Abstract
Many soil bacteria contain 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, which degrades ACC, a precursor of the phytohormone ethylene. In order to examine the regulation of the acdS gene encoding ACC deaminase in Mesorhizobium loti MAFF303099 during symbiosis with the host legume Lotus japonicus, we introduced the β-glucuronidase (GUS) gene into acdS so that GUS was expressed under control of the acdS promoter, and we also generated disruption mutants with mutations in a nitrogen fixation regulator gene, nifA. The histochemical GUS assay showed that there was exclusive expression of acdS in mature root nodules. Two homologous nifA genes, mll5857 and mll5837, were found in the symbiosis island of M. loti and were designated nifA1 and nifA2, respectively. Quantitative reverse transcription-PCR demonstrated that nifA2 disruption resulted in considerably diminished expression of acdS, nifH, and nifA1 in bacteroid cells. In contrast, nifA1 disruption slightly enhanced expression of the acdS transcripts and suppressed nifH to some extent. These results indicate that the acdS gene and other symbiotic genes are positively regulated by the NifA2 protein, but not by the NifA1 protein, in M. loti. The mode of gene expression suggests that M. loti acdS participates in the establishment and/or maintenance of mature nodules by interfering with the production of ethylene, which induces negative regulation of nodulation.
The formation of nitrogen-fixing root nodules is the result of a series of interactions between (Brady)rhizobium and its legume host plants (8). The host legumes have several mechanisms for regulating nodule formation (28, 36). The plant hormone ethylene is also known to have inhibitory effects on rhizobial infection and the formation of nodule primordia and to limit nodule number (21, 24, 31). Rhizobia often interfere with ethylene biosynthesis in the host plants by means of rhizobitoxine (25, 37, 38) or 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase (19, 33) and reduce host ethylene emission to overcome the negative regulation.
ACC deaminase (EC 4.1.99.4) catalyzes the degradation of an ethylene precursor, ACC, into ammonium and α-ketobutyrate (13). The ACC deaminase structural gene (acdS) has been found in many rhizosphere bacteria (10, 13), including fast- and slow-growing rhizobia such as Rhizobium leguminosarum bv. viciae 128C53K (19), Bradyrhizobium japonicum USDA110 (16), Mesorhizobium loti MAFF303099 (15), and M. loti R7A (32). In the rhizobacterium Enterobacter cloacae UW4, promoter analysis of the acdS gene showed that expression of this gene requires both ACC and a leucine-responsive regulatory protein (LRP)-like protein and that anaerobic conditions enhance the expression (10). In B. japonicum USDA110 and R. leguminosarum bv. viciae 128C53K, the acdS genes are also probably regulated by an LRP-like protein and a σ70 promoter (16, 19).
In M. loti MAFF303099, acdS was found in the symbiosis island (15). The enhancing effect of the acdS gene on nodulation of Lotus japonicus MG-20 Miyakojima roots was demonstrated by using an M. loti acdS disruption mutant (33). Furthermore, DNA macroarray analysis showed that a clone containing acdS (mlr5932) was upregulated in bacteroid cells. The DNA sequence upstream of acdS in M. loti includes a putative σ54 RNA polymerase sigma factor recognition site and a nitrogen fixation regulator NifA protein binding site known as the upstream activating sequence (4, 33). These lines of evidence suggest that the expression of acdS in M. loti might be coupled with symbiotic nitrogen fixation. In this study, we examined the expression of M. loti acdS during symbiosis using specifically manipulated bacteria; in particular, we focused on the regulation by nifA.
MATERIALS AND METHODS
Plant materials and growth conditions.
L. japonicus MG-20 Miyakojima (17) was grown and inoculated with rhizobia as described previously (22).
Bacterial strains, plasmids, transposon, and growth conditions.
