Abstract
Using the phage display technology, a protein can be displayed at the surface of bacteriophages as a fusion to one of the phage coat proteins. Here we describe development of this method for fusion of an intracellular carboxylesterase of Bacillus subtilis to the phage minor coat protein g3p. The carboxylesterase gene was cloned in the g3p-based phagemid pCANTAB 5E upstream of the sequence encoding phage g3p and downstream of a signal peptide-encoding sequence. The phage-bound carboxylesterase was correctly folded and fully enzymatically active, as determined from hydrolysis of the naproxen methyl ester with Km values of 0.15 mM and 0.22 mM for the soluble and phage-displayed carboxylesterases, respectively. The signal peptide directs the encoded fusion protein to the cell membrane of Escherichia coli, where phage particles are assembled. In this study, we assessed the effects of several signal peptides, both Sec dependent and Tat dependent, on the translocation of the carboxylesterase in order to optimize the phage display of this enzyme normally restricted to the cytoplasm. Functional display of Bacillus carboxylesterase NA could be achieved when Sec-dependent signal peptides were used. Although a Tat-dependent signal peptide could direct carboxylesterase translocation across the inner membrane of E. coli, proper assembly into phage particles did not seem to occur.
In the past decade, the most remarkable successes of protein engineering have been the result of combining random mutagenesis and screening by means of a high-throughput assay (6). Unfortunately, for many enzymes, such as esterases and lipases, no high-throughput methods are available; consequently, evaluation of the enantioselectivity of lipases and esterases is dependent on time-consuming assays (7). Thus, it would be highly advantageous if the screening process could be combined with a rapid selection method that limits the amounts of mutants to be assayed. Phage display (30) is a well-defined technique that has led to a breakthrough in selection methodology for enzymes with desirable properties from a pool of mutants.
Derivatives of M13 filamentous phages that are phage particles with a single-stranded genome encapsulated by the phage coat proteins (30) are most commonly used for display in Escherichia coli. Enzymes can be expressed as fusions to one of the M13 phage coat proteins, such as the g3p protein (30). As phage particles are assembled in the cell envelope of E. coli, translocation of the g3p fusion protein across the inner membrane of E. coli is a prerequisite for proper phage display. The g3p protein is synthesized with an 18-residue amino-terminal signal peptide that targets this protein to the E. coli Sec machinery for membrane insertion (26). Theoretically, any protein fused to the amino-terminal region of the g3p protein that is efficiently translocated across the inner membrane and that is able to enter the phage assembly site can be presented as a fusion protein on M13 phages (30).
Today, phage display can be used for selection of small peptides, antibody fragments, and enzymes. For instance, many enzymes, such as amylases (36), β-lactamases (35), lipases (13), and transferases (15), have been successfully displayed on bacteriophages. Most of these enzymes are extracellular enzymes exported from the cytoplasm of the homologous host in a signal peptide-dependent manner. An important exception is the gluthathione-S-transferase from Schistosoma japonicum (15).
In the general procedure for phage display, a gene of interest is cloned in a phagemid vector downstream of the signal sequence of g3p or pelB in order to direct the corresponding protein through the cell membrane via the Sec-dependent translocation pathway. In addition to the Sec pathway, which transports unfolded proteins over the inner membrane of E. coli (21, 32), the twin-arginine translocation (Tat) pathway can be distinguished. In sharp contrast to the Sec route, the Tat pathway of E. coli seems to accept only folded proteins for membrane translocation (9, 22). In this context, we wanted to determine whether translocation of a g3p fusion protein via the Tat pathway of E. coli results in productive phage display of a cytoplasmic protein. To examine this, we compared the capability and effectiveness of the Sec-specific signal peptides of E. coli TEM β-lactamase (SpBla) and g3p (SpG3p) and the capability and effectiveness of the Tat-specific signal peptide of the E. coli trimethylamine N-oxide reductase (TorA; the signal peptide is referred to as SpTor) in the export and phage display of Bacillus subtilis carboxylesterase NA (CesA) and lipase A (LipA). SpBla was used because its Sec specificity is very well documented (1), whereas SpTor was used because it was previously shown to direct the export of heterologous or truncated proteins (4, 29). Furthermore, LipA of B. subtilis, a lipase with a twin-arginine signal peptide that results in partial dependence on TatC (17), was used as a control protein for display, because functional phage display of this enzyme with the help of the g3p signal peptide was recently demonstrated (13). Importantly, the functional phage display of both CesA and LipA is of particular biotechnological interest as the corresponding displayed fusion proteins can be used for selection of improved variants for the enantioselective conversion of several interesting pharmaceutical compounds, as recently shown for selection of an S-(+)-1,2-O-ispropylidene-sn-glycerol-specific LipA mutant (10, 11).
MATERIALS AND METHODS
Plasmids, bacterial strains, and media.
