Abstract
Cladophora glomerata, a macrophytic green alga, is commonly found in the Great Lakes, and significant accumulations occur along shorelines during the summer months. Recently, Cladophora has been shown to harbor high densities of the fecal indicator bacteria Escherichia coli and enterococci. Cladophora may also harbor human pathogens; however, until now, no studies to address this question have been performed. In the present study, we determined whether attached Cladophora, obtained from the Lake Michigan and Burns Ditch (Little Calumet River, Indiana) sides of a breakwater during the summers of 2004 and 2005, harbored the bacterial pathogens Shiga toxin-producing Escherichia coli (STEC), Salmonella, Shigella, and Campylobacter. The presence of potential pathogens and numbers of organisms were determined by using cultural methods and by using conventional PCR, most-probable-number PCR (MPN-PCR), and quantitative PCR (QPCR) performed with genus- and toxin-specific primers and probes. While Shigella and STEC were detected in 100% and 25%, respectively, of the algal samples obtained near Burns Ditch in 2004, the same pathogens were not detected in samples collected in 2005. MPN-PCR and QPCR allowed enumeration of Salmonella in 40 to 80% of the ditch- and lakeside samples, respectively, and the densities were up to 1.6 × 103 cells per g Cladophora. Similarly, these PCR methods allowed enumeration of up to 5.4 × 102 Campylobacter cells/g Cladophora in 60 to 100% of lake- and ditchside samples. The Campylobacter densities were significantly higher (P < 0.05) in the lakeside Cladophora samples than in the ditchside Cladophora samples. DNA fingerprint analyses indicated that genotypically identical Salmonella isolates were associated with geographically and temporally distinct Cladophora samples. However, Campylobacter isolates were genetically diverse. Since animal hosts are thought to be the primary habitat for Campylobacter and Salmonella species, our results suggest that Cladophora is a likely secondary habitat for pathogenic bacteria in Lake Michigan and that the association of these bacteria with Cladophora warrants additional studies to assess the potential health impact on beach users.
Cladophora is a branching, filamentous, green alga (Chlorophyta, Cladophoraceae) that is found in both fresh and marine waters (17). While Cladophora grows primarily on rocky substrates, it often becomes detached and accumulates along the shoreline, forming large, foul-smelling algal mats. Recently, Cladophora spp. (mostly Cladophora glomerata L.) obtained from several southern and northern Lake Michigan beaches were shown to harbor high densities of Escherichia coli and enterococci (57). These bacteria may even grow on Cladophora under certain conditions (12). Algae, including Cladophora, have been reported to provide nutrients and to protect attached bacteria from environmental stresses, such as desiccation, predation, and harmful radiation (12, 35). These findings support the hypothesis that Cladophora can potentially harbor and enhance the survival of pathogenic bacteria released into the environment through point and nonpoint sources (12).
While it has been argued previously that growth of fecal bacteria in secondary habitats is limited, there have been numerous reports which have documented that suspended particles and sediments increase the survival time of bacteria in water (7, 10). Recently, Byappanahalli et al. (13) and Ishii and coworkers (23) demonstrated that E. coli can grow and persists in northern temperate soils exposed to extreme temperature conditions. Taken together, these studies indicate that many secondary habitats provide conditions that are conducive for the growth and survival of fecal bacteria that were once thought to be restricted to the gastrointestinal tracts of warm-blooded animals. This has obvious public health and regulatory implications.
Some Shiga toxin-producing E. coli (STEC), Salmonella, Shigella, and Campylobacter strains cause diarrheal diseases in humans. The STEC strains produce Shiga-like toxins and belong to a broad range of serotypes (45). While E. coli serotype O157:H7, most likely originating from cattle, is one of the most recognized serotypes and has received a lot of attention recently, other serotypes are also responsible for human disease and are prevalent in various host animals (45). Campylobacter has been found to be the leading cause of diarrhea in humans in the United States (46%), followed by Salmonella (28%), Shigella (17%), and E. coli O157:H7 (5%) (3). The worldwide rate of Campylobacter infections has been increasing over the last several years and frequently exceeds the rates of infections caused by Salmonella and Shigella (3, 40). Campylobacter jejuni and Campylobacter coli are the two Campylobacter species that are isolated most frequently from human patients (3, 9). Although Campylobacter was previously thought to survive poorly in the environment (26, 28), C. jejuni has been isolated from soil, surface water, beach sand, and waterborne protozoans (9, 16, 50). Protozoan-enabled survival of Salmonella and Shigella has also been reported previously (27).
Humans are the primary habitat for Shigella (19), while birds have been thought to be the major reservoir of Salmonella and Campylobacter (3, 31). These human pathogens, however, have also been reported to exist outside their primary hosts. For example, Salmonella has been reported to survive in nonmammalian environments, including fresh vegetables (41), reptiles (29), water, and macroorganisms (58), as well as in sands and soils (56, 58). Similarly, E. coli O157:H7 has been detected in fresh vegetables and apple cider, and recently, growth of this bacterium was observed in manure-rich soils (4). Since E. coli has been shown to survive and even grow in soils and on the macroalga Cladophora (11, 12, 23, 43, 57), pathogenic E. coli may also be able to survive and grow in these environments.
In the research described here, we investigated whether attached Cladophora at a southern Lake Michigan beach was a secondary habitat for the potential bacterial pathogens STEC, Salmonella, Shigella, and Campylobacter. The presence of pathogens and the numbers of bacteria were determined by cultural methods and by conventional PCR, most-probable-number PCR (MPN-PCR), and quantitative PCR (QPCR) performed with genus- and toxin-specific primers and TaqMan probes. Here we report that over a 2-year period, STEC, Salmonella, Shigella, and Campylobacter were readily isolated from Cladophora. DNA fingerprint analyses indicated that genotypically identical Salmonella isolates associated with geographically separated Cladophora algae. The association of these bacteria with Cladophora warrants additional studies to assess the risk to public health, as well as the impact on regulatory issues.
MATERIALS AND METHODS
Site description and sampling.
