Abstract
The role of okadaic acid (OA) in the defense system of the marine demosponge Suberites domuncula against symbiotic/parasitic annelids was examined. Bacteria within the mesohyl produced okadaic acid at concentrations between 32 ng/g and 58 ng/g of tissue (wet weight). By immunocytochemical methods and by use of antibodies against OA, we showed that the toxin was intracellularly stored in vesicles. Western blotting experiments demonstrated that OA also existed bound to a protein with a molecular weight of 35,000 which was tentatively identified as a galectin (by application of antigalectin antibodies). Annelids that are found in S. domuncula undergo apoptotic cell death. OA is one candidate inducer molecule of this process, since this toxin accumulated in these symbionts/parasites. Furthermore, we identified the cDNA encoding the multifunctional prosurvival molecule BAG-1 in S. domuncula; it undergoes strong expression in the presence of the annelid. Our data suggest that sponges use toxins (here, OA) produced from bacteria to eliminate metazoan symbionts/parasites by apoptosis.
Sponges, the evolutionarily oldest metazoan phylum (Porifera), share one common ancestor with the other metazoan phylum, the Urmetazoa (31, 35). Most sponges are marine sessile filter feeders; some species daily filter tons of water containing millions of bacteria or viruses through their bodies (48; reviewed in references 18 and 26). Due to their efficient chemical and biological defense systems, sponges have survived not only adverse climatic periods (24) but also biotic and anthropogenic threats (34).
Two strategies are used by sponges to eliminate attacking (infectious) bacteria: first, chemical strategies via the production of secondary metabolites (see references 45 and 51) and, second, humoral and cellular defense mechanisms (33). The hitherto studied molecular biological or cell biological defense systems used by sponges against microorganisms have been elucidated primarily in the demosponge Suberites domuncula. This species is able to recognize and defeat gram-positive as well as gram-negative bacteria by the antimicrobial lectin tachylectin (52). In addition, S. domuncula has developed a specific system to recognize gram-positive bacteria via the peptidoglycan coat; subsequently bacteria are endocytosed and/or extracellularly digested by lysozyme (59). The lipopolysaccharide (LPS) outer membrane of gram-negative bacteria induces in S. domuncula an adaptive antibacterial response by increasing the release of bioactive and antimicrobial lyso-platelet-activating factors (36). We have shown that in the aqueous environment also, fungi are recognized by sponges via a receptor, the (1→3)-β-d-glucan binding protein; after interaction of fungal (1→3)-β-d-glucans with the corresponding receptor, S. domuncula causes the expression of a series of defense molecules, e.g., a fibrinogen-like protein and an epidermal growth factor precursor (43).
Finally, the more general defense system against microorganisms should be highlighted; LPS activates the mitogen-activated protein kinase pathway which modulates the sponge immune system (8, 66). In conclusion, sponges are provided with a very efficient immune system (32) that involves an array of proteinaceous as well as nonproteinaceous defense molecules. In addition, sponges not only contain symbiotic/parasitic bacteria but also live in a symbiotic (mutualistic) relationship with prokaryotic microorganisms (reviewed in references 1, 8, 17, and 45).
Earlier it was shown that S. domuncula harbors bacteria that produce the secondary metabolite okadaic acid (OA) (66). OA was first isolated from the Caribbean marine sponge Halichondria melanodocia by Tachibana et al. (58); later, related OA polyethers were also isolated from other sponges (67). The polyether fatty acid derivative OA is rather toxic to mammals (55). The free-living microalgae Prorocentrum lima (39) is the prime producer of OA.
The biological activity of OA is linked with an inhibition of protein phosphatases (5), resulting in an induction of apoptosis (8, 41, 49). In S. domuncula OA was shown to augment the immune response against LPS and bacteria via increased phosphorylation of p38, a central kinase of the mitogen-activated protein kinase pathway, resulting in a differential phosphorylation of proapoptotic and antiapoptotic molecules (66).
S. domuncula lives together with the hermit crab Pagurites oculatus (Decapoda: Paguridea), which resides predominantly in shells of the mollusk Trunculariopsis trunculus (Gastropoda: Muricidae) (4, 23) on which the sponge grows. In the natural environment the sponge body harbors surprisingly few symbiotic/parasitic metazoans, which might indicate that effective defense systems eliminate such organisms. Only some annelids, e.g., Syllis sp., Haplosyllis sp., or Nereis sp., have been identified (3). The elucidation of the molecular and cellular mechanism(s) by which sponges protect themselves against parasitic metazoans has begun only recently. It could be demonstrated that sponge-associated bacteria produce 2-methylthio-1,4-napthoquinone, a compound that suppresses the CD36/LIMPII signal transduction pathway that modulates angiogenesis (in vertebrates) and canal formation (in S. domuncula) (37).
In the present study we show that in S. domuncula OA is accumulated in tissue, especially in those areas where annelids live in the sponge. The experiments were performed with antibody probes that were raised against the toxin and that had been successfully applied previously for an enzyme-linked immunosorbent assay (66). Our results indicate that the annelids die of apoptosis while—in parallel—the cells of the surrounding sponge tissue strongly express the multifunctional prosurvival molecule BAG-1 (see reference 63). We selected this gene for the studies because it exists only in metazoa (62). BAG-1 interacts with a series of cellular targets, e.g., the Bcl-2 apoptosis regulator, heat shock proteins, or components of the proteasome machinery (63). We assume that in S. domuncula OA causes an apoptogenic effect in response to invading symbionts/parasites.
MATERIALS AND METHODS
Chemicals, materials, and enzymes.
Restriction enzymes, a Total RNA Isolation kit, and reagents for the rapid amplification of cDNA ends (RACE) procedure were purchased from Invitrogen (Carlsbad, CA); TriplEx2 vector was from BD (Palo Alto, CA); TRIzol reagent was from GibcoBRL (Grand Island, N.Y.); Hybond-N+ nylon membrane was from Amersham (Little Chalfont, Buckinghamshire, United Kingdom); a PCR-DIG-Probe-Synthesis kit, cell death detection kit (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling [TUNEL] reaction; fluorescein isothiocyanate [FITC] based), and a BM Chemoluminescence Blotting Substrate kit were from Roche (Mannheim, Germany); aprotinin, sodium orthovanadate, Nonidet-P40, and 4′,6-diamidino-2-phenylindole-dilactate (DAPI) were from Sigma (St. Louis, MO).
Sponges and extracts.
Specimens of the marine sponge S. domuncula (phylum Porifera, class Demospongiae, order Hadromerida) were collected in the northern Adriatic Sea near Rovinj (Croatia) and then kept in aquaria in Mainz (Germany) at a temperature of 17°C. Extracts were prepared from tissue samples; they were homogenized in lysis buffer (Tris-buffered saline, pH 7.5, 1 mM EDTA, 1% Nonidet-P40, 10 mM NaF, 0.1 μM aprotinin, 1 mM sodium orthovanadate).