The plasmids and bacterial strains that were used in the present study are listed in Table 1. E. coli HB2151ΔtatC was constructed by P1 transduction of the tatC::Spec allele (31). Helper phage M13K07 and pCANTAB 5E were purchased from Pharmacia (Amersham Pharmacia Biotech, Uppsala, Sweden). Genencor International (Leiden, The Netherlands) provided fermentor broth of strain B. subtilis 1051 producing LipA (8). LB medium contained 1% (wt/vol) Bacto Tryptone, 0.5% (wt/vol) Bacto Yeast Extract, and 0.5% (wt/vol) sodium chloride, and 2× TY medium contained 1.6% (wt/vol) Bacto Tryptone, 1% (wt/vol) Bacto Yeast Extract, and 0.5% (wt/vol) sodium chloride. Antibiotic agents (Duchefa Biochemie, Haarlem, The Netherlands) were used at the following concentrations: ampicillin, 100 μg ml−1; and kanamycin, 50 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or properties | Source or reference | ||
|---|---|---|---|---|
| E. coli strains | ||||
| TG-1 | supE K-12 Δ(lac-pro) thi hsdD5/F′ traD36 proAB laqIqlacZΔM15 | Amersham Pharmacia Biotech, Uppsala, Sweden | ||
| HB2151 | K-12 Δ(lac-pro) ara Nalrthi/F′ proAB laqIqlacZΔM15 | Amersham Pharmacia Biotech, Uppsala, Sweden | ||
| HB2151ΔtatC | K-12 Δ(lac-pro) ara Nalrthi/F′ proAB laqIqlacZΔM15 tatC::Spec | This study | ||
| Plasmids
|
||||
| pCANTABSpBlaCesA | pCANTAB 5E derivative containing the B. subtilis 168 cesA gene downstream of the bla signal sequence | This study | ||
| pCANTABSpG3pCesA | pCANTAB 5E derivative containing the B. subtilis 168 cesA gene downstream of the g3p signal sequence | This study | ||
| pCANTABSpTorACesA | pCANTAB 5E derivative containing the B. subtilis 168 cesA gene downstream of the torA signal sequence | This study | ||
| pCANTABSpBlaLipA | pCANTAB 5E derivative containing the B. subtilis 168 lipA gene downstream of the bla signal sequence | This study | ||
| pCANTABSpG3pLipA | pCANTAB 5E derivative containing the B. subtilis 168 lipA gene downstream of the g3p signal sequence | This study | ||
| pCANTABSpTorALipA | pCANTAB 5E derivative containing the B. subtilis 168 lipA gene downstream of the torA signal sequence | This study | ||
Chemicals.
The methyl ester of (S)-naproxen was provided by H. V. Wikström (Department of Medicinal Chemistry, University of Groningen, Groningen, The Netherlands). Butyrate esters of both enantiomers of 1,2-O-ispropylidene-sn-glycerol (IPG) were provided by M. T. Reetz (Max-Planck-Institut für Kohlenforschung, Mülheim, Germany). p-Nitrophenyl caprylate was purchased from Sigma Chemical Co. (Axel, The Netherlands).
DNA techniques.
Recombinant DNA techniques were performed as described by Sambrook et al. (28). Plasmid DNA was prepared as described by Birnboim and Doly (3). DNA purification was performed using a Qiaquick gel extraction kit (QIAGEN, Hilden, Germany).
Construction of phagemids.
The LipA-encoding gene (lipA) was cloned in phagemid pCANTAB 5E downstream of a modified g3p signal sequence, as described previously (13). The cesA gene sequence was amplified from the chromosomal DNA of B. subtilis 168 using primers napfor1 (5′-GCATGAATCATAGGCCCAGCCGGCCATGGCACAAAACCATTCATCTAGTATTC C-3′) and naprev1 (5′-GATCGTTAGAATGCGGCCGCCCGTGAAATGCCTGTT-3′). PCR was performed using Pfu DNA polymerase (Stratagene, La Jolla, CA). The amplified gene fragment was cloned in E. coli TG-1 into the SfiI and Eco52I sites of a modified pCANTAB vector (37). To exchange the g3p signal sequence (VKKLLFAIPLVVPFYAAQPAMA) for the signal sequence of E. coli SpBla (MSIQHFRVALIPFFAAFCLPAMA) or E. coli SpTorA (MNNNDLFQASRRRFLAQLGGLTVAGMLGPSLLTPRRATAAMA), a PCR was performed using primers 5′-CCCAAGCTTGGTACCGTTGGAGCCTTTTTTTTGGAGATTTTCAACATGAAATTTGTAAAAAGAAGG-3′ and 5′-GCGGCCATGGCAGGAAGGCAAAATGC-3′ for amplification of SpBla from template pUC18 (23) and primers 5′-CCCAAGCTTGGTACCGTTGGAGCCTTTTTTTTGGAGATTTTCAACATGAAATTTGTAAAAAGAAGG-3′ and 5′-GCGGCCATGGCCGCAGTCGCACGTCGCGGCG-3′ for amplification of SpTorA from pJDT1 (34). The PCR program consisted of two steps. For the first step we used 4 min at 94°C, followed by eight cycles of 1 min at 94°C, 2 min 50°C, and 1 min at 72°C. Then 5 pmol of primer commonsmall (5′-CCCAAGCTTGGTACCGTTGG-3′) was added, and 22 cycles of 1 min at 94°C, 2 min 50°C, and 1 min at 72°C were performed. At the end of the reaction, the DNA synthesis mixtures were incubated for 10 min at 72°C. The amplified gene fragments were cloned in E. coli TG-1 at the HindIII and NcoI sites of pCANTABSpG3pCesA and pCANTABSpG3pLipA, respectively.
Isolation of the periplasmic fraction, spheroplasts, and whole-cell extracts.