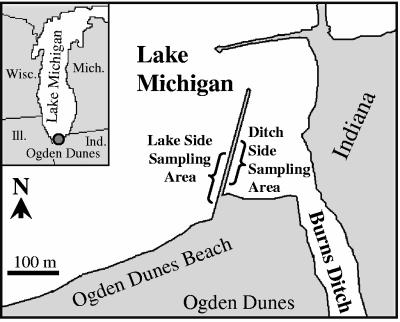
Cladophora samples were collected from a breakwater located within the Indiana Dunes National Lakeshore in northwest Indiana (Fig. 1). The breakwater is located between Lake Michigan and the Burns Ditch embayment at Ogden Dunes Beach in Ogden Dunes, Indiana. Water from the Little Calumet River feeds into Lake Michigan via the Burns Ditch outfall; the river receives a complex combination of rural and urban waste, including combined sewage overflows, septic field leachate, urban runoff, sludge or runoff from manure-enriched farm fields, significant sedimentation, and nutrient loads (42). Samples were collected in July and September 2004 and monthly between July and October 2005. Five Cladophora samples (thalli) were collected from each site at each time, except for the 2004 samples (n = 4) and the September 2005 ditchside samples (n = 3). Cladophora attached to submerged rocks was collected from below the waterline on the Lake Michigan side (less polluted) and on the Burns Ditch side (more polluted) of the breakwater. Rocks with attached Cladophora were generally well separated (lake side, 3 to 12 m apart; ditch side, 5 to 25 m apart). Cladophora samples were collected by hand, placed in Whirl-Pak bags, and transported to the laboratory at 4°C. The samples were immediately shipped to the University of Minnesota and were analyzed within 24 h of collection.
FIG. 1.
Locations of sampling sites at the Indiana Dunes National Lakeshore. Samples were taken near a breakwater located between Lake Michigan and the Burns Ditch embayment at Ogden Dunes Beach in Ogden Dunes, Indiana. Cladophora samples were taken from both ditch- and lakeside sampling areas.
Bacterial elutriation.
Algal samples (1 g) were shaken in 9 ml of phosphate-buffered water (pH 6.8) (PBW) as described previously (12, 57). To obtain greater numbers of bacteria, algal subsamples (10 to 25 g) were placed in sterile bottles and extracted with PBW containing 0.01% hydrolyzed gelatin (54) at a 1:4 (wt/vol) ratio. The bottles were shaken for 30 min using a wrist action shaker and allowed to stand for 20 min, and the upper phase was used as the initial dilution (2 × 10−1) for MPN analysis, for presence/absence analysis by conventional PCR, and for real-time QPCR assays as described below.
Most-probable-number analyses.
Five-tube MPN analyses (2) were used to quantify culturable, Cladophora-borne, pathogenic bacteria and E. coli. Aliquots (1 ml) of the initial dilution were serially diluted in 9 ml PBW to a 2 × 10−6 dilution. Aliquots (5 ml and 0.5 ml) of the initial dilution were added to selective broth media (45 and 4.5 ml, respectively). Aliquots (0.5 ml) of all subsequent dilutions were added to 4.5 ml of selective broth media. Lauryl tryptose broth (LTB) containing 4-methylumbelliferyl-β-d-glucuronide (MUG) (Difco, Detroit, MI), tetrathionate broth (TTB) (Difco), Hajna GN broth (Difco), and Bolton broth (Oxoid, Hampshire, United Kingdom) were used for selective growth of E. coli, Salmonella, Shigella, and Campylobacter, respectively. Most culture tubes were incubated at 37°C for 48 h; the exceptions were the Bolton broth tubes, which were incubated at 37°C for 4 h and then at 42°C for 44 h under microaerobic conditions created with the CampyPak Plus microaerophilic system with a palladium catalyst (Becton-Dickinson Microbiology Systems, Cockeysville, MD) (22). MPN counts were calculated based on previously published tables (2) and were expressed as log MPN per g (dry weight) of Cladophora. The means and ranges of MPN counts for five replicate samples are reported below on a dry weight basis.
Isolate confirmation and identification.
Selective and differential agar media (52) were used to confirm the presence of the pathogenic bacteria in MPN tubes. The presence of E. coli in MPN tubes containing LTB supplemented with MUG was confirmed by fluorescence under long-wave UV light (366 nm) (54), and the presence of STEC in tubes containing LTB supplemented with MUG, the presence of Salmonella in tubes containing TTB, the presence of Shigella in tubes containing GN broth, and the presence of Campylobacter in tubes containing Bolton broth were confirmed by PCR performed with toxin- or genus-specific primers as described below. Salmonella sp. strains in tubes containing TTB were isolated by streaking samples onto bismuth sulfide agar (Difco) and incubating the preparations at 37°C for 48 h. Well-isolated black colonies were spot inoculated onto MacConkey agar (Difco), Rambach agar (CHROMagar Microbiology, Paris, France) (48), and urea agar (Difco) plates and incubated at 37°C for 24 h. Colonies exhibiting responses typical of Salmonella on these media (transparent colonies on MacConkey agar, pink colonies on Rambach agar, and yellow growth on urea agar) were tested further using triple sugar iron agar (Difco) slants (52). Confirmed Salmonella isolates were stored in 50% glycerol at −70°C until they were used. Three isolates per MPN tube were obtained in this manner and were subsequently confirmed to be Salmonella sp. isolates by PCR using genus-specific primers.
Shigella sp. strains were isolated from LTB by streaking samples onto xylose lysine desoxycholate (Difco) and salmonella-shigella (Difco) agar plates and incubating the preparations at 37°C for 24 h. To isolate STEC strains, samples from MPN tubes were streak plated onto tryptose blood agar (Difco) supplemented with washed sheep blood cells (6), sorbitol MacConkey agar, and CHROMagar O157 plates (CHROMagar Microbiology, Paris, France).
Campylobacter sp. strains were isolated from Bolton broth by streaking samples onto Campylobacter blood-free agar plates (Oxoid); the plates were incubated at 42°C for 48 h under microaerobic conditions as described previously (22). E. coli strain ATCC 25922, E. coli O157:H7 strain ATCC 43895, Salmonella enterica subsp. enterica serovar Typhimurium strain ATCC 14028, Shigella flexneri ATCC 20170, and C. jejuni ATCC 33291 were used for quality control of growth media and PCR.
DNA extraction.
DNA was extracted either directly from initial Cladophora dilutions or from enrichment samples. For direct extraction, 10-ml aliquots of Cladophora washes were centrifuged at 10,000 × g for 10 min, the pelleted cells were saved, and each supernatant was filtered through a 0.2-μm membrane filter (Millipore, Billerica, MA). DNA from the combined pellet and membrane fractions was directly extracted by bead beating using a Bio 101 FastDNA mini kit (Qbiogene, Carlsbad, CA). The final DNA solution was used as the template for conventional PCR.