Preparation of antibodies against OA.
Polyclonal antibodies against OA (PcAb-OA) were raised in female rabbits (White New Zealand) as previously described (66). OA conjugate was injected at 4-week intervals into the animals; after three boosts serum was collected, and the antibodies were prepared (21). The titer of the antibodies was determined to be 1:2,000.
Histological analysis.
Tissue was fixed in paraformaldehyde, embedded in Technovit 8100 (Heraeus Kulzer, Wehrheim, Germany), and sectioned, essentially as previously described (7, 65). To remove the siliceous spicules, the tissue samples were incubated with HF/NH4F. The 4-μm-thick slices were incubated with PcAb-OA (1:500 dilution) overnight. Then the slides were treated with Cy3-conjugated goat anti-rabbit immunoglobulin G (IgG) for 2 h. The sections were inspected by immunofluorescence with an Olympus AHBT3 microscope. In parallel, slices were stained with 5 μg/ml of DAPI for 30 min to identify DNA, especially in bacteria. In the tissue of the sponge S. domuncula, the bacteria are almost exclusively found in clusters, which can easily be identified as such (8). Where indicated, the slices were stained with the trichrome stain ASTRIN (28) and inspected with an Olympus AHBT3 microscope. In addition, the specimens were inspected directly using Nomarsky interference contrast optics. For in situ detection of apoptosis, tissue samples were permeabilized and subjected to a TUNEL reaction (in situ cell death detection kit) according to the instructions of the manufacturer. The DNA was end labeled with FITC-dUTP, which was followed by direct analysis of fluorescent cells.
As outlined previously (66) tissue sections were reacted with PcAb-OA (1:300 dilution) overnight to also identify cellular structures that form immunocomplexes with the antibodies. Then the slides were incubated with FITC-conjugated goat anti-rabbit IgG for 2 h, and subsequently the sections were inspected for immunofluorescence (Olympus AHBT3 microscope).
Electron microscopy.
Transmission electron microscopy was performed as previously described (38). Briefly, sponge samples were cut into pieces, incubated in phosphate buffer, treated with OsO4, and finally dehydrated with ethanol. The dried samples were incubated with propylene oxide, fixed in propylene oxide-araldite, and covered with araldite before being cut into 60-nm ultrathin slices (Ultracut S; Leica, Wetzlar, Germany). The samples were transferred onto coated copper grids and analyzed with a Tecnai 12 microscope (FEI Electron Optics, Eindhoven, The Netherlands).
Immunogold labeling with detection by electron microscopy was performed with tissue samples treated in glutaraldehyde or paraformaldehyde (38). After 2 h the material was dehydrated in ethanol and embedded in LR White resin (London Rein Company, Berkshire, England); the samples were blocked with 5% bovine serum albumin in phosphate-buffered saline (PBS) and then incubated with the primary antibody PcAb-OA (1:500) for 12 h at 4°C. Preimmune serum was used as a control. After three washes with PBS-1% bovine serum albumin, sections were incubated with a 1:100 dilution of the secondary antibody (1.4-nm nanogold anti-rabbit IgG, diluted 1:200; Nanoprobes, Yapbank, NY) for 2 h. Sections were rinsed in PBS, treated with 1% glutaraldehyde-PBS for 5 min, washed, and dried. Subsequently, enhancing of the immunocomplexes was performed with silver, as previously described (12). The samples were examined with a Tecnai 12 microscope.
Western blotting.
Total cell extracts (10 μg/lane) were subjected to 0.1% sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (7). Western blotting experiments were performed using PcAb-OA (1:300 dilution). After incubation for 3 h, the blots were incubated with goat anti-rabbit IgG, peroxidase coupled (1:5,000 dilution; New England Biolabs). Detection of the immunocomplex was carried out using BM chemoluminescence blotting substrate. For one series of experiments a polyclonal antibody against the recombinant S. domuncula galectin (PcAb-rGAL) was used (H. C. Schröder et al., submitted for publication); the titer of this antibody preparation was 1:3,000.
Isolation of a cDNA for BAG-1 from S. domuncula.
The complete cDNA of the sponge BCL2-associated athanogene (BAG1_SUBDO) was isolated from the cDNA library (29) by PCR. The degenerate primers were designed against the conserved ubiquitin domain signature, which in the human BAG-1 sequence (CAH72742.1) spans the segment from amino acid 50 to 57. The forward primer 5′-CARAARCTIATHTTYAARGGIAAR-3′ (I, inosine) was successful together with the vector primer [5′-TAATACGACTCACTATAGGG-3′]. PCR was carried out at an initial denaturation at 95°C for 5 min, followed by 35 amplification cycles of 95°C for 30 s, 58°C for 45 s, and 74°C for 1.5 min, with a final extension step at 74°C for 10 min. Fragments of the expected sizes were obtained and cloned into the TOPO TA (pCRII) vector in Escherichia coli TOP10 cells. Sequencing was performed with primers directed to the SP6 promoter and the T7 promoter. The sequence was completed with insert-specific primers in combination with 5′ RACE primer or with 3′ RACE primer using the CapFishing full-length cDNA premix kit. The final sequence (SDBAG1) was confirmed by an additional PCR using primers directed against the nontranslated region of the cDNA, followed by sequencing.
Sequence comparison.
The BAG-1 deduced protein was analyzed using the computer programs BLAST (http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi) and FASTA (http://www.ebi.ac.uk/fasta33/). Multiple alignments were performed with CLUSTAL W, version 1.6 (61). Phylogenetic trees were constructed on the basis of amino acid sequence alignments by the neighbor-joining method, as implemented in the Neighbor program from the PHYLIP package (16). The distance matrices were calculated using the Dayhoff PAM matrix model as previously described (13). The degree of support for internal branches was further assessed by bootstrapping (16). The graphic presentations were prepared with GeneDoc (40).
RNA preparation and Northern blot analysis.
Tissue samples (1 g each) from the mesohyl were taken from regions of the sponge which contained no annelids and from tissue around parts in which these organisms had been identified. RNA was extracted from liquid-nitrogen-pulverized tissue with TRIzol reagent as previously described (19). Then 5 μg of total RNA was electrophoresed and blotted onto Hybond-N+ nylon membrane. Hybridization was performed with a 420-nucleotide (nt) fragment of the SDBAG1 cDNA; this region was within the open reading frame. The housekeeping gene β-tubulin, SDTUB (accession number AJ550806), of S. domuncula was used as an internal standard. The probes were labeled with a PCR-DIG Probe Synthesis Kit (Roche, Mannheim; Germany). After the slices were washed, digoxigenin (DIG)-labeled nucleic acid was detected with anti-DIG Fab fragments and visualized by chemiluminescence using CDP (Roche).
In situ localization studies.