E. coli HB2151 was grown in 50-ml tubes containing 10 ml of 2× TY medium, ampicillin, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cultures were incubated at 37°C and 250 rpm for 16 h. The optical density at 600 nm was recorded, and the cells were harvested and resuspended in 10 mM Tris HCl (pH 7.4). After centrifugation, the cells were resuspended in 200 μl buffer containing 10 mM Tris HCl (pH 8.0), 25% sucrose, 2 mM EDTA, and 0.5 mg ml−1 lysozyme. After incubation on ice for 20 min, 50 μl buffer containing 10 mM Tris HCl (pH 8.0), 20% sucrose, and 125 mM MgCl2 was added. The suspension was centrifuged at 12,000 × g for 10 min, and the supernatant, representing the periplasmic fraction, was isolated and used as an enzyme solution in the activity assay. The pellet was resuspended in 200 μl buffer containing 50 mM Tris HCl (pH 8.4) and 2 mM EDTA. The suspension, representing the spheroplasts, was used as an enzyme solution in the activity assay. The protein content of each fraction was determined by a Bradford assay in duplicate using bovine serum albumin (Pierce, Rockford, Ill.) as the standard. To obtain whole-cell extracts, cells were centrifuged at 12,000 × g and taken up in 200 μl buffer containing 50 mM Tris HCl (pH 8.4) and 2 mM EDTA.
Phage rescue.
A total of 1010 helper phage particles were added to exponential-phase growing E. coli TG-1 cells transformed with the plasmids shown in Table 1 (phage-to-bacterium ratio, 30:1), and this was followed by 16 h of growth at 28°C in glucose-depleted 2× TY medium containing ampicillin and kanamycin. Phage were precipitated by addition of 5% (wt/vol) polyethylene glycol 4000 in 2.5 M NaCl. After centrifugation, the phage were resuspended in 2 ml of 10 mM Tris HCl buffer (pH 7.4) containing 1 mM EDTA and filtered through a 0.45-μm filter (Millipore, Bedford, MA). The number of phage particles in the suspension was determined by absorption spectroscopy as described by Bonnycastle et al. (5) using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, Del.). The protein content was also determined by performing a Bradford assay using bovine serum albumin as the standard.
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with 11% separating and 4% stacking polyacrylamide gels for LipA (18) and with 12.5% separating and 3% stacking gels for CesA. Molecular mass markers were purchased from Bio-Rad. After electrophoresis, proteins were blotted onto nitrocellulose and immunostained with a rabbit antiserum against LipA or CesA or with mouse monoclonal antibodies against g3p (PSKAN3; MoBiTec, Göttingen, Germany). The antibody was detected with alkaline phosphatase-conjugated antibodies against rabbits (LipA and CesA antiserum) or mice (g3p antibody).
Enzyme kinetics.
LipA enzymatic activity was determined spectrophotometrically by the p-nitrophenyl caprylate assay as described previously (19).
Esterase activity was determined using a naproxen methyl ester assay (11). The Michaelis-Menten constants (Km), specific activities, and turnover numbers for soluble and phage-bound CesA (phages were produced using E. coli TG-1 transformed with pCANTABSpBlaCesA) were determined with (S)-naproxen methyl ester substrate concentrations between 0.25 mM and 0.75 mM. All data were expressed as means ± standard errors of the means (n = 3). The statistical significance of differences was tested at a P value of <0.05 using a two-tailed Student's t test.
For analysis of IPG butyrate ester hydrolysis, assays and gas chromatographic analyses were performed as described by Dröge et al. (12).
RESULTS
Construction of phagemids.
With the ultimate aim of displaying LipA and CesA of B. subtilis on M13 phages, we cloned the corresponding genes in phagemid pCANTAB 5E downstream of a modified g3p signal sequence and upstream of a collagenase cleavage site, six consecutive histidine residues (His tag), an amber stop codon, and the sequence encoding residues 3 to 406 of the g3p coat protein (13). The original g3p maturation site in this phagemid, SHS, was modified to AAQPAMA so that it resembled the consensus site for signal peptidase I cleavage (37). The His tag can be used for purification of wild-type and mutant enzymes, and the collagenase cleavage site can be used for phage rescue during phage display selection (13).
In order to compare the effects of different Sec- and Tat-specific signal peptides in phage display, the sequences encoding signal peptides SpBla and SpTor were used to replace the g3p signal sequence in pCANTABSpG3pLipA. The resulting constructs were stably maintained in E. coli HB2151 and E. coli HB2151ΔtatC.
Export and phage display of Bacillus LipA. (i) Processing of LipA.
The periplasmic fractions, spheroplasts, and whole-cell extracts of E. coli HB2151 and E. coli HB2151ΔtatC transformed with plasmid pCANTABSpBlaLipA, pCANTABSpG3pLipA, or pCANTABSpTorALipA were isolated to examine the processing of LipA and to determine the activity with p-nitrophenyl caprylate. As the TAG stop codon is not suppressed in these strains, the different forms of LipA encoded by the three plasmids are not fused to g3p. SDS-PAGE under reducing conditions and Western blot analysis with a rabbit antiserum against LipA resulted in detection of mature LipA at a molecular mass of approximately 21 kDa, corresponding to the molecular mass of the His-tagged enzyme (Fig. 1A, C, and E). The different precursor proteins were detected at apparent molecular masses of 23 to 27 kDa, depending on the signal peptide used (SpBlaLipA, 24 kDa; SpG3pLipA, 23 kDa; and SpTorALipA, 27 kDa). Notably, for some signal peptide-LipA fusions, particularly the SpTorALipA fusion, several distinct precursor forms were observed.