The bacterial strains, plasmids, and transposon used in this study are listed in Table 1. M. loti MAFF303099 and Escherichia coli cells were cultured as described previously (22). When required, the media were supplemented with appropriate antibiotics at the following concentrations: for M. loti, 50 mg liter−1 phosphomycine, 50 mg liter−1 streptomycin, 50 mg liter−1 spectinomycin, 20 mg liter−1 gentamicin, 10 mg liter−1 tetracycline, and 50 mg liter−1 kanamycin; and for E. coli, 100 mg liter−1 ampicillin, 50 mg liter−1 streptomycin, 100 mg liter−1 spectinomycin, 50 mg liter−1 kanamycin, and 15 mg liter−1 tetracycline. Phosphomycine was used for counterselection of M. loti against E. coli.
TABLE 1.
Bacterial strains, plasmids, and transposon used in this study
| Strain, plasmid, or transposon | Relevant characteristicsa | Reference or source |
|---|---|---|
| Mesorhizobium loti strains | ||
| MAFF303099 | Wild type, ACD+ Fix+ Pmr | 15 |
| ML-GUS | MAFF303099 labeled with gusA by mTn5SSgusA20, Pmr Spr Smr | This study |
| ACD-GUS | MAFF303099 labeled with gusA fused with the acdS gene under regulation of acdS promoter, Pmr Kmr | This study |
| nifA1 mutant | nifA1::Gm derivative of ML-GUS, Fix+ Pmr Spr Smr Gmr Kms | This study |
| nifA2 mutant | ΔnifA2::Tc derivative of ML-GUS, Fix− Pmr Spr Smr Tcr Kms | This study |
| Escherichia coli strains | ||
| DH5α | Cloning strain | Toyobo Inc., Tokyo, Japan |
| S17-1 | Strain used for conjugation and gene disruption | 30 |
| Plasmids | ||
| pBluescriptII KS(−) | Cloning vector, Apr | Takara Shuzo Co., Kusatsu, Japan |
| pBS-GUS | pBluescriptII KS(−) carrying gusA amplified from pKW107, Apr | This study |
| pMS246 | Plasmid carrying 1-kb gentamicin resistance cassette, Gmr Apr | 1 |
| p34S-Tc | Plasmid carrying 1.5-kb tetracycline resistance cassette, Tcr Apr | 3 |
| pmTn5SSgusA20 | Plasmid used for transposon mTn5SSgusA20 insertion, Apr Spr | 35 |
| pK18mob | Plasmid used for cloning and mating, mob, Kmr | 29 |
| pPD1 | pK18mob carrying 6.6-kb BamHI fragment containing acdS, Kmr | 33 |
| pPD3 | gusA inserted into acdS gene StuI site of pPD1, Kmr | This study |
| C229 | M. loti ordered cosmid clone no. 229 including mll5837 (nifA2), Tcr | 11 |
| C230 | M. loti ordered cosmid clone no. 230 including mll5857 (nifA1), Tcr | 11 |
| C232 | M. loti ordered cosmid clone no. 232 including mlr5905 (nifH), Tcr | 11 |
| pKnifA1 | pK18mob carrying 4.8-kb EcoRI fragment containing mll5857 (nifA1) from C230, Kmr | This study |
| pKnifA1::Gm | pK18mob carrying 5.7-kb nifA1::Gm, Kmr Gmr | This study |
| pKnifA2 | pK18mob carrying 6.7-kb SmaI fragment containing mll5837 (nifA2) from C229, Kmr | This study |
| pKnifA2::Tc | pK18mob carrying 7.2-kb ΔnifA2::Tc, Kmr Tcr | This study |
| pFAJ1702 | Plasmid used for complementation, Apr Tcr | 6 |
| pFAJnifA2 | pFAJ1702 containing 6.7-kb nifA2 fragment from pKnifA2, Apr Tcr | This study |
| Transposon | ||
| mTn5SSgusA20 | Minitransposon, gusA, Spr Smr | 35 |
ACD, ACC deaminase; Apr, ampicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Pmr, phosphomycine resistant; Smr, streptomycin resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant.
Construction of gusA reporter fused with acdS gene.
