Abstract
All cultivated isolates of the bacterial order Thermotogales are either thermophiles or hyperthermophiles, but Thermotogales 16S rRNA gene sequences have been detected in many mesophilic anaerobic and microaerophilic environments, particularly within communities involved in the remediation of pollutants. Here we provide metagenomic evidence for the existence of Thermotogales lineages, which we informally call “mesotoga,” that are adapted to growth at lower temperatures. Two fosmid clones containing mesotoga DNA, originating from a low-temperature enrichment culture that degrades a polychlorinated biphenyl congener, were sequenced. Phylogenetic analysis clearly puts this bacterial lineage within the Thermotogales order, with the rRNA gene trees and 21 of 58 open reading frames strongly supporting this relationship. An analysis of protein sequence composition showed that mesotoga proteins are adapted to function at lower temperatures than are their identifiable homologs from thermophilic and hyperthermophilic members of the order Thermotogales, supporting the notion that this bacterium lives and grows optimally at lower temperatures. The phylogenetic analysis suggests that the mesotoga lineage from which our fosmids derive has used both the acquisition of genes from its neighbors and the modification of existing thermophilic sequences to adapt to a mesophilic lifestyle.
Characterized members of the order Thermotogales are all either hyperthermophiles, growing at temperatures of >80°C, or thermophiles, with optimal growth between 45 and 80°C (17). No mesophilic Thermotogales strains (growing optimally at temperatures under 45°C) have so far been described. However, Thermotogales 16S rRNA genes have been detected in several culture-independent analyses of different mesophilic anaerobic environments. For instance, Holoman et al. (15) detected Thermotogales 16S rRNA genes in an anaerobic enrichment culture inoculated with sediments from Baltimore Harbor that ortho-dechlorinate 2,3,5,6-tetrachlorobiphenyl (incubation temperature was 30°C), Dollhopf et al. (6) detected Thermotogales rRNA genes in 34°C anaerobic bioreactors inoculated with sediments, Chouari et al. (4) reported the presence of Thermotogales rRNA genes in an anaerobic sludge digester, and Yan et al. (44) detected Thermotogales rRNA genes in sediment cultures capable of reductively dechlorinating 2,3,4,5-tetrachlorobiphenyl (Table 1). Interestingly, the mesophilic Thermotogales signal from Baltimore Harbor corresponded to the second most predominant 16S sequence detected during active polychlorinated biphenyl dechlorination (15). A similar type of rRNA gene restriction fragment length polymorphism was recovered by the same group in a similar enrichment culture inoculated with sediments from Charleston Harbor (41, 43). In addition to these published studies, several 16S rRNA sequences from similar environments are available in GenBank from unpublished studies (Table 1). It seems reasonable to infer from such observations that members of the order Thermotogales that are adapted for growth at less than 45°C, which we might informally call “mesotogas,” do exist. The metagenomic exercise described here confirms this conjecture.
TABLE 1.
Characteristics of Thermotogales rRNA genes from mesophilic habitats
| Accession no.a | Site of isolation | Reference(s) |
|---|---|---|
| AF338765 | Anaerobic bioreactors inoculated with sediments at 34°C | 6 |
| AF058000 (100%) | Enrichment culture on defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community inoculated with sediment from Baltimore Harbor. The same restriction fragment length polymorphism-type was detected in a polychlorinated biphenyl-dechlorinating consortium originally inoculated with sediments from Charleston Harbor | 15, 41, 43 |
| CR933200, CR933305 | Anaerobic sludge samples from anaerobic mesophilic digester | 4 |
| AB186798 | Polychlorinated dioxin-dechlorinating microcosmos community seeded by polluted river sediments | 45 |
| DQ080178, DQ080153, DQ080154 | Anaerobic enrichment cultures dechlorinating 2,3,4,5-chlorobiphenyl inoculated with sediments from Palos Verdes and Baltimore Harbor | 44 |
| AF482446, AF482447 | Granular sludge | |
| AY692052 | Anaerobic biofilms | |
| AB195914-AB195923 | Anaerobic reactors | |
| DQ447166, DQ447170, DQ447172 | Microbial community in a gas-fed bioreactor treating sulfate- and zinc-rich wastewater | |
| AY261392 98% identity | Polycyclic aromatic hydrocarbon-degrading methanogenic community initiated with sediments from Baltimore Harbor | |
| AB232569 (97%) | Methanogenic sludges treating wastewater containing a high strength of lipids | |
| DQ191738, DQ191695, DQ191698, DQ191720 (96 to 97%) | Psychrophilic anaerobic bioreactors treating solvent-containing pharmaceutical wastewater | |
| DQ325495 98% identity | Acidogenic sulfate-reducing reactor | |
| DQ306713, DQ306715 (97%) | Chlorophenol-degrading anaerobic fluidized bed bioreactor | |
| DQ443958, DQ443994 (98%) | Microbial communities from different bioreactors fed with the same seed sludge |
Partial sequences were not included in the tree in Fig. 2. For these sequences, the percent identity to the rRNA gene in the fosmid is given in parentheses.