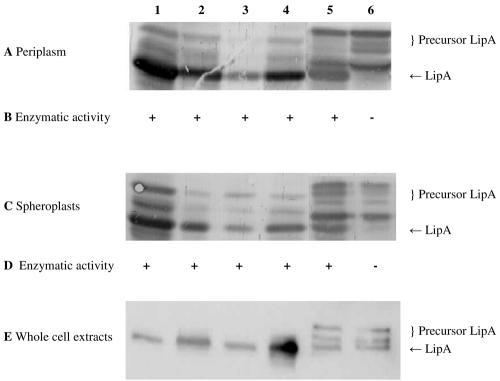
FIG. 1.
Detection of LipA in the periplasmic fraction, spheroplasts, and whole-cell extract (10 μg of protein per lane). SDS-PAGE (11% gel), Western blotting, and immunostaining with a rabbit antiserum against LipA were performed with the periplasmic fractions (A), spheroplasts (C) and whole-cell extracts (E) of E. coli HB2151 and E. coli HB2151ΔtatC transformed with pCANTABSpBlaLipA (lanes 1 and 2), pCANTABSpG3pLipA (lanes 3 and 4), or pCANTABSpTorALipA (lanes 5 and 6). Lanes 1, 3, and 5 contained samples from E. coli HB2151, and lanes 2, 4, and 6 contained samples from E. coli HB2151ΔtatC. Activities of LipA in the periplasmic fraction (B) and in the lysed spheroplasts (D) were determined using p-nitrophenyl caprylate as the substrate. +, enzyme activity; −, no enzyme activity.
During isolation of the periplasmic fractions, some cell lysis may have occurred as precursor proteins were detected in these fractions. This effect was most prominent in strains containing pCANTABSpTorALipA, suggesting that production of the SpTorALipA fusion protein makes the cells more prone to cell lysis or that the corresponding precursor forms are not efficiently retained in the inner membrane. Conversely, mature LipA was observed in the spheroplasts, which was obviously explained by the presence of membranes in these fractions and the lipophilic behavior of lipases in general. However, comparison of the periplasmic and spheroplast fractions revealed that with the exception of the SpTorALipA-producing E. coli strains, the relative amounts of the different precursor proteins were lower in the periplasmic fractions than in the spheroplasts. Most interestingly, LipA was present exclusively in precursor forms in the periplasmic and spheroplast fractions of the ΔtatC mutant transformed with plasmid pCANTABSpTorALipA. As the precise fusion between SpTorA and LipA, encoded by pCANTABSpTorALipA, resulted in accumulation of relatively high levels of SpTorALipA precursor forms in the wild-type E. coli strain, an alternative SpTorALipA fusion containing the original signal peptidase recognition site of SpTorA and the first three residues of the mature TorA protein (pCANTABSpTorA-AQAATD-LipA) was constructed (Fig. 2). This, however, did not result in reduced levels of the SpTorALipA precursor. Importantly, neither of the two SpTorALipA constructs was processed to mature LipA in the ΔtatC strain, indicating that they require functional Tat machinery for membrane translocation and subsequent processing by a signal peptidase.
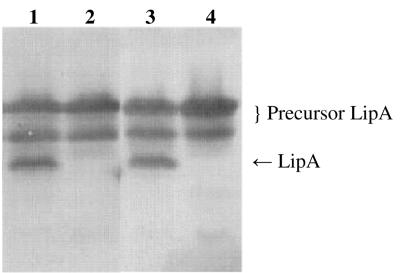
FIG. 2.
TatC-dependent processing of SpTorALipA fusions. SDS-PAGE (11% gel), Western blotting, and immunostaining with a rabbit antiserum against LipA were performed with cell lysates of E. coli HB2151 and E. coli HB2151ΔtatC transformed with pCANTABSpTorA-AQAATD-LipA (lanes 1 and 2, respectively) and cell lysates of E. coli HB2151 and E. coli HB2151ΔtatC transformed with pCANTABSpTorALipA (lanes 3 and 4, respectively).
To investigate the enzymatic activity of LipA in the periplasmic fraction and lysed spheroplasts of E. coli HB2151 and the ΔtatC mutant, the specific activity with p-nitrophenyl caprylate was determined (Fig. 1B and D). All fractions containing mature LipA were able to hydrolyze the caprylate ester of p-nitrophenol. Note that no LipA activity was detected in fractions derived from the ΔtatC mutant producing SpTorALipA and that the same was true for fractions of E. coli strains not producing LipA (data not shown).
Together, these results show that LipA precursors are translocated across the membrane and are processed to the mature form in E. coli HB2151 whether a Sec-dependent signal peptide or a Tat-dependent signal peptide is used.
(ii) Phage display of LipA.
E. coli TG-1 cells, transformed with pCANTABSpBlaLipA, pCANTABSpG3pLipA, or pCANTABSpTorALipA, were infected with M13K07 helper phages to produce phage particles containing the phagemid genome and a mixture of wild-type g3p and LipA-g3p fusion proteins. The LipA fusion to g3p resulted from partial suppression of the TAG stop codon in the E. coli TG-1 host cells. To visualize the presence of phage-bound LipA, phage particles were analyzed by SDS-PAGE under reducing conditions and by immunoblotting with a rabbit antiserum against LipA (Fig. 3A).
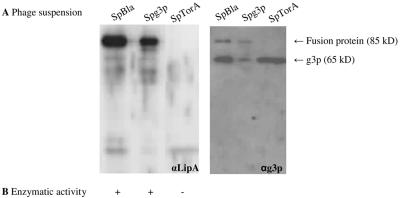
FIG. 3.