DNA was extracted from enrichment samples as follows. One milliliter of an initial Cladophora dilution was added to 9 ml of LTB or TTB and incubated at 35°C for 24 h. The cultures were centrifuged at 10,000 × g for 10 min, and the pellets were resuspended in 1 ml of 0.85% NaCl. Ten microliters of each solution was mixed with 90 μl of 0.05 M NaOH in a microcentrifuge tube, heated at 95°C for 15 min, and centrifuged at 10,000 × g for 1 min. The supernatant was diluted 10-fold in distilled H2O and used as a template for PCR.
For MPN-PCR analyses, DNA was extracted from each MPN tube except Bolton broth tubes as follows. Ten microliters of liquid culture from each MPN tube was mixed with 90 μl of 0.05 M NaOH in 96-well PCR plates (MJ Research, Waltham, Mass.), heated at 95°C for 15 min, and centrifuged at 640 rpm for 10 min. The supernatants were diluted 10-fold in distilled H2O and used as templates for PCR.
For colony PCR analyses of Campylobacter, cells (1 μl) from single colonies growing on Campylobacter blood-free agar were suspended in 100 μl of 0.05 M NaOH in 96-well PCR plates, heated at 95°C for 15 min, and centrifuged at 640 rpm for 10 min. The supernatants were diluted 10-fold in distilled H2O and used as templates for PCR.
The DNA used for real-time PCR was extracted using initial washes of Cladophora as described above. DNA was extracted by bead beating using a PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) according to the manufacturer's instructions, except that the silica spin column was washed twice with solution C5 and DNA was eluted from the spin column using 10 μl of elution buffer solution C6. Elution was repeated four times (total volume, 40 μl) to maximize the DNA concentrations.
Detection of pathogens using conventional PCR.
Toxin- and genus-specific primer pairs (Table 1) and conventional PCR were used to determine if pathogenic bacteria were associated with Cladophora. The reaction mixtures (25 μl) contained 10 mM Tris-HCl, 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 0.2 μg bovine serum albumin (Roche, Basel, Switzerland) per μl, 0.1% Triton X-100, 0.625 U of Taq DNA polymerase (Promega, Madison, WI), and 1 μl of DNA template. The concentrations of MgCl2 and primers, the annealing temperature, and extension time were different for different primer sets and are shown in Table 1. PCRs were performed using a PTC-100 thermal cycler (MJ Research) and the following conditions: initial denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, primer annealing at the annealing temperature for 30 s, and extension at 72°C for the extension time. The annealing temperature and DNA extension time were different for different primer pairs (Table 1). After a final extension at 72°C for 5 min, the PCR mixtures were held at 4°C. Positive and negative (no DNA) controls were included each time. The presence of target amplicons that were the correct size was recorded and used to calculate the MPN count.
TABLE 1.
Primers and conventional PCR conditions used to detect pathogenic bacteria
| Target | Primer | Sequence (5′-3′) | Amplification condition
|
Reference | |||
|---|---|---|---|---|---|---|---|
| MgCl2 concn (mM) | Primer concna (μM) | Annealing temp (°C) | Extension time (s) | ||||
| E. coli uidA | uidA 298F | AATAATCAGGAAGTGATGGAGCA | 1.5 | 0.5 | 60 | 45 | 47 |
| uidA 884R | CGACCAAAGCCAGTAAAGTAGAA | ||||||
| E. coli stx1 and stx2 | stx 300Fa | GAACGAAATAATTTATATGT | 1.5 | 0.5 | 53 | 60 | 32 |
| stx 1166Ra | TTTGATTGTTACAGTCAT | ||||||
| E. coli O157 rfb | O157PF8 | CGTGATGATGTTGAGTTG | 2 | 0.2 | 60 | 45 | 38 |
| O157PR8 | AGATTGGTTGGCATTACTG | ||||||
| Shigella sp. ipaH | ipaH-1 | GTTCCTTGACCGCCTTTCCGATACCGTC | 2.5 | 0.2 | 60 | 45 | 21 |
| ipaH-2 | GCCGGTCAGCCACCCTCTGAGAGTAC | ||||||
| Salmonella sp.-specific sequences | ST-11 | AGCCAACCATTGCTAAATTGGCGCA | 2.5 | 0.2 | 60 | 45 | 1 |
| ST-15 | GGTAGAAATTCCCAGCGGGTACTG | ||||||
| Campylobacter sp. 16S rRNA gene | C412F | GGATGACACTTTTCGGAGC | 2.5 | 0.4 | 55 | 45 | 33 |
| C1228R | CATTGTAGCACGTGTGTC | ||||||
Our primer designation.
Quantitative PCR analyses.
QPCR analyses were performed using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) and the primers and target probes listed in Table 2. For Salmonella, the ttr locus (36) was used as the target, whereas for Campylobacter, the genus-specific16S rRNA gene sequence was targeted (34). Each reaction mixture (25 μl) contained 1× Universal PCR Master Mix, 0.2 μg/μl bovine serum albumin (Roche, Basel, Switzerland), each primer at a concentration of 400 nM, each target probe (Integrated DNA Technologies, Coralville, IA) at a concentration of 250 nM, and 5 μl of DNA template. The PCRs were performed in MicroAmp optical 96-well reaction plates that were sealed with optical adhesive covers (Applied Biosystems). The optimized reaction conditions for Salmonella and Campylobacter were experimentally determined to be 50°C for 2 min (for activation of uracil-N-glycosylase), 95°C for 10 min (initial denaturation and Taq DNA polymerase activation), and 50 cycles of 95°C for 15 s and 60°C for 1 min. PCRs, subsequent monitoring, and data analyses were performed using an ABI PRISM 7000 sequence detection system (Applied Biosystems). The baseline and threshold cycles (Ct) were determined automatically using the sequence detection system software (Applied Biosystems). PCRs were considered valid when a typical exponential amplification curve was observed and the Ct value was between 20 and 42. Standard curves were generated by adding 0, 5, 50, 5 × 102, 5 × 103, 5 × 104, and 5 × 105 Salmonella and Campylobacter cells to Cladophora washes prior to DNA extraction. The sizes of final PCR products were confirmed by analysis on agarose gels. Numbers of cells were determined using plate count agar (Difco) for Salmonella and Campylobacter blood-free agar for Campylobacter. The Ct values were plotted against cell counts, and linear regression analysis was performed to generate standard curves. The detection limits of the method were 50 Salmonella CFU/g of Cladophora and 5 Campylobacter CFU/g of Cladophora, and the method linearities were 0.94 and 0.96 for Salmonella and Campylobacter, respectively. S. enterica subsp. enterica serovar Typhimurium ATCC 14028 and C. jejuni ATCC 33291 were used as standards. The statistical significance of results was determined using the R program software (version 2.0.1; http://www.r-project.org/).