We constructed a fosmid library from DNA isolated from one of the polychlorinated biphenyl-containing enrichment cultures described by Holoman et al. (15), and here we present an analysis of the sequences of two fosmids from this library. We conclude that these DNA fragments do indeed originate from a mesophilic member of the order Thermotogales, and we also suggest that the adaptation to a mesophilic lifestyle has occurred both through the acquisition of new genes and through the modification of sequences already present.
MATERIALS AND METHODS
DNA was extracted from a room temperature enrichment culture, BH459 3,5-ACE, previously analyzed by Holoman et al. (15), following the protocol of Rondon et al. (36) except that preparative pulsed-field gel electrophoresis was excluded since the isolated DNA was >25 kb and clean enough for ligation after isolation. The enrichment cultures were a gift from Joy Watts (Center of Marine Biotechnology, University of Maryland Biotechnology Institute, presently at Department of Biological Sciences, Towson University).
A fosmid library was constructed using the CopyControl Fosmid Library production kit from Epicentre (Madison, WI), following the protocol of the manufacturer. Fosmid clone DNA was extracted using the REAL Prep 96 plasmid kit (QIAGEN, Mississauga, ON, Canada). We screened six 96-well plates for the presence of a mesotoga 16S rRNA by PCR using the Thermotogales-specific primers Balt.SSU.D.42, ATC ACT GGG CGT AAA GGG AG, and Balt.SSU.D.494, GTG GTC GTT CCT CTT TCA AT.
Subcloning of fosmids was performed using the TOPO Shotgun subcloning kit (Invitrogen, Burlington, ON, Canada), and each fosmid was sequenced to more than 8× coverage. Low-quality regions and gaps were targeted by PCR. The fosmid sequences were assembled using phredPhrap and Consed (http://www.phrap.org/phredphrapconsed.html) (8, 9, 13). Open reading frames (ORFs) were identified using the run-glimmer2 script using the default settings provided in this script (5), and ORFs shorter than 100 bp were eliminated. If two overlapping ORFs were identified, we selected the one that had significant homologs in GenBank. tRNAs were identified with tRNAscan-SE (25). The ORFs were annotated using BLASTP searches (1) of GenBank at http://www.ncbi.nlm.nih.gov/BLAST/ and Pfam searches (2) at http://www.sanger.ac.uk/Software/Pfam/search.shtml. The ORFs were designated MES0001 and MES0058.
Phylogenetic analyses of the 23S rRNA fragment and 16S rRNA genes were performed in PAUP*, version 4.0b10 (39). A 16S rRNA gene alignment based on secondary structure containing several Thermotogales lineages as well as outgroup sequences (see Fig. 2) was obtained from http://www.psb.ugent.be/rRNA/ssu/. Additional Thermotogales sequences were added to this alignment manually. The 23S rRNA alignment contained only Thermotogales sequences and could be aligned by eye. Minimum evolution (ME) trees were constructed using LogDet distances, and maximum likelihood (ML) trees were constructed using a general time-reversible model with gamma-distributed rates with four categories and invariable sites (GTR + Γ + Ι). Ten random-addition cycles of the sequences and tree bisection and reconnection branch swapping were used in both cases.
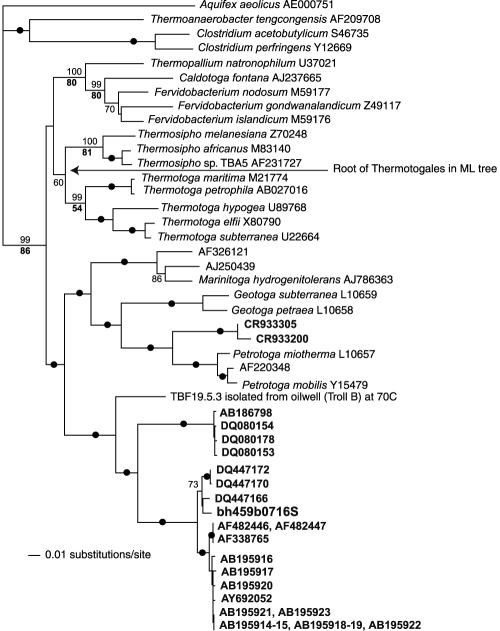
FIG. 2.