Detection of phage-bound LipA. (A) LipA-g3p fusion proteins in 0.25-μg phage suspensions were visualized by SDS-PAGE, Western blotting, and immunostaining with rabbit antisera against LipA (αLipA) (left panel) or mouse monoclonal antibodies against g3p (αg3p) (right panel). Phages were isolated from the growth media of E. coli TG-1 cells transformed with pCANTABSpBlaLipA (SpBla), pCANTABSpG3pLipA (SpG3p), or pCANTABSpTorALipA (SpTorA). (B) Hydrolysis of p-nitrophenyl caprylate by phage-bound LipA. +, hydrolysis; −, no hydrolysis.
A LipA-g3p fusion protein was detected in phages isolated from the growth media of E. coli TG-1 cells transformed with pCANTABSpBlaLipA or pCANTABSpG3pLipA. This fusion protein had an apparent molecular mass of approximately 85 kDa, which corresponds to the apparent molecular mass of LipA plus the apparent molecular mass of a g3p protein on SDS-PAGE gels (33). Notably, the LipA-g3p fusion protein was absent from the phage suspension derived from E. coli TG-1 cells transformed with pCANTABSpTorALipA.
Previously, it has been demonstrated by determination of the Michaelis-Menten constants and the turnover numbers of soluble and phage-bound LipA proteins (phages were produced using E. coli TG-1 transformed with pCANTABSpG3pLipA) that phage-bound LipA was correctly folded and fully enzymatically active (13). To assess whether the specific lipase activities in the different phage suspensions corresponded to the amounts of fusion protein, the activity with p-nitrophenyl caprylate was determined. Importantly, Fig. 3B shows that the presence of the fusion protein correlated with LipA activity. Overall, the highest lipase activity and largest amount of fusion protein were obtained when Sec-specific signal peptides were used. Although the Tat-specific signal peptide of TorA can be used to direct export of LipA of B. subtilis to the periplasmic space of E. coli, proper phage display of the TatC-dependent exported enzyme seems to be impaired.
Export and phage display of Bacillus CesA. (i) Processing of CesA.
To investigate the export and processing of CesA, the periplasmic fractions, spheroplasts, and whole-cell extracts of E. coli HB2151 and E. coli HB2151ΔtatC transformed with plasmid pCANTABSpBlaCesA, pCANTABSpG3pCesA, or pCANTABSpTorACesA were isolated, and SDS-PAGE under reducing conditions and Western blot analyses were performed with a rabbit antiserum against CesA (Fig. 4A, C, and E). Mature CesA with the predicted molecular mass (36 kDa) was detected in both strains. Furthermore, CesA precursor forms were detected at apparent molecular masses ranging from 38 to 40 kDa (SpBlaCesA, 38 kDa; SpG3pCesA, 38 kDa; SpTorACesA, 40 kDa) in most periplasmic fractions and spheroplasts. Again, precursor forms of CesA were detected in the periplasmic fractions, indicating either that the E. coli strains were subject to lysis upon spheroplasting or that the precursors are not effectively retained in the inner membrane. Importantly, Fig. 4 shows that cytoplasmic protein CesA of B. subtilis can be exported to the periplasm of E. coli using Sec-specific signal peptides. Mature CesA was detected in the periplasmic fraction of E. coli HB2151 cells producing SpTorACesA as well, although most of the mature CesA was present in the cytoplasmic fraction of these cells. In contrast, the SpTorACesA produced in the ΔtatC mutant remained mainly in the precursor form, which is consistent with the fact that SpTorA is a Tat-specific signal peptide. These observations suggest that CesA can be translocated across the membrane via the Tat machinery.
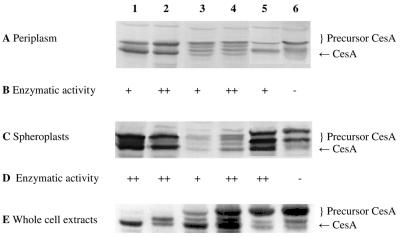
FIG. 4.
Detection of CesA in the periplasmic fraction, spheroplasts, and whole-cell extract (20 μg of protein per lane): SDS-PAGE (12.5% gel), Western blotting, and immunostaining with a rabbit antiserum against CesA of the periplasmic fractions (A), spheroplasts (C), and whole-cell extracts (E) of E. coli HB2151 and E. coli HB2151ΔtatC transformed with pCANTABSpBlaCesA (lanes 1 and 2), pCANTABSpG3pCesA (lanes 3 and 4), or pCANTABSpTorACesA (lanes 5 and 6). Lanes 1, 3, and 5 contained samples from E. coli HB2151, and lanes 2, 4, and 6 contained samples from E. coli HB2151ΔtatC. (B) Hydrolysis of racemic esters of 1,2-O-ispropylidene-sn-glycerol butyrate. (D) Hydrolysis of the methyl ester of (S)-naproxen. +, enzymatic activity; ++, higher enzymatic activity; −, no enzymatic activity.
Determination of the IPG-hydrolyzing activity of CesA in the periplasmic fraction and lysed spheroplasts of E. coli HB2151 and the ΔtatC mutant revealed that an endogenous IPG hydrolase was present in the cytoplasm of E. coli. This complicated determination of the specific CesA activity in the lysed spheroplast fractions. In contrast, no endogenous IPG hydrolase activity was detected in the periplasmic fractions of E. coli HB2151 and the ΔtatC mutant (Fig. 4B). Although the level of IPG conversion was very low, enzymatic activities were found in most of the periplasmic fractions of E. coli HB2151 and the ΔtatC mutant. However, no CesA activity was detected in the periplasmic fraction of E. coli HB2151ΔtatC transformed with plasmid pCANTABSpTorACesA.