TABLE 2.
Primers and TaqMan probes used for QPCR
| Target | Primer or probe | Sequence (5′-3′)a | Reference |
|---|---|---|---|
| Salmonella-specific ttr locus | ttr-6 (forward primer) | CTCACCAGGAGATTACAACATGG | 36 |
| ttr-4 (reverse primer) | AGCTCAGACCAAAAGTGACCATC | ||
| ttr-5 (probe) | FAM-CACCGACGGCGAGACCGACTTT-BHQ1 | ||
| Campylobacter sp. 16S rRNA gene | campF2 (forward primer) | CACGTGCTACAATGGCATAT | 34 |
| campR2 (reverse primer) | GGCTTCATGCTCTCGAGTT | ||
| campP2 (probe) | FAM-CAGAGAACAATCCGAACTGGGACA-BHQ1 |
FAM, 6-carboxyfluorescein; BHQ1, black hole quencher 1.
DNA fingerprint analyses.
DNA fingerprinting of Salmonella was done using the BOXA1R primer (55) and the horizontal fluorophore-enhanced repetitive extragenic palindromic PCR (HFERP) DNA fingerprinting technique, as previously described (23, 24), whereas the population structure of Campylobacter strains was investigated by repetitive extragenic palindromic PCR DNA fingerprinting with the ERIC 1R and ERIC 2 primers (55) (ERIC-PCR). The HFERP method produced an average of 30 bands with the Salmonella strains, while the ERIC-PCR method produced an average of eight bands with isolates of Campylobacter. The PCR conditions were those described by Rademaker et al. (46), except that an annealing temperature of 40°C was used (51). To ensure maximum genetic diversity among isolates, the initial dilutions, five replicates per dilution, three dilutions per sample, and four different samples were used to isolate Salmonella and Campylobacter from Cladophora samples. Electrophoresis, visualization, and analyses were performed as previously described (18, 24). DNA fingerprint similarities were calculated by using Pearson's product-moment correlation coefficient with 1% optimization, and dendrograms were generated using the unweighted pair group method with arithmetic means.
16S rRNA gene sequence and serotype analyses.
Nearly full-length genes encoding 16S rRNA from two representative Salmonella isolates and eight Campylobacter isolates were sequenced and analyzed as previously described (13, 30). Strains were chosen for 16S rRNA gene sequencing based on groups obtained by using HFERP and ERIC-PCR DNA fingerprints. Salmonella serogrouping and serotyping were performed at the Veterinary Diagnostic Laboratory of the University of Minnesota, St. Paul, and the National Veterinary Services Laboratories, Ames, IA, respectively.
RESULTS AND DISCUSSION
Conventional PCR analysis to detect pathogens associated with Cladophora.
Toxin- and genus-specific primer pairs (Table 1) and conventional PCR were used to determine if pathogenic bacteria were associated with Cladophora. The fluorescence of MUG in growth tubes and PCR performed with the uidA gene primers indicated that E. coli was present in all Cladophora samples obtained in July and September 2004. These results are similar to those reported by Byappanahalli et al. (12) and Whitman et al. (57), who used conventional microbiological methods. PCR analyses performed with primer pairs for the Shiga-like toxin genes stx1 and stx2 and for the invasion plasmid antigen H gene (ipaH) indicated that STEC and Shigella, respectively, were present in the September 2004 Cladophora samples collected from the Burns Ditch side of Lake Michigan. While Shigella was detected in all four (100%) alga samples obtained near Burns Ditch, STEC was found in only one of four (25%) of the samples. No STEC, Salmonella, or Shigella was detected in the lakeside Cladophora samples, and Salmonella was not detected in the ditchside samples taken in July and September 2004. Moreover, no PCR products were obtained when primers O157PF8 and O157PR8 were used, indicating that the STEC found in the ditchside sample was not E. coli serotype O157 (data not shown). The source of STEC and Shigella in the September 2004 samples is unknown. However, since Burns Ditch is exposed to contamination from combined sewer overflows (42, 44) and humans are the major reservoir of Shigella (19), these bacteria most likely originated from humans.
Despite the detection of STEC by PCR, isolation of STEC from LTB containing MUG inoculated with the September 2004 ditchside samples was not successful due, in part, to the growth of the background microflora and perhaps to the fact that STEC could not be clearly differentiated on CHROMagar O157 (5), sorbitol MacConkey agar (45), and tryptose blood agar. While Beutin et al. (6) proposed that the production of enterohemolysin is a useful marker for detecting STEC, Boczek et al. (8) recently reported that enterohemolysin can be produced by non-STEC environmental strains of E. coli. Thus, more studies are needed to define the relationship between enterohemolysin production and STEC more clearly.
Pathogen quantification by MPN-PCR.
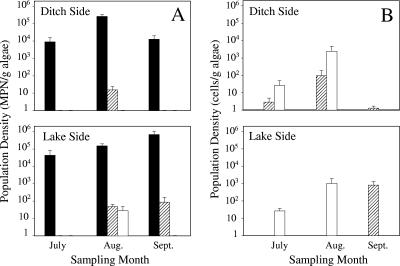
The detection by PCR of STEC and Shigella in the September 2004 Cladophora samples led us to use MPN-PCR, a more quantitative approach, in experiments conducted in 2005. For comparison purposes, analyses of E. coli were done in parallel. The MPN analyses indicated that Cladophora samples contained high numbers of E. coli cells in the summer months. In general, the numbers of Cladophora-associated E. coli cells increased as the summer progressed, and the greatest number of Cladophora-associated E. coli cells in lakeside samples occurred in August and September (5.2 ± 4.6 and 5.8 ± 5.5 log MPN E. coli per g algae, respectively) (Fig. 2A). By October, however, the Cladophora E. coli population in lakeside samples had decreased to barely detectable levels (0.9 ± 0.4 log MPN/g algae). In contrast, the greatest number of E. coli cells (5.4 ± 4.8 log MPN/g algae) in the ditchside samples occurred in August, while July and September samples contained 3.9 ± 3.8 and 4.1 ± 3.8 log MPN per g algae, respectively. The high E. coli densities and population variations among algal samples were generally comparable to those observed by Whitman et al. (57). Furthermore, the seasonal variation in the E. coli populations in lake- and ditchside samples was most likely due to a decrease in the water level and the greater exposure of algal mats to solar radiation. Whitman et al. (57) reported that sunlight significantly decreased the density of E. coli in Cladophora mats, although the extent of the reduction was dependent on mat thickness.