16S rRNA gene phylogeny of the order Thermotogales. Sequences in bold are 16S rRNA genes from mesophilic environments (see Table 1). The tree is a ME tree estimated in PAUP* using LogDet distances. The ML tree estimated using a GTR + Γ + I model was similar, the main difference being the placement of the root of the order Thermotogales (indicated by an arrow). Numbers on branches indicate the percentage of occurrence in 100 bootstrap replicates using LogDet distances (plain) and ML (bold). If the support was at least 90% in both analyses, a black circle is given on the branch.
Homologs of the ORFs in GenBank were identified and retrieved using BLASTP searches at http://www.ncbi.nlm.nih.gov/BLAST/. In addition, we retrieved ORFs from the unfinished genome of Thermosipho sp. TCF52B that we are currently sequencing.
Clusters of very similar sequences from the same or sister taxon were trimmed down to one representative sequence. We also removed sequences that were considerably shorter than the rest of the alignment as well as sequences that were difficult to align. The alignments were edited by deleting regions with many or large gaps. For each ORF, we made both simple neighbor-joining (NJ) trees with bootstrap analysis and a ME tree (with bootstrap analysis; 100 replicates with global rearrangements) estimated from ML distances (the substitution matrix was auto selected by TREE-PUZZLE 5.2 [37] + Γ, global rearrangements, and 10 random-addition replicates). In addition to these trees, we also constructed automated ML trees (WAG [42] + Γ + Ι model) using the PhyloGenie package (11) modified by Eric Bapteste (Department of Biochemistry and Molecular Biology, Dalhousie University). Thermosipho sp. TCF52B sequences were not included in these trees as this program performs an automated retrieval of sequences from GenBank. In cases where the automated ML tree differed significantly from both the NJ and ME trees or where the Thermosipho sp. TCF52B was the only close Thermotogales homolog, an ML tree was constructed from the same alignments as those of the NJ and ME trees using PMBML (40), a modified version of the PROML within the PHYLIP package, version 3.6a2 (10). For these analyses, we used a JTT (21) + Γ model, global rearrangements, and 10 random-addition replicates. In the ML bootstrap analyses, we performed only one random addition of sequences per bootstrap replicate.
The ratio of synonymous to nonsynonymous mutations (ds/dn ratio) was determined by using the Nei-Gojobori method (27) in SNAP (synonymous/nonsynonymous analysis program) (23) at http://hiv-web.lanl.gov/SNAP/WEBSNAP/SNAP.html.
The absolute difference between the frequency of charged and polar amino acid residues (charged versus polar [CvP] bias) (38) was calculated for each mesotoga ORF that was predicted from the fosmid sequences as well as for their Thermotoga maritima MSB8 and Thermosipho sp. TCF52B homologs (where available) using an in-house Perl script. Transmembrane helices were predicted using the TMAP program from the EMBOSS package (35). ORFs with at least two predicted transmembrane helices were considered putative transmembrane proteins. Since the correlation of the CvP value with the thermostability of a protein does not hold for transmembrane proteins (38), the predicted transmembrane proteins were excluded from further analyses. ORFs were further classified into three categories according to their CvP values: mesophilic (CvP value, <7.35), thermophilic (CvP value, >10.62) and intermediate (CvP value, >7.35 to <10.62). The categories were defined following the observed CvP values in different genomes (Table 1 in reference 38).
Thermotogales 16S rRNA stems were identified by comparing the 16S rRNA gene alignment used in the phylogenetic analysis to the T. maritima MSB8 secondary structure obtained from Konings and Gutell (22), available at the comparative RNA web site (3). We used only unambiguously aligned regions. Optimal growth temperatures for members of the order Thermotogales were obtained from Huber and Hannig (17).
Nucleotide sequence accession numbers.
The sequenced fosmids have been submitted to GenBank with accession numbers AM184115 and AM184116.
RESULTS AND DISCUSSION
Identification and sequencing of mesotoga fosmid clones.