The method applied is based on a previously described procedure (42, 44). Frozen cross-sections (8 μm thick) of tissue were prepared, fixed, treated with proteinase K, and subsequently fixed again with paraformaldehyde. After rehydration the sections were hybridized with the labeled probe, a 420-nt long part of the SDBAG1 cDNA portion. After blocking, the sections were incubated with an anti-DIG antibody conjugated with alkaline phosphatase. The dye reagent NBT/X-phosphate was used for visualization of the signals. Antisense and sense single-stranded DNA DIG-labeled probes were synthesized by PCR using a PCR DIG probe synthesis kit. Sense probes were used in parallel as negative controls in the experiments.
Analytical procedures.
The OA concentration present in sponge tissue was assessed by coupled high-performance liquid chromatography-mass spectrometry as described before (25, 66). The amount of OA detected was correlated with the weight of fresh tissue used for the analysis (n = 5; the mean values ± standard deviations [SD] are given). Protein concentrations were determined as previously described (30) using bovine serum albumin as a standard.
Nucleotide accession number.
The sequence reported here was deposited in the EMBL/GenBank database under accession number AM183251 for the S. domuncula BCL2-associated athanogene (SDBAG1).
RESULTS
Production of OA in bacteria from S. domuncula.
The OA concentration was determined in tissue from S. domuncula either immediately after collection from the sea or after maintenance in the aquarium for 12 months. Tissue from freshly collected sponges had an OA content of 58 ± 7 ng/g of wet weight (70 nM), while tissue from specimens which grew in the aquarium had a lower content of 32 ± 9 ng/g (39 nM).
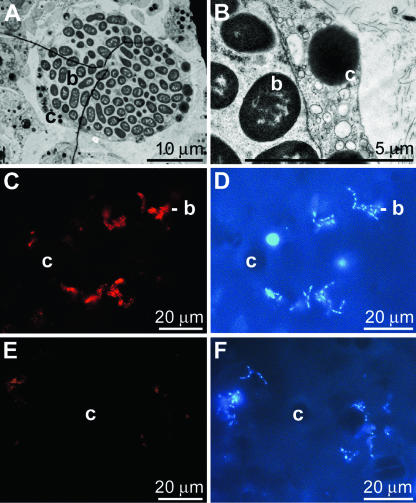
Tissue from S. domuncula contained bacteria either compartmented in cells, in bacteriocytes (Fig. 1A and B), or scattered within the mesohyl (Fig. 1C to F). The bacteriocytes completely surrounded the bulky clusters of bacteria (Fig. 1A and B). The bacteria within the mesohyl could be visualized by DAPI (Fig. 1D and F) and were detected immunohistochemically by applying PcAb-OA. With this tool the bacteria became brightly stained (Fig. 1C). Preimmune serum was used as a control and produced no significant fluorescence (Fig. 1E).
FIG. 1.
Localization of bacteria in S. domuncula. (A) Transmission electron microscopy through S. domuncula tissue showing one bacteriocyte (c) which is filled with bacteria (b). (B) A higher magnification shows that the cell body (c) surrounds the bacteria clusters (b). Besides these compartmented bacteria, microorganisms are also scattered throughout the mesohyl of the tissue. In the sections they can be seen by light microscopy after staining with DAPI (D and F). By using PcAb-OA and then a Cy3-conjugated anti-rabbit secondary antibody the bacteria become brightly stained (C). As a control, preimmune serum was used, which gave no signal (E).
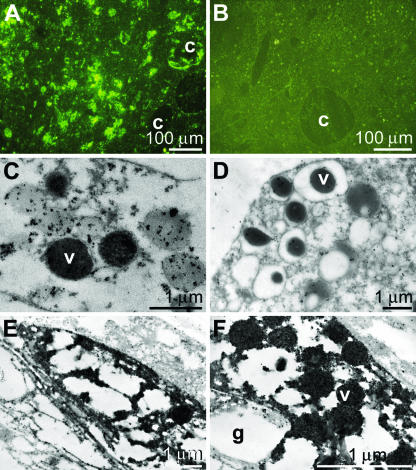
Storage of OA in tissue from S. domuncula.
In an earlier study we demonstrated that PcAb-OA reacted in S. domuncula with bacteria as well as with tissue structures (66). These antibodies were also applied in this study; the experiments revealed that especially the cells that surround aquiferous canals were brightly stained (Fig. 2A). Preimmune serum did not show clear reactions (Fig. 2B). Electron immunogold labeling was applied to identify the structures that were brightly stained by the antibodies (Fig. 2C to F). The results showed that PcAb-OA strongly accumulated as electron-dense grains in vacuoles which were localized within cells (Fig. 2C). In contrast, if preimmune serum was used, no significant reactions could be visualized (Fig. 2D). Lower magnification showed that these granules were stored in distinct cells, which were heavily filled with 200- to 300-nm vacuoles, positively reacting with PcAb-OA (Fig. 2E). It was striking that these cells were composed of large granules containing no electron-dense material, suggesting that these cells were archaeocytes.
FIG. 2.
Cellular structures reacting with PcAb-OA. (A) Tissue was sliced and reacted first with PcAb-OA and then with FITC-anti-rabbit IgG. The cells that surround the aquiferous canals (c) are especially well stained (light microscopy inspection). (B) Reaction of the tissue with preimmune serum. (C, E, and F) Application of the electron immunogold labeling technique (with detection by electron microscopy), using PcAb-OA, to identify OA in cells. The immunogold particles are accumulated in vesicles (v), which can be seen at a higher magnification (C and F). At lower magnification it becomes evident that the vesicles are arranged within cells around larger granules (g) containing no electron-dense material (E). (D) In a control the sections were treated with a preimmune serum.
The PcAb-OA showed a positive reaction in a protein extract from S. domuncula tissue. Total tissue extracts were prepared and subjected to SDS-PAGE (Fig. 3), as described in Materials and Methods. Western blotting experiments were performed using PcAb-OA. After incubation, the immunocomplexes on the blots were visualized by labeled secondary antibodies (Fig. 3, WB). One band with a molecular weight of 35,000 was prominently stained (lane a), strongly suggesting that in the sponge OA was (perhaps covalently) bound to a protein. In order to assess the nature of the postulated protein, a panel of antibodies that had been raised in our laboratory was screened. PcAb-rGAL raised earlier against S. domuncula galectin (53) was able to detect this band; it cross-reacted with this protein (molecular weight of 35,000), suggesting that OA was linked to galectin (lane b). Preimmune sera were used as controls after the blotting; they did not stain the protein species on the blot (lanes c and d).
FIG. 3.