FIG. 2.
Seasonal shifts in the population densities of E. coli and potential pathogenic bacteria in lake- and ditchside Cladophora samples determined by MPN-PCR (A) and QPCR (B). Solid bars, E. coli; striped bars, Campylobacter; open bars, Salmonella. Samples were obtained in 2005, and the bars and error bars indicate means and standard errors, respectively.
Despite the detection of STEC and Shigella in the 2004 samples, these pathogens were not detected in 2005 Cladophora samples. However, Salmonella was detected by MPN-PCR in 80% (four of five) of the August lakeside samples (Fig. 2A). The Salmonella population size in four replicate samples ranged from 0.6 to 1.9 log MPN per g algae, and the mean was 1.5 log MPN per g algae. Salmonella was not detected by MPN-PCR in other lake- or ditchside samples obtained on other sampling dates. The variation may have been due to changes in lake inputs (58), predation (49), or environmental factors such as temperature (49) and solar radiation (39), all of which have been shown to control Salmonella populations in aquatic systems.
In contrast, all of the ditch- and lakeside samples obtained in August and 60% (three of five) of the samples obtained from the lakeside in September contained Campylobacter (Fig. 2A). The mean densities of Campylobacter in August ditch- and lakeside samples were 1.2 ± 0.92 log MPN/g (range, 0.69 to 1.7 log MPN/g) and 1.7 ± 1.2 log MPN/g (range, 1.3 to 1.9 log MPN/g), respectively. The mean number of campylobacters in September lakeside samples was 1.9 log MPN/g algae (range, 1 to 2.4 log MPN/g). However, by October Campylobacter could not be detected in any sample using this method (data not shown).
While culturable Salmonella and Campylobacter were detected in the August and September 2005 Cladophora samples, the abundance was not related to the density of E. coli on this host. Similarly, Carter et al. (14) reported that there was not a significant correlation between Campylobacter counts and the density of total and fecal coliforms or fecal enterococci. In our studies, the Salmonella and Campylobacter densities were about 4 orders of magnitude less than the E. coli densities, suggesting that growth of these pathogens on Cladophora may be limited by host and environmental factors. Moreover, the occurrence and density of these pathogenic bacteria with Cladophora may be temporally related to their concentrations in input sources, sediments, sands, and water. Whitman and colleagues (56) reported high levels of S. enterica subsp. enterica serovar Typhimurium in southern Lake Michigan beach water. Interestingly, this bacterium was also recovered from West Beach waters, about 2 km downcurrent (west) of Burns Ditch, and the fingerprints of some of the isolates were similar to the fingerprints of strains isolated from Chicago beach water, sand, and gull feces. Moreover, the eaeA gene, indicative of enteropathogenic E. coli, has been detected by PCR in beach water samples from Ogden Dunes (Sheridan Haack, personal communication). In contrast, Chomeau et al. (15) did not detect Salmonella in the five Lake Michigan beach waters that they investigated in 2003 and 2004 in Door County, Wisconsin.
Quantitative PCR analysis.
The numbers of Salmonella and Campylobacter cells harbored by Cladophora collected in 2005 were also determined by QPCR using primers and TaqMan probes specific for these pathogens (Table 2). Salmonella was detected in 60 and 40% of July and August samples, respectively, collected from the lake- and ditchside sample areas. However, Salmonella was not detected in the September or October samples (Fig. 2B). QPCR analyses indicated that the greatest densities of Salmonella cells were present in August in the lake- and ditch-side samples, and these densities were 9.9 × 102 and 1.6 × 103 cells/g (dry weight) of Cladophora, respectively. QPCR was more sensitive than MPN-PCR for detecting Cladophora-borne Salmonella in lake and ditchside samples. While MPN-PCR was effective for identifying Salmonella only in August lakeside Cladophora samples, QPCR indicated that the July samples contained up to 3.9 ×102 Salmonella cells/g. Similarly, while MPN-PCR analyses did not detect Salmonella in any of the ditchside samples, QPCR analyses detected significant numbers of cells of this pathogen in both July and August Cladophora samples (3.7 × 102 and 1.6 × 103 Salmonella cells/g, respectively) (Fig. 2B).
Campylobacter was present in 60, 60, and 100% of ditchside samples taken in July, August, and September, respectively. The greatest cell density, 39 cells/g Cladophora, occurred in August samples (Fig. 2B), which corresponded to results obtained by MPN-PCR. While Campylobacter was not detected in August lakeside samples, perhaps due to PCR inhibition, this pathogen was detected in 60% of lakeside samples obtained in September and October, and the highest cell density, 5.4 ×102 cells/g of Cladophora, occurred in September samples. While significantly more Campylobacter cells were found in lakeside samples than in ditchside samples (P < 0.05), the numbers of Salmonella cells associated with Cladophora were not significantly different for samples obtained on the two sides of the embayment.
In general, QPCR analyses detected higher densities than MPN assays detected. For example, QPCR detected a nearly 36-fold-greater number of Salmonella cells in August 2005 lakeside samples than an MPN analysis detected. Moreover, while this pathogen was easily detected and enumerated in July and August ditchside samples, MPN analyses did not detect Salmonella in any sample. The ability of QPCR to detect viable but nonculturable (VBNC) and/or dead cells may also explain why this method detected higher densities of Salmonella and Campylobacter cells in some of the samples than MPN analysis detected. While it may be argued that enumerating dead or VBNC pathogen cells does not provide important information, such data are useful for assessing total pathogen loads in waterways, for determining differential die-off of the pathogens once they leave host animals, and for determining the potential health risks associated with the recovery of pathogens in the VBNC state (25). While quantitative real-time PCR assays alleviate some of the biases associated with cultivation, the accuracy and reliability of QPCR for pathogen quantification may still be biased by the presence of PCR inhibitors or the numerical dominance of specific genotypes (20).
DNA fingerprint analyses.