A fosmid library was constructed from DNA isolated from one of the enrichment cultures characterized by Holoman et al. (15). Two overlapping fosmid clones were sequenced. The first, bh459f1.4.b07, was identified by screening six 96-well plates with primers targeting the Thermotogales rRNA gene sequence already identified in the Baltimore Harbor enrichment culture. In addition to this clone, at least one more clone was identified that contained an identical 16S rRNA gene fragment. The second sequenced fosmid clone, bh459f1.5.c09, was identified by screening the library with primers matching the 5′ end of the first clone. The full contig is 72,913 bp. We identified 58 ORFs designated MES0001 and MES0058, one 16S, 23S, and 5S rRNA gene-containing operon, and four tRNA genes (Fig. 1 and see Table S1 in the supplemental material). The internal transcribed spacer did not contain the genes for tRNAIle and tRNAAla, which are found in the internal transcribed spacer of characterized Thermotoga strains (28, 29), and no intron was found in the 23S rRNA (observed in some Thermotoga strains [30]). Interestingly, in screening 367 end sequences by BLASTN searches for matches to the sequenced fosmid clones, one additional fosmid clone, bh459f1.2.e11, was identified (but not sequenced) that matched the 5′ part of the bh459f1.5.c09 clone, suggesting that our fosmid library is rich in mesotoga DNA.
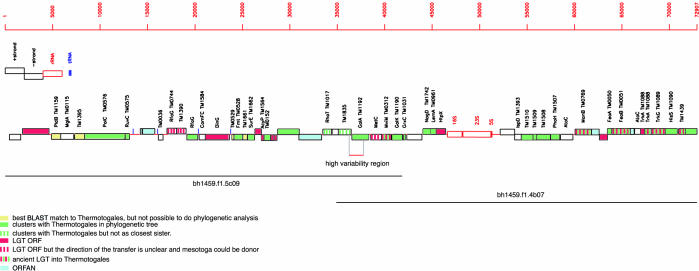
FIG. 1.
Overview of the sequenced mesotoga fosmids. Where a homolog is found in the Thermotoga maritima MSB8 genome, the locus tag is given. A white box represents an ORF for which none of the classifications listed in the figure applied, i.e., it does not have a Thermotogales sequence as a top-scoring BLAST hit, it does not cluster with Thermotogales sequences in phylogenetic trees, and the trees or the alignments were not reliable enough to conclude whether it is an instance of LGT.
We did not observe any indication that the fosmid clones sequenced here were chimeric in origin. The G+C content was very uniform across both fosmid clones (∼46%) (see Table S1 in the supplemental material). The end sequences obtained from the fosmid library varied in G+C content from 33 to 75%, with a median of 55.85%. Thus, we would expect to observe variations in G+C content if the clones were chimeric. We also observed only a single potential pseudogene (MES0053). Duplication or illegitimate recombination seems to be the most likely explanation for its origin (see below) since it has close neighbors on both sides that cluster strongly within the order Thermotogales (Fig. 1).
Annotation of the identified ORFs is given in Table S1 of the supplemental material, and an overview of the contig is given in Fig. 1. There were four cases in which adjacent ORFs were also neighbors in the T. maritima MSB8 genome and one case each where three and four ORFs were found in the same order in T. maritima MSB8. We examined the syntenic four-gene cluster in some detail. It comprises a hypothetical protein of unknown function, a predicted metal-dependent hydrolase, a predicted “HD” superfamily hydrolase, and a putative phosphate starvation-inducible protein. The three ORFs with no transmembrane domains were all predicted to be hyperthermophilic from their CvP values (CvP values of 15, 17.4, and 13.2, see below). Phylogenetic analyses showed that the mesotoga sequences clustered with at least one other member of the order Thermotogales for all four ORFs.
The two sequenced fosmid clones overlapped by 6,936 bp. The two clones were very similar in sequence in the overlapping region, except for a 1,524-bp region were 56 differences were observed, giving an uncorrected distance of 3.67% (see Table S1 in the supplemental material; Fig. 1). For the rest of the overlapping region, only five differences (0.1%) were observed. Thus, the two clones were likely obtained from two very similar strains, with the high-variability region representing genes subjected to diversifying selection or, as we think more likely, homologous recombination with a less closely related lineage. The ratio between synonymous to nonsynonymous substitutions for the two ORFs where the polymorphisms were observed suggested a purifying selection; ds/dn was 3.86 for MES0029 (a glycosidase) and 8.06 for MES0030 (alpha-galactosidase), in support of this interpretation.
