: des and P450-3 encode GA4 desaturase and the 13-hydroxylase, respectively. J. Biol. Chem. 278:28635-28643. [DOI] [PubMed] [Google Scholar]
- 52.Tudzynski, P., K. Hölter, T. Correia, C. Arntz, N. Grammel, and U. Keller. 1999. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 261:133-141. [DOI] [PubMed] [Google Scholar]
- 53.Vaughan, G. T., and B. V. Milborrow. 1988. The stability of the 1′,4′-diols of abscisic acid. Phytochemistry 27:339-343. [Google Scholar]
- 54.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: a hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]
- 55.Weiler, E. W. 1982. An enzyme-immunoassay for cis(+)-abscisic acid. Physiol. Plant. 54:510-514. [Google Scholar]
- 56.Yang, S. F., and N. E. Hoffman. 1984. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 35:155-189. [Google Scholar]