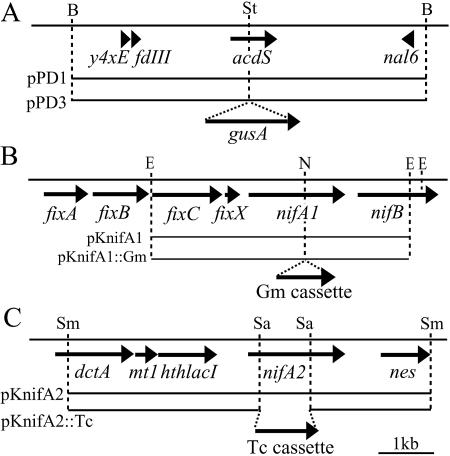
The gusA gene was amplified from plasmid pmTn5SSgusA20 with the following primers: gusA+StuIAAA-1F (5′-AAAAGGCCTATGTTACGTCCTGTAGAAAC-3′) and gusA+StuIAAA-1801R (5′-AAAAGGCCTTCATTGTTTGCCTCCCTGCT-3′). The PCR product was cloned and inserted into StuI-digested pPD1. The plasmid generated, pPD3, was introduced into M. loti MAFF303099 (Fig. 1A). Single-crossover mutants were selected. One of the mutants was designated ACD-GUS and was inoculated into the host plant.
FIG. 1.
Physical and restriction enzyme map of the acdS, nifA1, and nifA2 genes and the flanking region in M. loti MAFF303099. The arrows represent open reading frames. (A) Construction of gusA reporter fused with the acdS gene. A single-crossover mutation between pPD3 containing an acdS-gusA fusion and the genomic DNA generated the ACD-GUS strain harboring an intact genomic acdS gene and an acdS-gusA fusion. (B) Generation of the nifA1 mutant. A double-crossover mutation between pKnifA1::Gm and genomic DNA generated the nifA1 mutant. (C) Generation of the nifA2 mutant. A double-crossover mutation between pKnifA2::Tc and genomic DNA generated the nifA2 mutant. B, BamHI; E, EcoRI; N, NdeI; Sa, SalI; Sm, SmaI; St, StuI.
Construction of M. loti with constitutive gusA gene.
The constitutive gusA gene driven by the aminoglycoside phosphotransferase (aph) promoter from Tn5 was introduced into M. loti MAFF303099 by using minitransposon mTn5SSgusA20 (35). A β-glucuronidase (GUS)-labeled strain, designated ML-GUS, was used throughout this study because it had the same growth and nodulation capacities as the wild-type strain.
Histochemical localization of GUS activity.
At 5 and 10 days after inoculation (DAI), plant roots inoculated with strains ML-GUS or ACD-GUS were evaluated for GUS activity, as described previously (38). The nodulation events, including infection of the host roots, were observed with a stereomicroscope, as described previously (22).
Construction of nifA1 and nifA2 mutants.
The nifA1 mutant was constructed by insertion of the gentamicin resistance gene aacC1 (Gm cassette) (1) into an NdeI site in nifA1 (mll5857) (Fig. 1B). The nifA2 mutant was constructed by deletion and insertion of the tetracycline resistance gene tet(C) (Tc cassette) (3) into a SalI site in nifA2 (mll5837) (Fig. 1C). Double-crossover mutants were selected by screening for resistance and sensitivity to appropriate antibiotics. The validity of the mutants was confirmed by reverse transcription PCR (RT-PCR); the nifA1and nifA2 transcripts from the mutants were not detected using primer sets designed on the basis of the sequence downstream of the inserted antibiotic cassette in the nifA homologues.
Complementation of the nifA2 mutant.
A 6.7-kb DNA fragment carrying the nifA2 gene from pKnifA2 was ligated into pFAJ1702 (6). The resulting plasmid, pFAJnifA2, was introduced into the nifA2 mutant by conjugation. Restoration of nifA2 transcription in the nifA2 mutant was confirmed by RT-PCR.
RNA extraction.
Total RNA was extracted from L. japonicus roots that included nodule primordia at 5 DAI and from roots with mature nodules at 10 DAI by using TRIzol (QIAGEN, Hilden, Germany). In all cases, genomic DNA was removed by DNase I treatment.