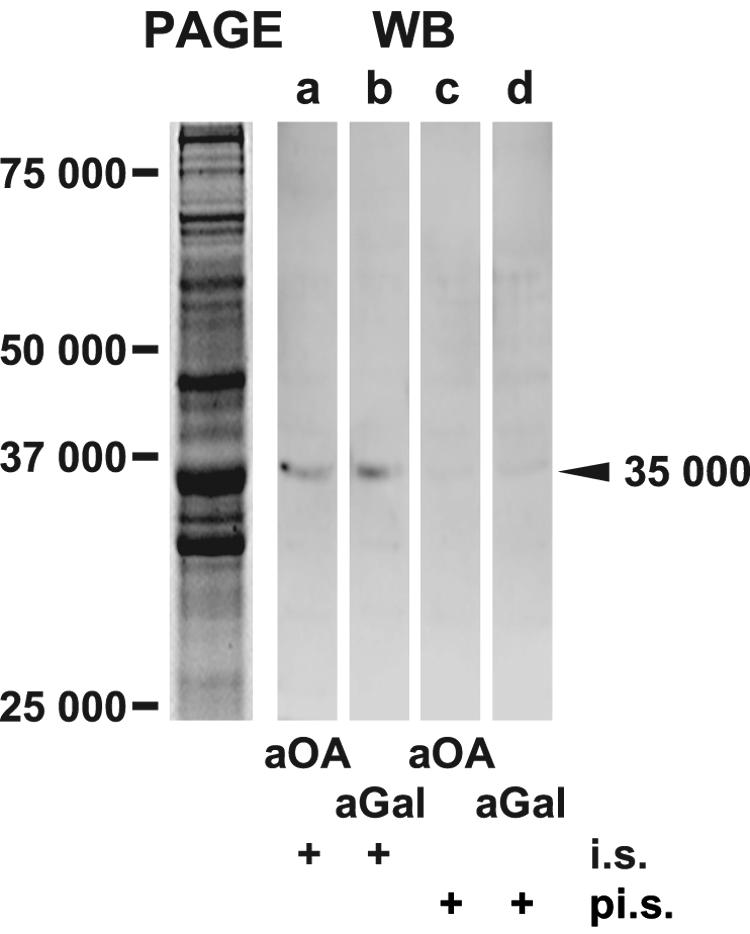
Identification of a macromolecule, reacting with antibodies against OA. Extracts were prepared from tissue of S. domuncula and separated by 0.1% SDS-10% PAGE (PAGE). The crude extract was size separated and stained with Coomassie brilliant blue. In a parallel series of experiments, the proteins were blot transferred, and the Western blots (WB) were reacted with PcAb-OA (aOA; lane a) or PcAb-aGAL (aGal; lane b). In both blots a protein with a molecular weight of 35,000 was recognized. In addition to immune sera (i.s.), preimmune sera (pi.s.; lanes c and d) were also applied. Size markers are given.
Annelid in S. domuncula tissue.
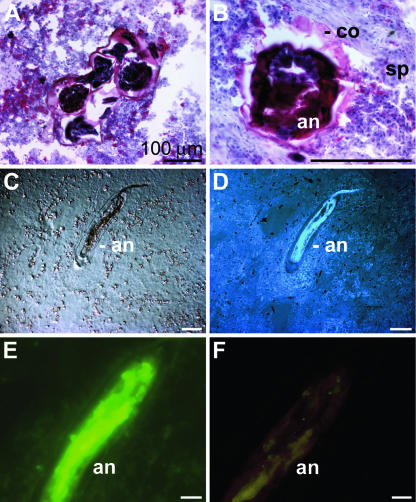
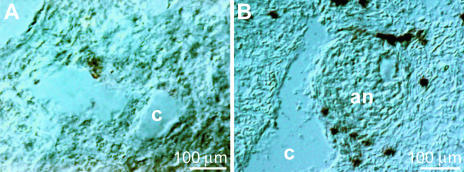
Cross-sections of the tissue occasionally disclosed annelids in the sponge (Fig. 4). The annelid, which could not at this point be identified or determined, was intensively stained with the trichrome stain ASTRIN (Fig. 4A and B). This staining also showed that the annelid was encapsulated by a coat that contained cells (Fig. 4B). In sections, the symbiont/parasite could also be identified by Nomarsky interference contrast optics (Fig. 4C). The annelid was characterized by a higher cell density than the sponge host, as was seen after DAPI staining (Fig. 4D). The size of the annelids in S. domuncula varied between 2 and 4 mm in length with diameters between 0.3 and 0.8 mm.
FIG. 4.
Annelid in S. domuncula. (A and B) Cross-sections of S. domuncula were stained with the trichrome stain ASTRIN to distinguish between the sponge tissue (sp), which appears bluish, and the more reddish annelid (an). Around the annelid an encapsulating coat (co) can be identified. (C and D) Longitudinal sections through the symbiont/parasite by Nomarsky interference contrast optics (C) or by fluorescence after DAPI staining (D). (E) Apoptotic cell death of the annelid. The section was subjected to a TUNEL enzymatic labeling assay and subsequently analyzed by fluorescent light microscopy. (F) No fluorescence was seen when the specimens were subjected to the assay lacking the FITC-labeled nucleotides. Scale bars, 100 μm (A, B, E, and F) and 25 μm (C and D).
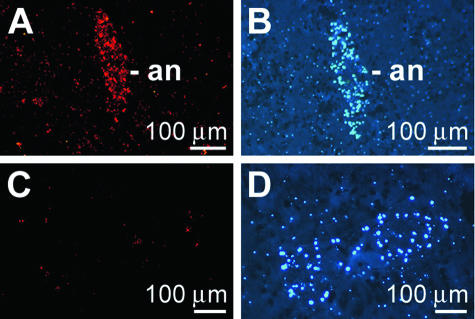
Enrichment of OA in the annelid.
The high cell density of the annelid was used as a marker to identify this symbiont/parasite within sponge tissue (Fig. 5B and D). If slices were reacted with PcAb-OA, the cells of the annelid were also prominently and brightly stained (Fig. 5A). No immunocomplexes were seen if the slices were stained with preimmune serum (Fig. 5C).
FIG. 5.
Accumulation of OA in the annelid (an) living in S. domuncula. (A) The sections were reacted with PcAb-OA to show the highly fluorescent signals coming from the cells of the symbiont/parasite. (B) In parallel, the sections were stained with DAPI. (C) When the sections were treated with preimmune serum, no signals were seen. (D) Parallel image after DAPI staining. Scale bars, 100 μm.
Induction apoptosis in the annelid.
A cell death detection kit was used to determine if the sponge (host) eliminated the symbiont/parasite by inducing it to programmed cell death (apoptosis). During the TUNEL assay procedure both single- and double-stranded DNA breaks were labeled with fluorescent nucleotides. Inspection by fluorescence microscopy showed that the annelids were stained green (Fig. 4E). In a parallel series the sections were not treated with the labeled nucleotides, and these specimens did not show any significant staining (Fig. 4F).
The S. domuncula BAG-1 molecule.