In another attempt to determine the activity of CesA in lysed spheroplasts, the specific activity with the methyl ester of (S)-naproxen was examined (Fig. 4D). Although the rate of conversion to (S)-naproxen was low, CesA activity was observed in most of the lysed spheroplast fractions. However, no (S)-naproxen methyl ester hydrolysis was detected in the lysed spheroplast fraction of the ΔtatC mutant transformed with pCANTABSpTorACesA. These findings imply that the CesA precursor forms detected in the spheroplasts and periplasmic cell fractions were not enzymatically active.
(ii) Phage display of CesA.
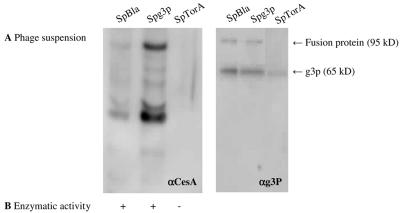
Phage particles were produced using E. coli TG-1 cells transformed with pCANTABSpBlaCesA, pCANTABSpG3pCesA, or pCANTABSpTorACesA. SDS-PAGE under reducing conditions and Western blot analysis with mouse monoclonal antibodies against g3p and with a rabbit antiserum against CesA were performed. Two specific protein bands reacting with the antibodies against the g3p coat protein were detected (Fig. 5B). These bands corresponded to g3p (apparent molecular mass, 65 kDa [33]) and to a CesA-g3p fusion protein. The apparent molecular mass of the CesA- g3p fusion protein was approximately 95 kDa, which corresponds to the molecular mass of CesA plus the molecular mass of a g3p protein. This fusion protein was also detected with the CesA antibody at 95 kDa (Fig. 5A). Notably, the CesA-g3p fusion protein was not detected in the phage suspension derived from cells producing SpTorACesA. The reaction of anti-CesA with bands around 65 kDa and 35 kDa most likely reflected the presence of degradation products of the CesA-g3p fusion protein. Despite the fact that only very low levels of (S)-naproxen methyl ester hydrolysis were detected, CesA activity was observed for both phage suspensions derived from cells producing CesA-g3p with Sec-specific signal peptides.
FIG. 5.
Detection of phage-bound CesA. (A) CesA-g3p fusion proteins in 0.25-μg phage suspensions were visualized by SDS-PAGE, Western blotting, and immunostaining with rabbit antisera against CesA (αCesA) (left panel) or mouse monoclonal antibodies against g3p (αg3P) (right panel). Phages were isolated from the growth media of E. coli TG-1 cells transformed with pCANTABSpBlaCesA, pCANTABSpG3pCesA, or pCANTABSpTorACesA. (B) Hydrolysis of the methyl ester of (S)-naproxen by phage-bound CesA. +, enzymatic activity; −, no enzymatic activity.
Enzymatic activity of soluble and phage-bound Bacillus CesA.
To investigate whether the kinetic properties of the phage-bound CesA were altered, the Km and specific activities were determined for both soluble and phage-bound CesA (Table 2). The steady-state hydrolysis of (S)-naproxen methyl ester showed that the Km of the enzyme remained unchanged (no significant difference at a P value of >0.05), suggesting that the protein was correctly folded and fully enzymatically active. In contrast, the specific activity of the phage-bound CesA was reduced. The observed difference was likely due to the different individual weights of the soluble CesA and a phage particle (5.98 × 10−17 and 2.36 × 10−14 mg, respectively).
TABLE 2.
Enzyme kinetics of CesA from B. subtilis (n = 3)a
| Enzyme | Sp act (U mg−1) | Km (mM) |
|---|---|---|
| Soluble CesA | 12.8 ± 11.3 | 3.71 ± 4.38 |
| Phage-bound CesA | 0.22 ± 0.07 | 0.22 ± 0.13 |
Statistical significance of differences, P < 0.05.
Taken together, our findings show that both LipA and CesA can be effectively exported from the cytoplasm of E. coli and displayed on M13 phages with the help of Sec-specific signal peptides, but not with the Tat-specific signal peptide of TorA.
DISCUSSION
The present study showed for the first time functional phage display of cytoplasmic protein CesA of B. subtilis as a fusion to the phage M13 minor coat protein g3p.
It was tempting to speculate that phage display of heterologous cytoplasmic proteins, such as Bacillus CesA, in E. coli might be enhanced if the fusion proteins were exported from the cytoplasm via the Tat pathway, as the Tat pathway is used for export of intracellularly folded proteins (2, 27). Remarkably, our results show that functional phage display of the CesA-g3p and LipA-g3p fusion proteins could be achieved only if Sec-specific signal peptides (SpBla and SpG3p) were used for translocation of the fusion protein across the inner membrane of E. coli. In marked contrast, the use of the Tat-specific signal peptide SpTorA did not result in functional phage display of these g3p fusion proteins. These results are in accordance with the recently described results of Paschke and Höhne. These authors demonstrated that fusion proteins containing mutated green fluorescent protein and the C-terminal domain of g3p, using a TorA or PelB signal sequence, could not be displayed sufficiently on phages. However, phage display was ultimately achieved by transporting g3p and green fluorescent protein to the periplasm independently, followed by combination using a coiled coil/disulfide strategy. Paschke and Höhne suggested that the unfolded g3p domain is not suitable for Tat-dependent export (24).