Phylogenetic analyses.
Phylogenetic analyses confirmed that the DNA fragments sequenced here do indeed originate from a Thermotogales bacterium (Fig. 2 and see Table S2 in the supplemental material). In the 16S rRNA gene tree, the sequence from the fosmid clone clusters with the majority of other Thermotogales 16S rRNA genes found in mesophilic environments. The closest relative of this mesotoga clade is a thermophilic strain, TBF19.5, that we recently isolated at 70°C from production water from the Troll oil field in the North Sea (C. L. Nesbø et al., unpublished data). TBF19.5 and the mesotoga sequences cluster together as a sister clade to the clade containing Marinitoga, Geotoga, and Petrotoga, of which all known isolates are thermophiles. Notably, there is a second clade containing mesophilic Thermotogales strains clustering as a sister to Petrotoga, indicating that adaptation to mesophily has occurred at least twice among these bacteria. Importantly, the topology of the tree suggests that members of the ancestral order Thermotogales were thermophiles or hyperthermophiles and that mesophily is a derived trait. A similar topology was obtained from the 23S rRNA tree, with the bh459.f1.4.b07 sequence clustering as a sister to Petrotoga and Marinitoga (no sequence is available from TBF.19.5 or other mesotoga lineages).
The majority of protein-coding genes also put mesotoga sequences within the order Thermotogales in phylogenetic analyses. For 21 of the ORFs, phylogenetic analysis strongly supported a monophyletic clade containing the mesotoga sequence and one or more other Thermotogales sequences (see Table S1 in the supplemental material). Mesotoga and other Thermotogales bacteria also formed a monophyletic clade, but with no bootstrap support, in an additional five gene trees. Finally, for two ORFs, mesotoga was found in the same clade as other members of the order Thermotogales interspersed with sequence from another bacterial group, suggesting lateral gene transfer (LGT) from Thermotogales bacteria to various lineages.
Among the 15 ORFs for which we have sequence data from T. maritima MSB8, Thermosipho sp. TCF52B, and mesotoga and for which these three Thermotogales lineages appear to form a monophyletic clade, 12 displayed our mesotoga lineage as sister to a monophyletic Thermotoga-Thermosipho clade, which was in agreement with the rRNA result. For two ORFs, Thermosipho sp. TCF52B clustered as sister to a monophyletic mesotoga-Thermotoga clade, and for one ORF, T. maritima MSB8 clustered as sister to a monophyletic mesotoga-Thermosipho clade.
The second most common pattern grouped mesotoga or the Thermotogales clade containing mesotoga with Firmicutes, which are low GC gram positives (this pattern was observed for 14 of the ORFs) (see Table S2 in the supplemental material). Omelchenko et al. (32) have previously shown that T. maritima clusters with Clostridium acetobutylicum within a Firmicutes clade in a gene content tree based on the presence and absence of clusters of orthologous groups. These observations could be due to a high level of LGT between members of the order Thermotogales and low GC gram positives, a specific relationship between these groups, or perhaps more likely, a combination of these factors. The phylogenetic analyses of mesotoga ORFs were consistent with LGT in most cases (at least nine ORFs) (see Table S2 in the supplemental material), both recent and likely ancient events involving all three Thermotogales lineages, where members of both Thermotogales and Firmicutes act as donors of the sequence (see Table S2 in the supplemental material). Mesotoga and anaerobic Firmicutes such as Clostridia are found in similar environments, providing ample opportunities for LGT (4, 6, 15, 45). For a few trees, a sister relationship between low G+C gram positives and Thermotogales appeared most likely. This was, for instance, the case for MES0017 encoding a methionyl-tRNA formyltransferase where the order Thermotogales clustered at the base of the phylum Firmicutes. For the potential phylogenetic marker MES0006, encoding a gram-positive DNA polymerase alpha subunit (PolC), the Thermotogales sequences (together with a sequence from Fusobacterium) clustered as sister to Clostridium and Thermoanaerobacter and, in trees containing more distantly related homologs to root the tree, the Thermotogales sequences are found at the base of the clade containing the PolC sequences.