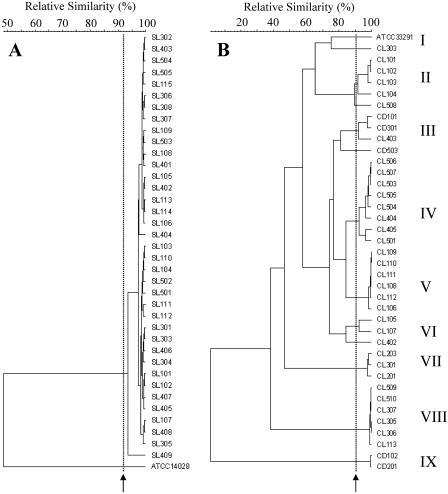
HFERP and ERIC-PCR DNA fingerprint analyses were performed to characterize 37 Salmonella strains and 38 Campylobacter strains isolated from Cladophora. The 37 Salmonella isolates were obtained from August lakeside Cladophora samples attached to four different rocks. The Campylobacter strains were isolated from August ditchside (5 isolates), August lakeside (18 isolates), and September lakeside (15 isolates) Cladophora samples attached to four, five, and three different rocks, respectively. Figure 3A shows that the majority of Salmonella strains examined were monophyletic, with similarity values ranging from 94 to 100%. Strains having HFERP DNA fingerprints with 92 to 95% similarity were previously shown to be genetically identical (23, 24) The high degree of genetic relatedness found among Salmonella isolates was unexpected, given that the strains were obtained independently from high and low dilutions and from five replicates per dilution, three dilutions per sample, and four different samples. The collection procedure ensured that both numerically dominant and less prevalent strains were isolated from Cladophora. Moreover, the genotypically identical Salmonella isolates were obtained from Cladophora samples attached to different rocks that were as much as 25 m apart on the two sides of the embayment. This result suggests either that a large number of nearly identical Salmonella cells simultaneously became attached to different Cladophora thalli, which seems unlikely, or that certain strains of Salmonella gain some advantage by associating with Cladophora. The latter hypothesis implies that there is strain-specific growth or survival. In contrast to these findings, Whitman et al. (56) described recovery of genetically diverse S. enterica subsp. enterica serovar Typhimurium isolates from Lake Michigan water and sediments. Similarly, diverse serovars of Salmonella have been isolated previously from a marine environment (37) and beach sand (9). Taken together, these results indicate that the salmonellae in water, sediment, and sand may be more diverse than the salmonellae that are associated with Cladophora. The potential source(s) of the Cladophora-associated Salmonella is unknown. However, Whitman et al. (56) reported that Salmonella isolated from Lake Michigan near Chicago, IL, may have originated from gulls.
FIG. 3.
Dendrograms based on HFERP DNA fingerprints of Salmonella isolates obtained using the BOXA1R primer (A) and on ERIC-PCR DNA fingerprints of Campylobacter isolates obtained using the ERIC 1R and ERIC 2 primers (B). Dendrograms were generated using Pearson's product-moment correlation coefficient and the unweighted pair group method with arithmetic means clustering method. Salmonella strains SL101 to SL115, SL301 to SL308, SL401 to SL409, and SL501 to SL505 were isolated from lakeside Cladophora samples obtained from four different rocks in August 2005. Campylobacter strains CD101, CD102, CD201, CD301, and CD503 were isolated from ditchside Cladophora attached to four different rocks in August 2005. Campylobacter strains CL101 to CL104, CL201 to CL203, CL301 to CL303, CL402 to CL405, and CL501 to CL507 were isolated from lakeside Cladophora attached to five different rocks in August 2005. Campylobacter strains CL105 to CL113, CL305 to CL307, and CL508 to CL510 were isolated from lakeside Cladophora attached to three different rocks in September 2005. The dashed lines and arrows indicate the cutoff values used to define identical strains, which were 92% for HFERP DNA fingerprints (A) and 90% for ERIC-PCR DNA fingerprints (B).
Since HFERP studies performed with the BOXA1R primer did not generate sufficient numbers of DNA fragments for adequate characterization of Campylobacter strains (data not shown), ERIC-PCR DNA fingerprinting was used to characterize the Campylobacter isolates obtained from Cladophora. Unlike the Salmonella strains, the Campylobacter strains were genetically diverse and could be divided into nine groups with genetic similarity values ranging from 2 to 98% (Fig. 3B). Steinbrueckner et al. (51) reported that a single Campylobacter strain repeatedly produced ERIC-PCR DNA fingerprints that were at least 80% similar. In our reproducibility studies, however, ERIC-PCR DNA fingerprints of strain ATCC 33291 were >90% similar when the strain was analyzed multiple times. Consequently, we considered the Campylobacter isolates that we examined to be genetically identical if their ERIC-PCR fingerprints were >90% similar. Based on this criterion, several strains were isolated from Cladophora. However, a few identical strains were isolated from different Cladophora samples obtained from both sampling locations. For example, CL509, CL307, and CL113 were isolated from different Cladophora samples attached to different rocks, yet their ERIC-PCR DNA fingerprints were >98% similar (group VIII) (Fig. 3B). Since Campylobacter isolates were found to be diverse, while Salmonella isolates were nearly identical genetically, our results suggest that enrichment preference and bias were most likely not issues with our isolation scheme. Environmental factors, such as nutrients, temperature, and predation (28, 53), have been shown to influence the survival of Campylobacter, and Cladophora may enhance the survival of campylobacters by providing a protective and nutritive niche. The potential origin of Cladophora-associated Campylobacter is still unclear. Since some waterfowl have been reported to harbor Campylobacter (31), this source may be responsible for the initial release of Campylobacter into Lake Michigan. Moreover, since we examined only submerged Cladophora growing on rocks below the water line, the initial inoculation of the algae most likely occurred in an indirect manner.
Taxonomic identities of Cladophora-associated Salmonella and Campylobacter strains.
Thirty-seven Salmonella strains were isolated from PCR-positive MPN tubes for subsequent DNA fingerprint analyses. A series of biochemical tests confirmed that these bacteria were Salmonella strains, and two representative strains (SL302 and SL409) were confirmed to be Salmonella strains based on 16S rRNA gene sequence analysis. BLASTn analysis of the nearly full-length 16S rRNA genes indicated that both of these strains exhibited 99.9% identity (1,496/1,497 bases) with S. enterica subsp. enterica serovar Typhimurium strain LT2. Serotype analysis revealed that isolate SL302 was an S. enterica subsp. enterica serovar Newport strain, strongly suggesting that the Salmonella strains associated with Cladophora were human pathogens. S. enterica subsp. enterica serovar Typhimurium has previously been reported to be present in Lake Michigan sand, sediment, and water (56).