At present, the reason why Tat-specific export of the g3p fusion proteins tested did not result in their incorporation into phages remains unclear. Assembly of M13 phages occurs at sites in the cell envelope where the inner and outer membranes are in close contact (20). Prior to incorporation into the phage particle, all phage proteins are assembled in the inner membrane (14, 25). Specifically, the g3p protein requires the Sec pathway for inner membrane assembly (26). Thus, there are at least two possible explanations for the ineffectiveness of SpTorA in phage display. First, the bacterial Tat machinery seems to accept only folded proteins for translocation (9), which may have a negative impact on assembly of g3p fusion proteins into phages. Possibly the CesA-g3p and LipA-g3p fusion proteins are competent for assembly into phages only if they are translocated via the Sec machinery in an unfolded state. Translocation in a folded state via the Tat machinery might render them incompetent for phage assembly. Second, the Tat system may not be able to sort proteins to the specific sites where phage assembly takes place. For example, the Tat pathway may export the g3p fusion proteins to the periplasm. This would hamper the assembly of these fusion proteins into phages, because they need to remain attached to the inner membrane for this purpose. However, missorting of g3p fusion proteins to the periplasm seems somewhat unlikely as it has been demonstrated recently that integral membrane proteins with a carboxyl-terminal membrane anchor (like g3p) can be inserted into the membrane by a Tat-dependent mechanism (16). Nevertheless, at least in the case of the CesA-g3p fusion, a missorting event seems to be a plausible explanation for the lack of phage incorporation upon Tat-dependent membrane translocation, because some of the mature CesA (not fused to g3p) that resulted from SpTorACesA processing was released into the periplasm. However, most of the mature protein was detected in the spheroplasts. In contrast, large amounts of the mature forms of CesA that resulted from SpBlaCesA or SpG3pCesA processing were detected in the periplasmic cell fraction.
Remarkably, cell fractionation experiments revealed that a significant proportion of all hybrid CesA and LipA precursor proteins analyzed in this study was readily released from spheroplasts into the spheroplast supernatant (i.e., the periplasmic fraction). This suggests either that spheroplasts of cells producing these precursor proteins were sensitive to cell lysis or that the precursor proteins were not effectively retained in the inner membrane of E. coli HB2151 and its ΔtatC mutant derivative. Although it is difficult to distinguish between these two possibilities at this time, we favor the idea that the precursors were poorly retained. This preference is related to the observation that the endogenous IPG hydrolase activity of E. coli HB2151 was detected only in cytoplasmic cell fractions and not in periplasmic cell fractions. However, we cannot rule out the possibility that this hydrolase activity was not present in the periplasmic fractions due to the presence of an as-yet-unidentified periplasmic inhibitor. Conversely, the presence of mature LipA and CesA in the cytoplasmic fractions can be explained by the presence of membranes in these fractions and the lipophilic behavior of these lipases and esterases. Finally, the CesA and LipA activity assays with periplasmic and cytoplasmic fractions of the E. coli HB2151ΔtatC mutant revealed that the SpTor precursor forms of these proteins are enzymatically inactive. Thus, it seems that fusion of CesA and LipA to the SpTor signal peptide not only precludes display of these proteins on M13 phages but also affects their ability to fold into an enzymatic active form. This could imply that these proteins do not reach the relevant folding catalysts when they are targeted in the Tat pathway. In fact, this may be true not only for mature CesA and LipA but also for the CesA-g3p and LipA-g3p fusion proteins. If this is true, the inability of SpTorA to direct functional phage display may be related both to targeting and to folding problems.
In conclusion, functional display of cytoplasmic protein CesA of B. subtilis can be achieved when Sec-dependent signal peptides are used for this purpose. Although the use of a Tat-dependent signal peptide, SpTorA, can result in CesA precursor processing, the mature form of this protein remains membrane bound. Proper phage display using SpTorA seems to be impossible for this substrate. It will be a major challenge in future phage display research to elucidate the molecular mechanisms underlying these observations. It is anticipated that such studies will provide novel insights concerning the mechanism of g3p assembly into M13 phages.
Acknowledgments
This project was funded by the European Commission under proposals QLK3-CT-2001-00519 and QLK3-CT-1999-00917.
We thank all our partners for discussions and contributions leading to generation of this project. We thank Tracy Palmer for providing P1 phages required to introduce the tatC::Spec cassette into E. coli HB2151.
REFERENCES
- 1.Beha, D., S. Deitermann, M. Müller, and H. G. Koch. 2003. Export of beta-lactamase is independent of the signal recognition particle. J. Biol. Chem. 278:22161-22167. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C., T. Palmer, and F. Sargent. 2003. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microbiol. Physiol. 47:187-254. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim, H., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 5.Bonnycastle, L. L. C., A. Menendez, and J. K. Scott. 2001. General phage methods, p. 15.17-15.18. In C. F. Barbas III, D. R. Burton, J. K. Scott, and G. J. Silverman (ed.), Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Bornscheuer, U. T. 2002. Methods to increase enantioselectivity. Curr. Opin. Biotechnol. 13:543-547. [DOI] [PubMed] [Google Scholar]
- 7.Bornscheuer, U. T., and R. J. Kazlauskas. 1999. Hydrolases in organic chemistry. Regio- and stereoselective biotransformations. Wiley-VCH Verlag GmbH, Weinhein, Germany.