In total, the phylogenetic analyses suggested that 15 ORFs had been acquired by LGT (Fig. 1 and see Table S2 in the supplemental material). This is clearly an underestimate since we consider only ORFs for which robust phylogenetic analyses appeared possible to perform. Five of these are ancient LGTs that involve all three Thermotogales lineages analyzed here. MES0032 is an oxidoreductase of eukaryotic origin that has again been transferred from a member of the order Thermotogales to the order Thermococcales. MES0046 is a phosphomannomutase for which the sequences in mesotoga and Thermosipho show patterns consistent with frequent transfer between Firmicutes and Thermotogales. MES0050, MES0054, and MES0055 all appear to be ancient LGTs from Archaea. Interestingly, both MES0053 and MES0054 encode TrkA (K+ transport systems, NAD-binding component). MES0053 is shorter than other TrkA homologs, being only 135 amino acids, and is probably a pseudogene, possibly the result of a duplication of MES0054. However, in phylogenetic trees, this ORF clusters consistently with Actinobacteria, suggesting instead that it is a result of a recent LGT and possibly illegitimate recombination upstream of MES0054. The eight remaining recent LGTs appear to have been acquired from a wide range of sources, Archaea, Eukarya, and Bacteria (see Tables S1 and S2 in the supplemental material). Most of the recent transfers appear to be from mesophilic organisms (see Table S2 in the supplemental material). Two exceptions are MES0020 and MES0021 that likely have been acquired from an archaeon, as they are found branching at the base of archaeal clades. These archaeal clades contain both mesophilic and thermophilic organisms; therefore, it is not possible to say whether the donor was a thermophile or a mesophile.
These recently transferred genes encode a range of different functions. Interestingly, one of the recently obtained genes, MES0002, encodes a protein with diguanylate cyclase and phosphodiesterase activity (DGC-PDE). This protein has four distinct domains, a periplasmic binding domain, a PAS-PAC domain, a GGDEF domain, and a metal-dependent phosphohydrolase (HDc) domain (see Table S1 in the supplemental material). Such proteins have been shown to be involved in control of the cellular levels of c-diGMP, a secondary messenger involved in modulating bacterial growth on surfaces by regulating cellular adhesion components (19). The PAS-PAC domain is likely to be involved in signal sensing, while the GGDEF domain converts GTP to c-diGMP (19). The HDc is likely to be responsible for the hydrolytic cleavage of c-diGMP into GMPs (19). Thus, the GGDEF domain and the HDc domain probably have opposing effects on the cellular level of c-diGMP. Biofilm formation regulated by c-diGMP has been shown for T. maritima MSB8 (20). The presence of a DGC-PDE protein suggests that this also may be the case for mesotoga. The phylogenetic analyses show that the mesotoga DGC-PDE likely was acquired from a Clostridium bacterium, perhaps providing mesotoga with the ability to sense changes specific to its current environment. The fact that both c-diGMP regulators are present in the same protein probably facilitated the integration of such a new sensory system. Notably, there are two other genes, MES0045 and MES0052, which encode response regulator proteins involved in a two-component signal transduction system in bacteria, that function to detect and respond to environmental changes. These proteins show the highest similarity to organisms outside the order Thermotogales, and mesotoga sequences did not cluster consistently with any other sequences in phylogenetic trees. It is likely that these genes were also acquired through LGT. Thus, it appears that mesotoga has acquired several genes involved in interactions with its new environment.
Adaptation to mesophily.
One interesting question is how mesotoga has changed as a result of its mesophilic lifestyle. The phylogenetic analyses suggest that both the modification of existing sequences and the acquisition of new genes from mesophilic organisms have played a role. The most prominent signature of hyperthermophilic lifestyle at the proteome level has been shown to be the large difference between the proportion of charged (Asp, Glu, Lys, and Arg) versus polar (noncharged) (Asn, Gln, Ser, and Thr) amino acids in soluble proteins, abbreviated as the CvP-bias (38). A plot of CvP values for each mesotoga ORF and its Thermotogales homolog (if present) is shown in Fig. 3. Mesotoga shows a lower average CvP value (excluding predicted transmembrane proteins) than do both of the other Thermotogales lineages analyzed, with 9 for mesotoga, 11.2 for Thermosipho, and 15.4 for T. maritima. The CvP values correlate well with the growth temperature of these lineages, where T. maritima has the highest growth temperature (55 to 90°C, with a 80°C optimum) (18) and Thermosipho is a more moderate thermophile (37 to 75°C, with a 65°C optimum [Birkeland and Dahle, University of Bergen, personal communication]).