Quantification of transcripts.
The transcripts of gusA, acdS (mlr5932), nifH (mlr5905), nifA1 (mll5857), and nifA2 (mll5837) in symbiotic cells were determined by real-time RT-PCR. For quantification of the transcripts, the assay was performed with an ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA). The sequences of the gusA, acdS, nifH, nifA1, and nifA2 genes were used to design primers and TaqMan probes specific for these genes. The nifH gene was used as the representative gene; this gene is induced by the NifA protein under nitrogen-fixing conditions in many diazotrophic bacteria, including rhizobia (34). The PCR primers and TaqMan probes used are listed in Table 2. Relative levels of gene expression were determined using gusA as the reference gene. Three independent experiments were performed.
TABLE 2.
Primers and probes used for quantitative real-time PCR
| Primer or probe | Sequence |
|---|---|
| Primers | |
| gusA-1257F | 5′-AGGTGCACGGGAATATTTCG-3′ |
| gusA-1317R | 5′-ACGCGTCGGGTCGAGTT-3′ |
| nifA1-933F | 5′-CGGCCGGTTCGAATTG-3′ |
| nifA1-992R | 5′-GAAATCTCGCCGATTTCATCA-3′ |
| nifA2-122F | 5′-TCCCTATTGTGCCGTTGCA-3′ |
| nifA2-185R | 5′-GCGGTGAGCGCTATCGA-3′ |
| nifH-602F | 5′-CCGCCAGACTCAATTCCAA-3′ |
| nifH-662R | 5′-GCGTGCTGGACGATGTT-3′ |
| acdS-299F | 5′-GCTGGGTTCCACATGAGGAT-3′ |
| acdS-365R | 5′-CCCAAGATGCGGCTCAAG-3′ |
| TaqMan probes | |
| gusA-1279TM | 5′-CCACTGGCGGAAGCAACGCG-3′ |
| nifA1-950TM | 5′-CGAATGGCGGAACCCTGCTGC-3′ |
| nifA2-143TM | 5′-AGAGGGCGCCGGACATCTTTGAAAC-3′ |
| nifH-625TM | 5′-ATCCACTTCGTGCCGCGC-3′ |
| acdS-323TM | 5′-TCTACGACCGGGTCGGCAACATTC-3′ |
RESULTS
ACC deaminase gene expression.
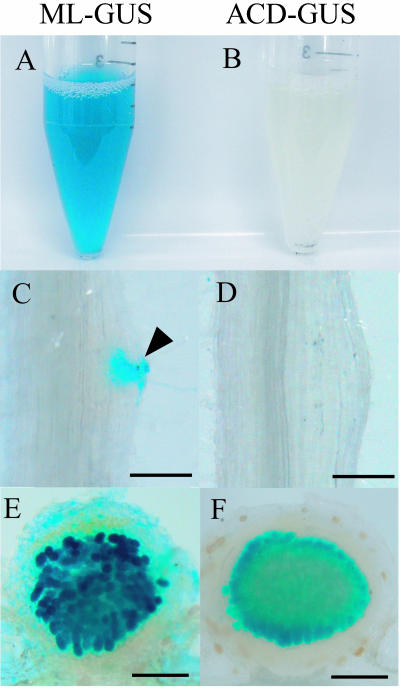
The GUS activity was monitored during nodulation in L. japonicus roots inoculated with the ACD-GUS strain at 5 and 10 DAI. At 5 DAI, no significant GUS activity was detected in either infection threads or nodule primordia (Fig. 2B and D). At 10 DAI, GUS activity was detected in bacteroids in the mature nodules (Fig. 2F) but not in infection threads, nodule primordia, or immature white nodules (data not shown). The free-living cells of strain ACD-GUS grown aerobically never exhibited GUS activity. In contrast, the ML-GUS strain in free-living cultures and the infection threads, nodule primordia, and immature and mature nodules of L. japonicus inoculated with the same strain consistently exhibited GUS activity (Fig. 2A, C, and E).
FIG. 2.