The above results indicate that OA is accumulated in the symbiont/parasite (Fig. 5), resulting in induction of apoptosis in the symbiotic/parasitic animal (Fig. 4E). To clarify whether the host (S. domuncula) responds to the annelid with a prosurvival reaction, the expression of the prosurvival molecule BAG-1 was studied.
The complete cDNA (SDBAG1) of the sponge BCL2-associated athanogene (BAG1_SUBDO) was isolated. The 791-nt sequence has an open reading frame from nt 6 to 8 and nt 780 to 782 (stop). The deduced 258-amino-acid protein (BAG1_SUBDO) (Fig. 6A) has a predicted molecular weight of 29,139. It displays (i) the ubiquitin domain signature ranging from amino acid 29 to amino acid 54 and (ii) the BAG domain from amino acid 101 to amino acid 181 (Motif Scan [http://myhits.isb-sib.ch/cgi-bin/motif_scan]).
FIG. 6.
(A) BAG-1 from S. domuncula (BAG1_SUBDO) was aligned with the human BAG-1 (BAG1_HUMAN; accession no. CAH72742.1). The two characteristic domains are marked: the ubiquitin domain (UBI) signature and the BAG domain. Amino acids that are similar in both sequences are shown boxed and in white type. (B) A rooted tree was constructed after alignment of these two sequences with the predicted protein from D. melanogaster (DMUB52AA_DROME; CAA37227.1), the Bag-1 (human) homolog protein 1 from C. elegans (BAG1_CAEEL; AAB96728.1), the predicted protein from S. cerevisiae (SC8021X_YEAST; CAA89774.1), and the putative protein from A. thaliana (ATT28J14_ARATH; CAB87278.1). After alignment the tree was built (1,000 replicates). The plant sequence was used as an outgroup. The scale bar indicates an evolutionary distance of 0.1 amino acid substitutions per position in the sequence.
In the database, the highest sequence similarity of BAG1_SUBDO was found to the human BAG-1 protein (accession number CAH72742.1) with a probability score E (11) of 1e−23. Only low similarity exists to predicted proteins from Drosophila melanogaster (1e−3), the Bag-1 (human) homolog protein Caenorhabditis elegans (1e−9), the predicted protein from Saccharomyces cerevisiae (2e−4), and the putative protein from Arabidopsis thaliana (1e−6). The phylogenetic tree, which was rooted with the plant sequence, reflects these relationships. One branch comprises the sponge protein and the human BAG-1 and shares a common ancestor together with the C. elegans Bag-1-like molecule (Fig. 6B).
Defense system of S. domuncula: expression of BAG-1.
Northern blotting experiments were performed to determine if the expression level of SDBAG1 changes in response to the symbiosis/parasitism of the annelid with the sponge. In the regions that contained annelids, a high expression level was seen, while in tissue that did not contain annelids, no expression could be detected by Northern blotting (Fig. 7). Four animals were analyzed in parallel. The housekeeping gene β-tubulin cDNA (SDTUB) was used as a control; its expression level did not vary among the four animals (Fig. 7).
FIG. 7.
Expression of BAG-1 in four sponge specimens (1 to 4); tissue samples from areas that did not contain symbionts/parasites (−) and from regions where the annelids were present (+) were analyzed. Following RNA isolation, the same amounts were loaded onto gels and subsequently size separated. After blot transfer the membranes were hybridized with the SDBAG1 probe (BAG1). The housekeeping gene α-tubulin (TUB) was used as a control to show that the same amounts of RNA were loaded onto the gels.
Expression and localization of BAG-1.
To support the assumption that BAG-1 is involved in the defense of S. domuncula against the metazoan symbiont/parasite, in situ hybridization was performed with the SDBAG1 cDNA probe. In tissue that did not harbor an annelid, no cells were positively stained with the antisense probe (Fig. 8A). However, if sections were reacted with the SDBAG1 probe, a layer of cells around the symbiont/parasite was brightly stained (Fig. 8B). No reaction could be observed if the sense SDBAG1 probe was used (not shown).
FIG. 8.
In situ hybridization analysis applying the antisense probe for BAG-1 (SDBAG1). With this probe no signals are detected in regions where no annelid is seen (A), while in regions that contain the annelid several cells frame the symbiont/parasite (an) (B). Two aquiferous canals are marked (c).
DISCUSSION
The first illustrated description of a symbiotic relationship between metazoans and a demosponge (Halichondria panicea) was given by Ellis (15), who reported on the frequent occurrence of an annelid on the surface of this sponge. Sponges have developed several strategies to resist or eliminate such symbionts/parasites; some species secrete mucins on their surfaces as a protective shield (54). Another efficient defense against sponge-attacking metazoans was thoroughly studied by Cimino et al. and Proksch (9, 10, 45). They demonstrated that secondary metabolites, produced by the sponges or their associated microorganisms, are transferred from them to the predator(s). When taken up by the predators, the original secondary metabolites from sponges can undergo biotransformation (10). In particular, the sponge Aplysina aerophoba, which is rich in brominated tyrosine derivatives, was found to display an efficient protection system against metazoan predators (14, 46). These secondary metabolites are taken up by symbiotic/parasitic metazoans and then may also act as defense molecules (60).
The polyether OA studied here is toxic for vertebrates (20) due to its inhibitory effect on protein phosphatases 1 and 2A (22). This algal toxin is an inducer of apoptotic cell death in vertebrates (6) and also in sponges (66). It was first found in sponges of the genus Halichondria and later in dinoflagellates, especially in benthic species of the taxon Prorocentrum. Furthermore it was reported that OA is accumulated in mussels, e.g., Mytilus edulis, to high concentrations (2 to 2,000 ng/g) (64). Hence, OA displays a potential threat to human health not only because it causes acute toxicity but also because it acts as a tumor promoter (56).
The occurrence of OA is not restricted to Halichondria; in addition to the evidence of the present study, it has previously been described in a second sponge, S. domuncula, at a level of 50 ng/g (66) as well as in Geodia cydonium (unpublished data). S. domuncula uses OA as a metabolite to modulate or augment its immune status against LPS-mediated bacterial effects (66). The data shown here demonstrate that S. domuncula harbors bacteria in special cells, bacteriocytes, and in cells scattered throughout the mesohyl. The bacteria are the producers of the toxin as demonstrated by immunohistological techniques. Now we show that in S. domuncula the protein which is presumptively galectin (Mr of 35,000) is linked to OA. It appears to be most likely that OA is transferred to the protein via an acetyltransferase, with ATP and acetyl-coenzyme A as the coenzymes (2). Future studies must also show if OA is deposited in the protein-bound form and released, if required, by deacetylase (27). Several cDNAs of such enzymes exist in the S. domuncula expressed sequence tag database at a higher copy number (http://spongebase.genoserv.de/).