Thirty-eight strains were isolated from Bolton broth MPN tubes, and all of these isolates were confirmed to be Campylobacter strains by 16S rRNA-based genus-specific PCR. The taxonomic identities of eight representative strains, one in each repetitive extragenic palindromic PCR group (CD101, CD102, CL101, CL105, CL109, CL203, CL506, and CL509), were ascertained by sequencing the 16S rRNA genes. Sequence and BLASTn analyses indicated that 87.5% of the strains (seven of eight strains) had 16S rRNA genes with 99.9% nucleotide identity (based on 1,419 bases) to the C. jejuni 16S rRNA genes. However, the 16S rRNA gene of one strain, CD102, exhibited 100% nucleotide identity (based on 1,419 bases) to the 16S rRNA gene of Campylobacter lari. This strain belongs to an outlying ERIC-PCR group, group IX (Fig. 2B). These results are similar to those of Bolton et al. (9), who reported that C. jejuni and C. lari were the Campylobacter species that were isolated most frequently from wet sands of United Kingdom beaches that did not meet the EEC Bathing Water Directive. Since C. jejuni and C. coli are the major campylobacters that are pathogenic to humans (3, 9), our results indicate that the majority of the Campylobacter strains that we isolated from Cladophora may be potential human pathogens.
Conclusions.
In this study we used cultural, physiological, and molecular approaches to determine if pathogenic bacteria associate with the green alga Cladophora. Taken as a whole, our data indicate that Cladophora in Lake Michigan serves as a reservoir for potential pathogens, such as STEC, Shigella, Salmonella, and Campylobacter. While both S. enterica subsp. enterica serovar Newport and C. jejuni were routinely isolated from Cladophora from culturally affected and relatively clean sites, detection of Shigella and STEC was time dependent, suggesting that the presence of these organisms on Cladophora was mostly related to increased inputs into the lake. While the ultimate source(s) of these pathogens is unknown, evidence from other studies suggests that they may have originated from gulls. Our results indicate that Cladophora may be an important reservoir for potential pathogens in Lake Michigan and that the association of these organisms warrants additional studies to assess the risk of the pathogens to public health.
Acknowledgments
We thank Chang-Hun Lee, John Ferguson, Katarzyna Przybyla-Kelly, Mesfin Tesfaye, and Deborah Samac for help and technical assistance. We also thank Francisco Diez-Gonzalez and Jerry Torrison for providing reference strains.
This study was supported in part by grants from Minnesota Sea Grant (to M.J.S. and R.H.) and from the University of Minnesota Agricultural Experiment Station (to M.J.S.).
Footnotes
Contribution number 1370 of the USGS Great Lakes Science Center.
REFERENCES
- 1.Aabo, S., O. F. Rasmussen, L. Roseen, P. D. Sorensen, and J. E. Olsen. 1993. Salmonella identification by the polymerase chain reaction. Mol. Cell. Probes 7:171-178. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, M. 1982. Most probable number method for microbial populations, p. 815-820. In A. L. Page, R. H. Miller, and D. R. Keeney (ed.), Methods of soil analysis, part 2, 2nd ed. American Society for Agronomy and Soil Science Society of America, Madison, Wis.
- 3.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry, E. D., and D. N. Miller. 2005. Cattle feedlot soil moisture and manure content. II. Impact on Escherichia coli O157. J. Environ. Qual. 34:656-663. [DOI] [PubMed] [Google Scholar]
- 5.Bettelheim, K. A. 1998. Reliability of CHROMagar O157 for the detection of enterohaemorrhagic Escherichia coli (EHEC) O157 but not EHEC belonging to other serogroups. J. Appl. Microbiol. 85:425-428. [DOI] [PubMed] [Google Scholar]
- 6.Beutin, L., M. A. Montenegro, I. Ørskov, F. Ørskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogosian, G., L. E. Sammons, P. J. Morris, J. P. O'Neil, M. A. Heitkamp, and D. B. Weber. 1996. Death of the Escherichia coli K-12 strain W3110 in soil and water. Appl. Environ. Microbiol. 62:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boczek, L. A., C. H. Johnson, E. W. Rice, and B. K. Kinkle. 2006. The widespread occurrence of the enterohemolysin gene ehlyA among environmental strains of Escherichia coli. FEMS Microbiol. Lett. 254:281-284. [DOI] [PubMed] [Google Scholar]
- 9.Bolton, F. J., S. B. Surman, K. Martin, D. R. Wareing, and T. J. Humphrey. 1999. Presence of campylobacter and salmonella in sand from bathing beaches. Epidemiol. Infect. 122:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brettar, I., and M. G. Hofle. 1992. Influence of ecosystematic factors on survival of Escherichia coli after large-scale release into lake water mesocosms. Appl. Environ. Microbiol. 58:2201-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byappanahalli, M., and R. Fujioka. 2004. Indigenous soil bacteria and low moisture may limit but allow faecal bacteria to multiply and become a minor population in tropical soils. Water Sci. Technol. 50:27-32. [PubMed] [Google Scholar]
- 12.Byappanahalli, M. N., D. A. Shively, M. B. Nevers, M. J. Sadowsky, and R. L. Whitman. 2003. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiol. Ecol. 46:203-211. [DOI] [PubMed] [Google Scholar]
- 13.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 14.Carter, A. M., R. E. Pacha, G. W. Clark, and E. A. Williams. 1987. Seasonal occurrence of Campylobacter spp. in surface waters and their correlation with standard indicator bacteria. Appl. Environ. Microbiol. 53:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomeau, V., G. Kleinheinz, R. Kolberg, C. McDermott, M. Nevers, W. Schuster, and R. L. Whitman. February. 2005, posting date. Door County beach contamination source identification interim report. [Online.] Door County Soil and Water Conservation Department, Sturgeon Bay, WI. http://www.doa.state.wi.us/ docs_view2.asp?docid=4779.