GUS activity in M. loti MAFF303099 expressing gusA under control of the constitutively active aph promoter (ML-GUS) or the acdS promoter (ACD-GUS) in free-living conditions and during nodulation. Constitutively gusA-expressing strain ML-GUS (A, C, and E) was used as the positive control for ACD-GUS (B, D, and F). Free-living cell cultures (A and B) and root nodules on L. japonicus at 5 DAI (C and D) and 10 DAI (E and F) were incubated with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc). Invasion of cortical cells of nodule primordia by the ML-GUS strain in panel C is indicated by an arrowhead. On the other hand, ACD-GUS showed GUS activity exclusively within mature nodules (F). (C and D) Bars = 100 μm. (E and F) Bars = 300 μm.
Construction of nifA1 and nifA2 mutants and their symbiotic phenotypes.
The genome of M. loti MAFF303099 was found to contain two nifA homologue genes, designated nifA1 and nifA2. Therefore, we constructed nifA1 and nifA2 mutants (Fig. 1B and C). Plants inoculated with the nifA1 mutant exhibited a wild-type phenotype (green plants and normal red nodules) (Fig. 3B), like ML-GUS-inoculated plants (Fig. 3A). On the other hand, nifA2 mutant-inoculated plants had tiny white nodules on the roots and yellow leaves (Fig. 3C). The typical Fix− phenotype indicated that nifA2 is essential for symbiotic nitrogen fixation.
FIG. 3.
Phenotypes of 56-day-old L. japonicus plants inoculated with ML-GUS (A), the nifA1 mutant (B), and the nifA2 mutant (C) on nitrogen-free medium. Root nodules cut in half are shown in the lower panels. Plants inoculated with the nifA1 mutant exhibited a wild-type phenotype (B), like ML-GUS-inoculated plants (A). The nifA2 mutant-inoculated plants had tiny white nodules on the roots and yellow leaves (C). Bars = 600 μm.
Transcription of acdS, nifH, nifA1, and nifA2 genes during infection.
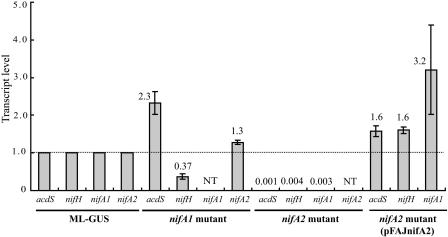
To investigate the expression profiles of the acdS and nitrogen fixation (nif) genes, the transcript levels of these genes in symbiotic cells of M. loti MAFF303099 were determined by quantitative RT-PCR. The analysis with the ML-GUS strain showed that transcripts of all four genes had accumulated at 10 DAI (Fig. 4), when mature nodules were present, but not at 5 DAI, when the roots lacked mature nodules (data not shown). The acdS expression in mature nodules was consistent with our previous findings (33). The patterns of nifA homologues and nifH were also consistent with the well-known regulation of nif genes dependent on nitrogen fixation.
FIG. 4.
Quantitative real-time RT-PCR analyses for the acdS, nifH, nifA1, and nifA2 genes in M. loti MAFF303099 for roots at 10 DAI. The relative transcript level for each target gene was normalized to the gusA copy number, and the transcript levels for ML-GUS were defined as 1.0. The values are means of three independent experiments, and the error bars indicate standard errors. NT, not tested.
As shown in Fig. 4, the nifA2 mutant was not able to transcribe acdS, nifA1, or nifH at 10 DAI. On the other hand, complementation of the nifA2 mutant with pFAJnifA2 restored acdS, nifH and nifA1expression, indicating that the nifA2 gene is required for transcription of these genes. In contrast, inoculation with the nifA1 mutant resulted in a significant increase (2.3-fold) in the acdS transcript level and a slight increase (1.3-fold) in the nifA2 transcript level, whereas there was a significant decrease (0.37-fold) in the nifH transcript level (Fig. 4). This result implies that NifA1 may play a secondary role compared with the role of NifA2.
DISCUSSION
Mature nodule-specific and nifA-dependent expression of acdS in M. loti.