- 8.Dartois, V., J. Y. Coppée, C. Colson, and A. Baulard. 1994. Genetic analysis and overexpression of lipolytic activity in Bacillus subtilis. Appl. Environ. Microbiol. 60:1670-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeLisa, M. P., D. Tullman, and G. Georgiou. 2003. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA 100:6115-6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dröge, M. J., Y. L. Boersma, G. van Pouderoyen, T. E. Vrenken, C. J. Rüggeberg, M. T. Reetz, B. W. Dijkstra, and W. J. Quax. 2006. Directed evolution of Bacillus subtilis lipase A by use of enantiomeric phosphonate inhibitors: crystal structures and phage display selection. Chembiochemistry 7:149-157. [DOI] [PubMed] [Google Scholar]
- 11.Dröge, M. J., R. Bos, and W. J. Quax. 2001. Paralogous gene analysis reveals a highly enantioselective 1,2-O-isopropylideneglycerol caprylate esterase of Bacillus subtilis. Eur. J. Biochem. 268:3332-3338. [DOI] [PubMed] [Google Scholar]
- 12.Dröge, M. J., R. Bos, H. J. Woerdenbag, and W. J. Quax. 2003. Chiral gas chromatography for the determination of 1,2-O-isopropylidene-sn-glycerol stereoisomers. J. Sep. Sci. 26:1-6. [Google Scholar]
- 13.Dröge, M. J., C. J. Rüggeberg, A. M. van der Sloot, J. Schimmel, D. S. Dijkstra, R. M. D. Verhaert, M. T. Reetz, and W. J. Quax. 2003. Binding of phage displayed Bacillus subtilis lipase A to a phosphonate suicide inhibitor. J. Biotechnol. 101:19-28. [DOI] [PubMed] [Google Scholar]
- 14.Feng, J., P. Model, and M. Russel. 1999. A trans-envelope protein complex needed for filamentous phage assembly and export. Mol. Microbiol. 34:745-755. [DOI] [PubMed] [Google Scholar]
- 15.Hansson, L. O., M. Widersten, and B. Mannervik. 1997. Mechanism based phage display and selection of active-site mutants of human glutathione transferase A1-1 SNAr reactions. Biochemistry 36:11252-11260. [DOI] [PubMed] [Google Scholar]
- 16.Hatzixanthis, K., T. Palmer, and F. Sargent. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 49:1377-1390. [DOI] [PubMed] [Google Scholar]
- 17.Jongbloed, J. D., H. Antelmann, M. Hecker, R. Nijland, S. Bron, U. Airaksinen, F. Pries, W. J. Quax, J. M. van Dijl, and P. G. Braun. 2002. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277:44068-44078. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lesuisse, E., K. Schanck, and C. Colson. 1993. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 216:155-160. [DOI] [PubMed] [Google Scholar]
- 20.Lopez, J., and R. E. Webster. 1985. Assembly site of bacteriophage f1 corresponds to adhesion zones between the inner and outer membranes of the host cell. J. Bacteriol. 163:1270-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manting, E. H., and A. J. Driessen. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 22.Muller, M. 2005. Twin-arginine-specific protein export in Escherichia coli. Res. Microbiol. 156:131-136. [DOI] [PubMed] [Google Scholar]
- 23.Norrander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 24.Paschke, M., and W. Höhne. 2005. A twin-arginine translocation (Tat)-mediated phage display system. Gene 350:79-88. [DOI] [PubMed] [Google Scholar]
- 25.Rakonjac, J., J. Feng, and P. Model. 1999. Filamentous phage are released from the bacterial membrane by a two step mechanism involving a short C-terminal fragment of pIII. J. Mol. Biol. 289:1253-1265. [DOI] [PubMed] [Google Scholar]
- 26.Rapoza, M. P., and R. E. Webster. 1993. The filamentous bacteriophage assembly proteins require the bacterial Sec protein for correct localization to the membrane. J. Bacteriol. 175:1856-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Santini, C.-L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L.-F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 30.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 31.Stanley, N. R., T. Palmer, and B. C. Berks. 2000. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275:11591-11596. [DOI] [PubMed] [Google Scholar]
- 32.Stephenson, K. 2005. Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway. Mol. Membr. Biol. 22:17-28. [DOI] [PubMed] [Google Scholar]
- 33.Tesar, M., C. Beckmann, P. Röttgen, B. Haase, U. Faude, and K. N. Timmis. 1995. Monoclonal antibody against pIII of filamentous phage: an immunological tool to study fusion protein expression in phage display systems. Immunotechnology 1:53-64. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 35.Vanwetswinkel, S., J. Marchand-Brynaert, and J. Fastrez. 1996. Selection of the most active enzymes from a mixture of phage-displayed β-lactamase mutants. Bioorg. Med. Chem. Lett. 6:789-792. [Google Scholar]
- 36.Verhaert, R. M. D., J. Beekwilder, R. Olsthoorn, J. van Duin, and W. J. Quax. 2002. Phage display selects for amylases with improved low pH starch-binding. J. Biotechnol. 96:103-118. [DOI] [PubMed] [Google Scholar]
- 37.Verhaert, R. M. D., J. van Duin, and W. J. Quax. 1999. Processing and functional display of the 86 kDa heterodimeric penicillin G acylase on the surface of phage fd. Biochem. J. 342:415-422. [PMC free article] [PubMed] [Google Scholar]