As shown by the studies presented here, OA exists in sponges (covalently) bound to protein. Therefore, it may be postulated that the toxin could also act as an antigen; this assumption is supported by our results showing that OA also exists in mussels in a protein-bound form (unpublished data). The size of the free toxin (Mr of 804) is only large enough to act as a hapten; however, together with a macromolecule (here the protein with a molecular weight of 35,000), it has the potential to act as an immunogen. This finding might also have implications for human health since until now OA had not been detected bound to proteins; previously only extracts were prepared from potential toxic food by methanolic extraction, which allowed the isolation of only unbound OA (47). Future studies must show if humans produce, e.g., after eating toxin-containing mussels, antibodies against this toxin resulting in subchronic symptoms. It should be stressed here that OA occurs in the producer and/or the host in an ester form also, covalently linked to low-molecular-weight, nonproteinaceous organic moieties (57).
It had been demonstrated that OA may initiate a signal transduction pathway which differentially influences apoptotic cell death via a protein phosphatase (50). Based on this finding we investigated whether OA functions in S. domuncula as an inducer of apoptosis against foreign metazoan invaders. The antibodies against OA were used to demonstrate that this toxin accumulates in the annelid and settles in the sponge tissue. With the in situ cell death detection technique (TUNEL reaction) a high level of single- and/or double-stranded DNA breaks could be demonstrated in the cells of the symbiont/parasite, indicating the (early) stages of apoptosis. This strongly suggests that sponges, which harbor the bacteria producing OA, eliminate their metazoan symbionts/parasites by intoxication with this secondary metabolite.
To support this notion we studied whether the host, S. domuncula, is protected against the apoptogenic effect caused by the increased levels of OA in the cell layers surrounding the symbiont/parasite. BAG-1 has been selected as a molecular marker and as a molecule of choice. This prosurvival molecule is a multifunctional protein protecting cells against apoptotic cell death (see references 62 and 63). BAG-1 physically interacts particularly with heat shock proteins in response to stress factors during morphogenesis or to toxic effectors.
Two lines of evidence indicate that the host responds to a settling of annelids in S. domuncula with an upregulation of the expression of the prosurvival gene BAG-1. First, it had been demonstrated that only tissue that is adjacent to the symbiont/parasite expresses detectable amounts of BAG-1 protein. Secondly, it is strikingly shown, by application of the in situ hybridization technique, that the cells that are in direct contact with the annelid strongly express BAG-1.
Taken together, our data indicate that in S. domuncula OA is a candidate apoptogenic molecule to eliminate invading symbionts/parasites. This study underscores the fact that sponges use secondary metabolites, including those that are synthesized by symbiotic microorganisms, as a chemical defense system against metazoan invaders.
Acknowledgments
This work was supported by grants from the European Commission (FOOD-CT-2004-513967), the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung (Center of Excellence BIOTECmarin project), and the International Human Frontier Science Program (RG-333/96-M).
Footnotes
This article is dedicated to R. A. Lewin (Scripps Institution of Oceanography, La Jolla, CA), who 30 years ago discovered Prochloron, an algal species that stimulated our present-day view on endosymbiosis between blue-green algae and tunicates/sponges.
REFERENCES
- 1.Althoff, K., C. Schütt, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536. [Google Scholar]
- 2.Archer, S. Y., and R. A. Hodin. 1999. Histones acetylation and cancer. Curr. Opin. Genet. Dev. 9:171-174. [DOI] [PubMed] [Google Scholar]
- 3.Arndt, W. 1930. Porifera (Schwämme, Spongien), p. 30-120. In C. Oppenheimer, and L. Pincussen (ed.), Tabulae biologicae, vol. VI. W. Junk, Berlin, Germany. [Google Scholar]
- 4.Arndt, W. 1933. Die biologische Beziehung zwischen Schwämmen und Krebsen. Mitt. Zool. Mus. Berl. 19:221-305. [Google Scholar]
- 5.Bagu, J. R., B. D. Sykes, M. M. Craig, and C. F. B. Holmes. 1997. A molecular basis for different interactions of marine toxins with protein phosphatase-1. J. Biol. Chem. 272:5087-5097. [DOI] [PubMed] [Google Scholar]
- 6.Bøe, R., B. T. Gjertsen, O. K. Vintermyr, G. Houge, M. Lanotte, and S. O. Doskeland. 1991. The protein phosphatase inhibitor okadaic acid induces morphological changes typical of apoptosis in mammalian cells. Exp. Cell Res. 195:237-246. [DOI] [PubMed] [Google Scholar]
- 7.Böhm, M., I. M. Müller, W. E. G. Müller, and V. Gamulin. 2000. The mitogen-activated protein kinase p38 pathway is conserved in metazoans: cloning and activation of p38 of the SAPK2 subfamily from the sponge Suberites domuncula. Biol. Cell 29:95-104. [DOI] [PubMed] [Google Scholar]
- 8.Böhm, M., U. Hentschel, A. Friedrich, L. Fieseler, R. Steffen, V. Gamulin, I. M. Müller, and W. E. G. Müller. 2001. Molecular response of the sponge Suberites domuncula to bacterial infection. Mar. Biol. 139:1037-1045. [Google Scholar]
- 9.Cimino, G., and G. Sodano. 1994. Transfer of sponge secondary metabolites to predators, p. 459-472. In R. W. M. van Soest, T. M. G. van Kempen, and J. C. Braekman (ed.), Sponges in time and space. A. A. Balkema, Rotterdam, The Netherlands.
- 10.Cimino, G., and M. T. Ghiselin. 2001. Marine natural products chemistry as an evolutionary narrative, p. 115-154. In J. B. McClintock, and B. J. Baker (ed.), Marine chemical ecology. CRC Press, Boca Raton, Fla.
- 11.Coligan, J. E., B. M. Dunn, H. L. Ploegh, D. W. Speicher, and P. T. Wingfield (ed.). 2000. Current protocols in protein science, p. 2.0.1-2.8.17. John Wiley & Sons, New York, N.Y.
- 12.Danscher, G. 1981. Histochemical demonstration of heavy metals. A revised version of the sulphide silver method suitable for both light and electronmicroscopy. Histochemistry 71:1-16. [DOI] [PubMed] [Google Scholar]
- 13.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. A model of evolutionary change in protein, p. 345-352. In M. O. Dayhoff (ed.), Atlas of protein sequence and structure. Nat. Biomed. Res. Foundation, Washington, D.C.
- 14.Ebel, R., M. Brenzinger, A. Kunze, H. J. Gross, and P. Proksch. 1997. Wound activation of protoxins in marine sponge Aplysina aerophoba. J. Chem. Ecol. 23:1451-1462. [Google Scholar]
- 15.Ellis, J. 1755. An Assay Towards a Natural History of the Corralines and other Marine products of the like Kind Commonly Found on the Coasts of Great Britain and Ireland. B. White, London, United Kingdom.
- 16.Felsenstein, J. 1993. PHYLIP (phylogenetic inference package), version 3.5. University of Washington, Seattle, Wash.