In this study, we examined the transcriptional regulation and nodulation stage-specific transcription of the acdS gene of M. loti MAFF303099 under symbiotic conditions. The ACD-GUS construct clearly demonstrated that acdS is exclusively transcribed in bacteroids (Fig. 2). Consistent with the other results, quantitative real-time RT-PCR analysis showed that acdS transcription occurred in roots with mature nodules at 10 DAI (Fig. 4) but not in roots at 5 DAI (data not shown), which lacked mature nodules. These results are also consistent with our previous report which showed that the level of transcription of the acdS gene was higher under symbiosis conditions than in free-living cells (33).
Using nifA mutants, we also demonstrated that the transcriptional regulation of the acdS gene of M. loti MAFF303099 is dependent on a functional nifA2 gene under symbiotic conditions (Fig. 4). This result is consistent with the DNA sequence upstream of acdS, which contains two −24/−12 type promoters (5) and one upstream activating sequence. The acdS gene is present in many soil bacteria, and this gene is known to be regulated mainly by an LRP-like protein, which is an important regulator for amino acid metabolism and related processes (2), even in rhizobia (16, 19). Therefore, the mature nodule-specific and nifA-dependent mechanism of acdS regulation in M. loti is quite different from the mechanism of acdS regulation in other bacteria. To the best of our knowledge, this is the first report of the molecular mechanism of symbiosis-specific suppression of ethylene biosynthesis leading to enhancement of rhizobial nodulation.
M. loti acdS seems to have acquired symbiosis-specific functions during its coevolution with the host legume L. japonicus. Two physiological effects of ethylene biosynthesis suppressers, such as ACC deaminase and rhizobitoxine, in the establishment of symbiosis have been proposed. One of these effects is enhancement of early infection events on host roots (for reviews see references 24 and 31). The other effect is a positive effect on the establishment and/or maintenance of mature nodules (7, 23). In Glycine max, ACC deaminase protein was reported to accumulate in the bacteroid (12). In canola roots, ACC deaminase-producing E. cloacae changed the expression of host genes involved in the defense response and cell division (14). Moreover, ethylene application suppressed nitrogenase activity in legume nodules (9). Taken together, the data show that although the detailed physiological functions of ACC deaminase in nodule bacteroids are not clear, ACC deaminase may control the host defense response, mitosis, and nitrogenase activity by lowering the ethylene level around the bacteroids.
Duplicated nifA genes.
Since two nifA genes were found in the M. loti MAFF303099 genome, we analyzed the accumulation of acdS and nifH transcripts in each nifA mutant. nifA2-dependent nifH regulation suggests that nifA2 encodes the canonical NifA protein, which may regulate other nif genes by collaborating with RNA polymerase σ54, similar to NifA proteins of other nitrogen-fixing bacteria (4). In contrast, regulation by nifA1 is complicated, and NifA1 may have roles different from those of NifA2 (Fig. 3 and 4). It is interesting that nifA1 is located in a gene cluster consisting of nif and fix genes (nifS, nifW, fixA, fixB, fixC, fixX, nifA1, nifB) (Fig. 1B), and this structure is similar to that of R. leguminosarum bv. viciae (20) and the symbiosis island in M. loti R7A (32). On the other hand, nifA2 is located 18 kb from nifA1, and it seems to be unaccompanied by other nif and fix genes. These gene structures suggest that nifA1 is the original nifA gene in M. loti.
Two bacteria other than M. loti have been reported to possess two nifA genes. A nonsulfur phototropic purple bacterium, Rhodobacter capsulatus, has two functional copies of nifA encoding NifA proteins. However, the amino acid identities and functions of the two NifA proteins are almost the same (26, 27). In Frankia sp. strain EuIK1, which forms nitrogen-fixing nodules on an actinorhizal plant, Elaeagnus umbellatata, nifA1 encodes a typical NifA protein, whereas the NifA encoded by nifA2 is rather similar to AnfA, the alternative nitrogen fixation regulator (18). Our findings and previous reports suggest that symbiotic interactions with host plants may have led to differentiation of the duplicated nifA genes.