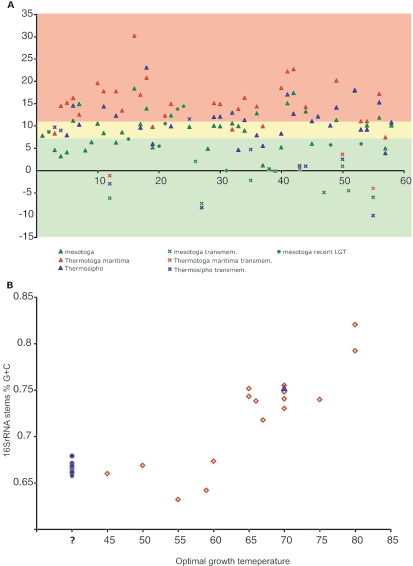
FIG. 3.
(A) Plot of CvP values of the ORFs along the mesotoga contig. CvP values are given on the y axis, and ORF numbers are along the x axis. The red area corresponds to ORFs classified as thermophilic (CvP value, >10.62), the yellow area corresponds to ORFs classified as intermediate (7.35 < CvP value < 10.62), and the green area corresponds to ORFs classified as mesophilic (CvP value, <7.35). The categories were defined following the observed CvP values in different genomes (Table 1 in reference 38). The CvP value was calculated for all mesotoga ORFs and for their closest T. maritima MSB8 and Thermosipho sp. TCF52B homologs (where available). (B) Plot of percent G+C content of 16S rRNA stems against optimal growth temperature. Thermotogales strains with known optimal growth temperature are represented by red diamonds. The strain TBF19.5.1, isolated at 70°C, is represented by a blue triangle. Mesotoga rRNA genes are represented by blue circles.
For those 37 mesotoga ORFs with homologs in the two other members of the order Thermotogales, the former had a lower CvP value than T. maritima did in 29 of 31 comparisons and lower than Thermosipho did in 20 of 29 comparisons (counting only nontransmembrane proteins) (Fig. 3A). It is difficult to say whether these are the result of adaptation to a mesophilic lifestyle or reflect neutral changes in a more relaxed selection regime in a mesophilic environment. However, as the closest relative, TBF19.5, shows little or no growth at room temperature (C. L. Nesbø et al., unpublished data), some adaptation to growing at lower temperatures must exist. One notable protein that does not fit this trend is MES0007, which carries a Holliday junction resolvasome endonuclease subunit, where the CvP values are reversed and the mesotoga homolog appears more thermophilic (Fig. 3A). The short length of the protein, together with structural and evolutionary constraints, is likely to be a contributing factor.
When we classified all 44 mesotoga nontransmembrane proteins into mesophilic (CvP value, <7.35), thermophilic (CvP value, >10.62), and intermediate, 17 appeared mesophilic, 14 intermediate, and 13 thermophilic. Among these, 10 of the mesophilic proteins, 12 of the thermophilic, and 9 of the intermediate could be used in phylogenetic analyses. From these trees, recent LGT was inferred for five of the mesophilic proteins (50%), while LGT was suggested for only two of the thermophilic proteins and one of the intermediate proteins, suggesting that mesotoga is using LGT to adapt to a mesophilic environment. Notably, the two thermophilic proteins that are suggested to have been acquired by LGT appear to have mesophilic donors. However, as observed for MES0007, both of these proteins are short (Fig. 1 and see Table S1 in the supplemental material) and the high CvP value might be due to structural and evolutionary constraints. LGT, as an important means to adapt to new environmental niches, high temperature, and high levels of radiation, was also shown for Thermus thermophilus and Deinococcus radiodurans (32).
Another adaptation to living at high temperatures is the higher G+C content of the stems in rRNA secondary structures (12). A plot of the percent G+C content of predicted stems of Thermotogales 16S rRNA genes from strains with known optimal growth temperature is shown in Fig. 3B. In addition, we plotted the G+C content of mesotoga's closest relative in 16S rRNA gene trees, the strain we isolated at 70°C (Fig. 3B), and the percent G+C content of all the rRNA genes from mesophilic environments, arbitrarily positioned at 40°C. As observed earlier (12), the variation in percent G+C at temperatures below the 60 to 65°C optimal growth temperature is considerable, likely because there is low or no selective pressure for lower G+C contents at lower temperatures, and thus it is not possible to predict mesotoga's optimal growth temperature from such plots. However, Fig. 3B shows that the percent G+C content of mesotoga 16S rRNA stems is considerably lower than that of its closest relative TBF19.5, which likely has an optimal growth temperature of ∼70°C. The fact that the mesotoga 16S rRNA stem percent G+C is somewhat higher than what is observed for strains with optimum growth temperatures much higher than the environments in which the mesotoga rRNA genes have been detected could be due to the fact that it has just recently, in terms of 16S rRNA gene evolution, moved into more temperate environments. Alternatively, it might thrive in a range of mildly thermophilic and mesophilic environments. We favor the first explanation, since mesotoga rRNA genes have repeatedly been isolated from similar environments all over the world (Table 1).