- 17.Friedrich, A. B., I. Fischer, P. Proksch, J. Hacker, and U. Hentschel. 2001. Temporal variation of the microbial community associated with the Mediterranean sponge Aplysina aerophoba. FEMS Microbiol. Ecol. 38:105-113. [Google Scholar]
- 18.Gonzales, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grebenjuk, V. A., A. Kuusksalu, M. Kelve, J. Schütze, H. C. Schröder, and W. E. G. Müller. 2002. Induction of (2′-5′)oligoadenylate synthetase in the marine sponges Suberites domuncula and Geodia cydonium by the bacterial endotoxin lipopolysaccharide. Eur. J. Biochem. 269:1382-1392. [DOI] [PubMed] [Google Scholar]
- 20.Hallegraeff, G. M., D. M. Anderson, and A. D. Cembella (ed.). 1995. Manual on harmful marine microalgae, p. 95-111. UNESCO, Paris, France.
- 21.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Haystead, T. A. J., A. T. R. Sinn, D. Carling, R. C. Honnor, Y. Tsukitani, P. Cohen, and D. G. Hardie. 1989. Effects of the tumor promoter okadaic acid on intracellular protein phosphorylation and metabolism. Nature 337:78-81. [DOI] [PubMed] [Google Scholar]
- 23.Herland-Meewis, H. 1948. La gemmulation chez Suberites domuncula (Olivi) Nardo. Arch. Anat. Microsc. 37:289-322. [Google Scholar]
- 24.Hoffman, P. F., A. J. Kaufman, G. P. Halverson, and D. P. Schrag. 1998. A Neoproterozoic snowball earth. Science 281:1342-1346. [DOI] [PubMed] [Google Scholar]
- 25.Hummert, C., S. Kastrup, K. Reinhardt, M. Reichelt, and B. Luckas. 2000. Use of gel permeation chromatography for automatic and rapid extract clean-up for the determination of diarrhetic shellfish toxins (DSP) by liquid chromatography-mass spectrometric. Chromatographia 51:397-403. [Google Scholar]
- 26.Kennish, M. J. 1994. Practical handbook of marine science. CRC Press, Boca Raton, Fla.
- 27.Klockendler-Yeivin, A., and Y. Moshe. 2001. Chromatin modifiers and tumor suppression. Biochim. Biophys. Acta 1551:M1-M10. [DOI] [PubMed] [Google Scholar]
- 28.Kovács, P., G. Csaba, and G. Balogh. 1982. Detection of different functional states of the cellular nucleus with the new trichrome staining technique. Acta Histochem. 71:73-75. [DOI] [PubMed] [Google Scholar]
- 29.Kruse, M., I. M. Müller, and W. E. G. Müller. 1997. Early evolution of metazoan serine/threonine and tyrosine kinases: identification of selected kinases in marine sponges. Mol. Biol. Evol. 14:1326-1334. [DOI] [PubMed] [Google Scholar]
- 30.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 31.Müller, W. E. G. 2001. How was metazoan threshold crossed: the hypothetical Urmetazoa. Comp. Biochem. Physiol. A 129:433-460. [DOI] [PubMed] [Google Scholar]
- 32.Müller, W. E. G., and I. M. Müller. 2003. Origin of the metazoan immune system: identification of the molecules and their functions in sponges. Integr. Comp. Biol. 43:281-292. [DOI] [PubMed] [Google Scholar]
- 33.Müller, W. E. G., B. Blumbach, and I. M. Müller. 1999. Evolution of the innate and adaptive immune systems: relationships between potential immune molecules in the lowest metazoan phylum (Porifera) and those in vertebrates. Transplantation 68:1215-1227. [DOI] [PubMed] [Google Scholar]
- 34.Müller, W. E. G., C. Koziol, M. Wiens, and H. C. Schröder. 2000. Stress response in marine sponges: genes and molecules involved and their use as biomarkers, p. 193-208. In K. B. Storey and J. Storey (ed.), Environmental stressors and gene responses. Elsevier Science, Amsterdam, The Netherlands.
- 35.Müller, W. E. G., H. C. Schröder, A. Skorokhod, C. Bünz, I. M. Müller, and V. A. Grebenjuk. 2001. Contribution of sponge genes to unravel the genome of the hypothetical ancestor of Metazoa (Urmetazoa). Gene 276:161-173. [DOI] [PubMed] [Google Scholar]
- 36.Müller, W. E. G., M. Klemt, N. L. Thakur, H. C. Schröder, A. Aiello, M. D'Esposito, M. Menna, and E. Fattorusso. 2003. Molecular/chemical ecology in sponges: evidence for an adaptive antibacterial response in Suberites domuncula. Mar. Biol. 144:19-29. [Google Scholar]
- 37.Müller, W. E. G., N. L. Thakur, H. Ushijima, A. N. Thakur, A. Krasko, G. Le Pennec, M. M. Indap, S. Perović-Ottstadt, H. C. Schröder, G. Lang, and G. Bringmann. 2004. Matrix-mediated canal formation in primmorphs from the sponge Suberites domuncula involves the expression of a CD36 receptor-ligand system. J. Cell Sci. 117:2579-2590. [DOI] [PubMed] [Google Scholar]
- 38.Müller, W. E. G., M. Rothenberger, A. Boreiko, W. Tremel, A. Reiber, and H. C. Schröder. 2005. Formation of siliceous spicules in the marine demosponge Suberites domuncula. Cell Tissue Res. 321:285-297. [DOI] [PubMed] [Google Scholar]
- 39.Murakami, Y., Y. Oshima, and T. Yasumoto. 1982. Identification of okadaic acid as a toxic component of a marine dinoflagellate Prorocentrum lima. Nihon Sisan Gakkaishi 48:69-72. [Google Scholar]
- 40.Nicholas, K. B., and H. B. Nicholas, Jr. 1997. GeneDoc: a tool for editing and annotating multiple sequence alignments, version 1.1.004. [Online.]cris.com/∼ketchup/genedoc.shtml.
- 41.Perović, S., C. Wetzler, F. Brümmer, M. Elbrächter, L. Tretter, A. Wichels, W. E. G. Müller, and H. C. Schröder. 1999. Changes of ICE protease activities caused by toxic supernatants of dinoflagellates (Prorocentrum species) from marine algal blooms. Eur. J. Protistol. 35:267-274. [Google Scholar]
- 42.Perović, S., H. C. Schröder, S. Sudek, V. A. Grebenjuk, R. Batel, M. Štifanić, I. M. Müller, and W. E. G. Müller. 2003. Expression of one sponge Iroquois homeobox gene in primmorphs from Suberites domuncula during canal formation. Evol. Dev. 5:240-250. [DOI] [PubMed] [Google Scholar]
- 43.Perović-Ottstadt, S., T. Adell, P. Proksch, M. Wiens, M. Korzhev, V. Gamulin, I. M. Müller, and W. E. G. Müller. 2004. A (1→3)-β-d-glucan recognition protein from the sponge Suberites domuncula: mediated activation of fibrinogen-like protein and epidermal growth factor gene expression. Eur. J. Biochem. 271:1924-1937. [DOI] [PubMed] [Google Scholar]
- 44.Polak, J. M., and J. D. McGee. 1998. In situ hybridization. Oxford University Press, Oxford, United Kingdom.