Acknowledgments
We thank K. Saeki (Nara Women's University, Nara, Japan) for providing ordered cosmid clones and H. Mitsui (Tohoku University) for providing several plasmids. We are also grateful to M. Sugawara (Tohoku University), Y. Kawaharada (Tohoku University), and N. Shimada (Nihon University) for valuable comments.
This work was supported by the 21st century COE program “Bioresource Utilization Based on Microbial Symbiotic Systems” from the Ministry of Education, Culture, Sports, Sciences and Technology of Japan. This work was also supported by Grant-in-Aid for Scientific Research on Priority Areas “Comparative Genomics” and by grants 15380002 and 17380046 to K.M. from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Becker, A., M. Schmidt, W. Jäger, and A. Pühler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 2.Brinkman, A. B., T. J. G. Ettema, W. M. de Vos, and J. van der Oost. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48:287-294. [DOI] [PubMed] [Google Scholar]
- 3.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon, R., and D. Kahn. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621-631. [DOI] [PubMed] [Google Scholar]
- 5.Dombrecht, B., K. Machal, J. Vanderleyden, and J. Michiels. 2002. Prediction and overview of the RpoN-regulation in closely related species of the Rhizobiales. Genome Biol. 3:research0076.1-0076.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 7.Duod, S., T. V. Bhuvaneswari, T. J. W. Stokkermans, and N. K. Peters. 1999. A positive role for rhizobitoxine in Rhizobium-legume symbiosis. Mol. Plant-Microbe Interact. 12:1082-1089. [Google Scholar]
- 8.Fisher, R. F., and S. R. Long. 1992. Rhizobium-plant signal exchange. Nature 357:655-660. [DOI] [PubMed] [Google Scholar]
- 9.Goodlass, G., and K. A. Smith. 1979. Effects of ethylene on root extension and nodulation of pea (Pisum sativum L.) and white clover (Trifolium repens L.). Plant Soil 51:387-395. [Google Scholar]
- 10.Grichko, V. P., and B. R. Glick. 2000. Identification of DNA sequences that regulate the expression of the Enterobacter cloacae UW4 1-aminocyclopropane-1-carboxylic acid deaminase gene. Can. J. Microbiol. 46:1159-1165. [DOI] [PubMed] [Google Scholar]
- 11.Hattori, Y., H. Omori, M. Hanyu, N. Kaseda, E. Mishima, T. Kaneko, S. Tabata, and K. Saeki. 2002. Ordered cosmid library of the Mesorhizobium loti MAFF303099 genome for systematic gene disruption and complementation analysis. Plant Cell Physiol. 43:1542-1557. [DOI] [PubMed] [Google Scholar]
- 12.Hoa, L. T. P., M. Nomura, and S. Tajima. 2004. Characterization of bacteroid proteins in soybean nodules formed with Bradyrhizobium japonicum USDA110. Microbes Environ. 19:71-75. [Google Scholar]
- 13.Honma, M., and T. Shimomura. 1978. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42:1825-1831. [Google Scholar]
- 14.Hontzeas, N., S. S. Saleh, and B. R. Glick. 2004. Changes in gene expression in canola roots induced by ACC-deaminase-containing plant-growth-promoting bacteria. Mol. Plant-Microbe Interact. 17:865-871. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi, M. 2000. Lotus japonicus ‘Miyakojima’ MG-20: an early-flowering accession suitable for indoor handling. J. Plant Res. 113:507-509. [Google Scholar]
- 18.Lee, H., S. B. Sung, H. B. Kim, and C. S. An. 2000. Sequence analysis and expression patterns of two nifA genes from Frankia EuIK1. Aust. J. Plant Physiol. 28:939-949. [Google Scholar]
- 19.Ma, W., F. C. Guinel, and B. R. Glick. 2003. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 69:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez, M., J. M. Palacios, J. Imperial, and T. Ruiz-Argueso. 2004. Symbiotic autoregulation of nifA expression in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 186:6586-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nukui, N., H. Ezura, K. Yuhashi, T. Yasuta, and K. Minamisawa. 2000. Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 41:893-897. [DOI] [PubMed] [Google Scholar]