- 16.Colles, F. M., K. Jones, R. M. Harding, and M. C. J. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodds, W. K., and D. A. Gudder. 1992. The ecology of Cladophora. J. Phycol. 28:415-427. [Google Scholar]
- 18.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DuPont, H. L. 1988. Shigella. Infect. Dis. Clin. N. Am. 2:599-605. [PubMed] [Google Scholar]
- 20.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garcés, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman, A. B., M. Vankatesan, E. V. Oaks, and J. M. Buysse. 1990. Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of Shigella flexneri. J. Bacteriol. 172:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, J. M., C. Abeyta, and T. Tran. 2001. Chapter 7. Isolation of Campylobacter species from food and water. In Food and Drug Administration bacteriological analytical manual. Association of Official Analytical Chemists International, Arlington, Va. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-7.html.
- 23.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. M., E. M. Sutcliffe, and A. Curry. 1991. Recovery of viable but non-culturable Campylobacter jejuni. J. Gen. Microbiol. 137:2477-2482. [DOI] [PubMed] [Google Scholar]
- 26.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 27.King, C. H., E. B. Shotts, Jr., R. E. Wooley, and K. G. Porter. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korhonen, L. K., and P. J. Martikainen. 1991. Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J. Appl. Bacteriol. 71:379-382. [DOI] [PubMed] [Google Scholar]
- 29.Kourany, M., and S. R. Telford. 1981. Lizards in the ecology of salmonellosis in Panama. Appl. Environ. Microbiol. 41:1248-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 31.Levesque, B., P. Brousseau, F. Bernier, E. Dewailly, and J. Joly. 2000. Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality. Water Res. 34:1089-1096. [Google Scholar]
- 32.Lin, Z., H. Kurazono, S. Yamasaki, and Y. Takeda. 1993. Detection of various verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol. Immunol. 37:543-548. [DOI] [PubMed] [Google Scholar]
- 33.Linton, D., R. J. Owen, and J. Stanley. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res. Microbiol. 147:707-718. [DOI] [PubMed] [Google Scholar]
- 34.Lund, M., S. Nordentoft, K. Pedersen, and M. Madsen. 2004. Detection of Campylobacter spp. in chicken fecal samples by real-time PCR. J. Clin. Microbiol. 42:5125-5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malinsky-Rushansky, N. Z., and C. Legrand. 1996. Excretion of dissolved organic carbon by phytoplankton of different sizes and subsequent bacterial uptake. Mar. Ecol. Prog. Ser. 132:249-255. [Google Scholar]
- 36.Malorny, B., E. Paccassoni, P. Fach, C. Bunge, A. Martin, and R. Helmuth. 2004. Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70:7046-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Urtaza, J., M. Saco, J. de Novoa, P. Perez-Pineiro, J. Peiteado, A. Lozano-Leon, and O. Garcia-Martin. 2004. Influence of environmental factors and human activity on the presence of Salmonella serovars in a marine environment. Appl. Environ. Microbiol. 70:2089-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer, J. J., D. Schmidt, P. Petrosko, S. Sanchez, L. Bolton, and M. D. Lee. 1999. Development of primers to O-antigen biosynthesis genes for specific detection of Escherichia coli O157 by PCR. Appl. Environ. Microbiol. 65:2954-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCambridge, J., and T. A. McMeekin. 1981. Effect of solar radiation and predacious microorganisms on survival of fecal and other bacteria. Appl. Environ. Microbiol. 41:1083-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natvig, E. E., S. C. Ingham, B. H. Ingham, L. R. Cooperband, and T. R. Roper. 2002. Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soils with incorporated bovine manure. Appl. Environ. Microbiol. 68:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nevers, M. B., and R. L. Whitman. 2005. Nowcast modeling of Escherichia coli concentrations at multiple urban beaches of southern Lake Michigan. Water Res. 39:5250-5260. [DOI] [PubMed] [Google Scholar]
- 43.Olapade, O. A., M. M. Depas, E. T. Jensen, and S. L. McLellan. 2006. Microbial communities and fecal indicator bacteria associated with Cladophora mats on beach sites along Lake Michigan shores. Appl. Environ. Microbiol. 72:1932-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olyphant, G. A., J. Thomas, R. L. Whitman, and D. Harper. 2003. Characterization and statistical modeling of bacterial (Escherichia coli) outflows from watersheds that discharge into southern Lake Michigan. Environ. Monit. Assess. 81:289-300. [PubMed] [Google Scholar]
- 45.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rademaker, J. L. W., F. J. Louws, and F. J. de Bruijn. 1998. Characterization of the diversity of ecologically important microbes by rep-PCR genomic fingerprinting, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, supplement 3. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 47.Ram, J. L., R. P. Ritchie, J. Fang, F. S. Gonzales, and J. P. Selegean. 2004. Sequence-based source tracking of Escherichia coli based on genetic diversity of β-glucuronidase. J. Environ. Qual. 33:1024-1032. [DOI] [PubMed] [Google Scholar]
- 48.Rambach, A. 1990. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl. Environ. Microbiol. 56:301-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhodes, M. W., and H. Kator. 1988. Survival of Escherichia coli and Salmonella spp. in estuarine environments. Appl. Environ. Microbiol. 54:2902-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schallenberg, M., P. J. Bremer, S. Henkel, A. Launhardt, and C. W. Burns. 2005. Survival of Campylobacter jejuni in water: effect of grazing by the freshwater crustacean Daphnia carinata (Cladocera). Appl. Environ. Microbiol. 71:5085-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taras, M. J., A. E. Greenberg, R. D. Hoak, and M. C. Rand. 1971. Standard methods for the examination of water and wastewater, 13th ed. American Public Health Association, Washington, D.C.
- 53.Terzieva, S. I., and G. A. McFeters. 1991. Survival and injury of Escherichia coli, Campylobacter jejuni, and Yersinia enterocolitica in stream water. Can. J. Microbiol. 37:785-790. [DOI] [PubMed] [Google Scholar]
- 54.Turco, R. F. 1994. Coliform bacteria, p. 145-158. In R. W. Weaver, J. S. Angle, and P. J. Bottomley (ed.), Methods of soil analysis. Soil Science Society of America, Madison, WI.
- 55.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitman, R. L., T. G. Horvath, M. L. Goodrich, M. B. Nevers, M. J. Wolcott, and S. K. Haack. 2001. Characterization of E. coli levels at 63rd Street Beach. Report to the City of Chicago Department of the Environment and the Chicago Park District. [Online.] U.S. Geological Survey, Porter, IN. http://www.glsc.usgs.gov/_files/reports/63rdStreetBeachReport.pdf.
- 57.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]