- 45.Proksch, P. 1994. Defensive role for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon 32:639-655. [DOI] [PubMed] [Google Scholar]
- 46.Puyana, M., W. Fenical, and J. R. Pawlik. 2003. Are there activated chemical defenses in sponges of the genus Aplysina from the Carribbean? Mar. Ecol. Progr. Ser. 246:127-135. [Google Scholar]
- 47.Quilliam, M. A., and J. L. C. Wright. 1995. Medods for diarrhetic shellfish poison, p. 95-111. In G. M. Hallegraeff, D. M. Anderson, and A. D. Cembella (ed.), Manual on harmful marine microalgae. UNESCO, Paris, France.
- 48.Reiswig, H. M. 1974. Water transport, respiration and energetics of three tropical marine sponges. J. Exp. Mar. Biol. Ecol. 14:231-249. [Google Scholar]
- 49.Rossini, G. P., C. Pinna, and R. Viviani. 1997. Inhibitors of phosphoprotein phosphatases 1 and 2A cause activation of a 53 kDa protein kinase accompanying the apoptotic response of breast cancer cells. FEBS Lett. 410:347-350. [DOI] [PubMed] [Google Scholar]
- 50.Sandal, T., R. Ahlgren, J. Lillehaug, and S. O. Døskeland. 2001. Establishment of okadaic acid resistant cell clones using a cDNA expression library. Cell Death Diff. 8:754-766. [DOI] [PubMed] [Google Scholar]
- 51.Sarma, A. S., T. Daum, and W. E. G. Müller. 1993. Secondary metabolites from marine sponges. Akademie gemeinnütziger Wissenschaften zu Erfurt, Ullstein-Mosby Verlag, Berlin, Germany.
- 52.Schröder, H. C., H. Ushijima, A. Krasko, V. Gamulin, J. Schütze, I. M. Müller, and W. E. G. Müller. 2003. Emergence and disappearance of an immune molecule, an antimicrobial lectin, in basal metazoan. A tachylectin-related protein in the sponge Suberites domuncula. J. Biol. Chem. 278:32810-32817. [DOI] [PubMed] [Google Scholar]
- 53.Schröder, H. C., A. Boreiko, M. Korzhev, M. N. Tahir, W. Tremel, C. Eckert, H. Ushijima, I. M. Müller, and W. E. G. Müller. Co-expression and functional interaction of silicatein with galectin: matrix-guided formation of siliceous spicules in the marine demosponge Suberites domuncula. J. Biol. Chem. 281:12001-12009. [DOI] [PubMed]
- 54.Simpson, T. L. 1984. The cell biology of sponges. Springer-Verlag, New York, N.Y.
- 55.Stonik, V. A., and G. B. Elyakov. 1988. Structure and biological activities of sponge and sea cucumber toxins, p. 107-120. In A. T. Tu (ed.) Handbook of natural toxins, vol. 3. Marcel Dekker, New York, N.Y. [Google Scholar]
- 56.Suganuma, M., H. Fujiki, H. Suguri, S. Yoshizwa, M. Hirota, M. Nakayasu, M. Ojika, K. Wakamatsu, K. Yamada, and T. Sugimura. 1990. Okadaic acid: an additional non-phorbol-12-tetradecanoate-13-acetate type tumor promoter. Proc. Natl. Acad. Sci. USA 85:1768-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki, T., V. Beuzenberg, L. Mackenzie, and M. A. Quilliam. 2004. Discovery of okadaic esters in the toxic dinoflagellate Dinophysis acuta from New Zealand using liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 18:1131-1138. [DOI] [PubMed] [Google Scholar]
- 58.Tachibana, K., P. J. Scheuer, Y. Tsukitani, H. Kikushi, D. V. Engen, J. Clardy, Y. Gopichand, and F. J. Schmitz. 1981. Okadaic acid, a cytotoxic polyether from the marine sponges of the genus Halichondria. J. Am. Chem. Soc. 103:2469-2471. [Google Scholar]
- 59.Thakur, N. L., S. Perović-Ottstadt, R. Batel, M. Korzhev, B. Diehl-Seifert, I. M. Müller, and W. E. G. Müller. 2005. Innate immune defense of the sponge Suberites domuncula against gram-positive bacteria: induction of lysozyme and AdaPTin. Mar. Biol. 146:271-282. [DOI] [PubMed] [Google Scholar]
- 60.Thomas, C., M. Wolff, K. Padmakumar, R. Ebel, and P. Proksch. 2004. Chemical defense of Mediterranean sponge Aplysina cavernicola and Aplysina aerophoba. Z. Naturforsch. C 59:113-122. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Townsend, P. A., R. I. Cutress, A. Sharp, M. Brimmell, and G. Packham. 2003. BAG-1: a multi-functional regulator of cell growth and survival. Biochim. Biophys. Acta 1603:83-98. [DOI] [PubMed] [Google Scholar]
- 63.Townsend, P. A., A. Stephanou, G. Packham, and D. S. Latchman. 2005. BAG-1: a multi-functional pro-survival molecule. Int. J. Biochem. Cell Biol. 37:251-259. [DOI] [PubMed] [Google Scholar]
- 64.Tubaro, A., C. Florio, E. Luxich, S. Sosa, R. Della Loggia, and T. Yasumoto. 1996. A protein phosphatase 2A inhibition assay for a fast and sensitive assessment of okadaic acid contamination in mussels. Toxicon 34:743-752. [DOI] [PubMed] [Google Scholar]
- 65.Wagner, C., R. Steffen, C. Koziol, R. Batel, M. Lacorn, H. Steinhart, T. Simat, and W. E. G. Müller. 1998. Apoptosis in marine sponges: a biomarker for environmental stress (cadmium and bacteria). Mar. Biol. 131:411-421. [Google Scholar]
- 66.Wiens, M., B. Luckas, F. Brümmer, M. S. A. Ammar, R. Steffen, R. Batel, B. Diehl-Seifert, H. C. Schröder, and W. E. G. Müller. 2003. Okadaic acid: a potential defense molecule for the sponge Suberites domuncula. Mar. Biol. 142:213-223. [Google Scholar]
- 67.Yasumoto, T., M. Murata, Y. Oshima, M. Sano, G. K. Matsumoto, and J. Clardy. 1985. Diarrhetic shellfish toxin. Tetrahedron 41:1019-1025. [Google Scholar]