- 22.Nukui, N., H. Ezura, and K. Minamisawa. 2004. Transgenic Lotus japonicus with an ethylene receptor gene Cm-ERS1/H70A enhances formation of infection threads and nodule primordia. Plant Cell Physiol. 45:427-435. [DOI] [PubMed] [Google Scholar]
- 23.Okazaki, S., K. Yuhashi, and K. Minamisawa. 2003. Quantitative and time-course evaluation of nodulation competitiveness of rhizobitoxine-producing Bradyrhizobium elkanii. FEMS Microbiol. Ecol. 45:155-160. [DOI] [PubMed] [Google Scholar]
- 24.Okazaki, S., N. Nukui, M. Sugawara, and K. Minamisawa. 2004. Rhizobial strategies to enhance symbiotic interactions: rhizobitoxine and 1-aminocyclopropane-1-carboxylate deaminase. Microbes Environ. 19:99-111. [Google Scholar]
- 25.Okazaki, S., M. Sugawara, and K. Minamisawa. 2004. Bradyrhizobium elkanii rtxC gene is required for expression of symbiotic phenotypes in the final step of rhizobitoxine biosynthesis. Appl. Environ. Microbiol. 70:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paschen, A., T. Drepper, B. Masepohl, and W. Klipp. 2001. Rhodobacter capsulatus nifA mutants mediating nif gene expression in the presence of ammonium. FEMS Microbiol. Lett. 200:207-213. [DOI] [PubMed] [Google Scholar]
- 27.Pawlowski, A., K. U. Riedel, W. Klipp, P. Dreiskemper, S. Gross, H. Bierhoff, T. Drepper, and B. Masepohl. 2003. Yeast two-hybrid studies on interaction of proteins involved in regulation of nitrogen fixation in the phototrophic bacterium Rhodobacter capsulatus. J. Bacteriol. 185:5240-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penmetsa, R. V., and D. R. Cook. 1997. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275:527-530. [DOI] [PubMed] [Google Scholar]
- 29.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:789-791. [Google Scholar]
- 31.Sugawara, M., S. Okazaki, N. Nukui, H. Ezura, H. Mitsui, and K. Minamisawa. 2006. Rhizobitoxine modulates plant-microbe interactions by ethylene inhibition. Biotechnol. Adv. 24:382-388. [Epub ahead of print.] [DOI] [PubMed]
- 32.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. De Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiumi, T., T. Ohwada, M. Itakura, H. Mitsui, N. Nukui, P. Dawadi, T. Kaneko, S. Tabata, T. Yokoyama, K. Tejima, K. Saeki, H. Omori, M. Hayashi, T. Maekawa, R. Sriprang, Y. Murooka, S. Tajima, K. Simomura, M. Nomura, A. Suzuki, Y. Shimoda, K. Sioya, M. Abe, and K. Minamisawa. 2004. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186:2439-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valderrama, B., A. Davalos, L. Girard, E. Morett, and J. Mora. 1996. Regulatory proteins and cis-acting elements involved in the transcriptional control of Rhizobium etli reiterated nifH genes. J. Bacteriol. 178:3119-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, K. J. 1995. Molecular techniques for the study of rhizobial ecology in the field. Soil Biol. Biochem. 27:501-514. [Google Scholar]
- 36.Wopereis, J., E. Pajuelo, F. B. Dazzo, Q. Jiang, P. M. Gresshoff, F. J. De Bruijn, J. Stougaard, and K. Szczyglowski. 2000. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 23:97-114. [DOI] [PubMed] [Google Scholar]
- 37.Yasuta, T., S. Satoh, and K. Minamisawa. 1999. New assay for rhizobitoxine based on inhibition of 1-aminocyclopropane-1-carboxylate synthase. Appl. Environ. Microbiol. 65:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuhashi, K., N. Ichikawa, H. Ezura, S. Akao, Y. Minakawa, N. Nukui, T. Yasuta, and K. Minamisawa. 2000. Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl. Environ. Microbiol. 66:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]