Mesotoga habitats.
The habitats in which mesotoga rRNA genes have been observed are listed in Table 1. Where reported, all of these environments are anaerobic or microaerophilic, consistent with all previously described members of the order Thermotogales being anaerobes. Moreover, these sequences are frequently recovered from communities involved in the remediation of environmental contamination of toxic chemicals, in particular, organohalide pollutants (Table 1). None of the published studies suggested that the Thermotogales bacterium was directly involved in the bioremediation. However, the consistent finding of mesotoga in such communities does suggest that a role in such processes is possible. The fosmid clones sequenced here contain two genes that might be involved in such processes. Both MES0001 and MES0039 encode proteins similar to predicted dienelactone hydrolases. These enzymes use substrate-assisted catalysis to degrade aromatic compounds (14) and have been shown to be involved in the degradation of several chloroaromatic compounds (7). These genes have no match in T. maritima MSB8 or Thermosipho sp. TCF52B, suggesting that they may have been acquired by LGT. Unfortunately, we were not able to construct reliable alignments of these ORFs and their homologs in GenBank.
It is interesting that mesotoga's closest relative, TBF19.5, was isolated from the production waters from an oil well. Similarly, the second clade of sequences obtained from mesophilic environments is of sisters to another Thermotogales group, Petrotoga, which has so far been found only in oil wells (17, 24, 31). Thus, it is possible that both mesophilic Thermotogales lineages started out as inhabitants of oil reservoirs and have adapted to living at lower temperatures, perhaps as the temperature in the oil well lowered or by following petroleum leaking out of the reservoir. As these environments contain chemicals similar to those found in the environments where mesotoga rRNA genes have been detected (e.g., various aromatic hydrocarbons) (16, 33), this might have prepared mesotoga to live in environments with anthropogenic pollution.
Conclusion.
We used a metagenomic approach to get a first glimpse of the genome of a Thermotogales bacterium lineage living at mesophilic temperatures. Our analyses show that mesotoga may have acquired several genes from mesophilic bacteria living in the same types of environments, while many of its original proteins have secondarily adapted to function at lower temperatures. It is not uncommon to obtain, from environmental DNA samples, 16S rRNA sequences of taxa that are not expected to occur there. For instance, Marchant et al. (26) and Rahman et al. (34) recently described the isolation of highly thermophilic bacteria from mesophilic soil environments and they show that these bacteria are present in significant numbers in the soils studied. Whether such sequences are “contaminants” or identify organisms that are actually growing at the sampled site will always be a question, especially if isolation and cultivation are difficult. We have shown that metagenomics and comparative bioinformatics can provide an answer. It is particularly compelling that of the 31 ORFs that have matches to the hyperthermophile Thermotoga maritima, 29 appear (through CvP analysis) to be adapted to function at lower temperatures. We are currently attempting to obtain a pure mesotoga culture by inoculating standard Thermotogales media with the enrichments we used to construct the fosmid library and isolating single colonies. Such an isolate will help us further determine how this lineage has adapted to living at lower temperatures and should also be useful in uncovering thermophilic and hyperthermophilic determinants in other members of the order Thermotogales.
Supplementary Material
Acknowledgments
This work was supported by funds from the Canadian Institutes for Health Research (MOP 4467) and Genome Canada (Genome Atlantic). O.Z. is supported by a CIHR postdoctoral fellowship.
We thank Joy Watts (Center of Marine Biotechnology, University of Maryland Biotechnology Institute, presently at the Department of Biological Sciences, Towson University) for providing enrichment cultures. We also thank E. Bapteste (Department of Biochemistry and Molecular Biology, Dalhousie University) for help with PhyloGenie.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannone, J. J., S. Subramanian, M. N. Schnare, J. R. Collett, L. M. D'Souza, Y. Du, B. Feng, N. Lin, L. V. Madabusi, K. M. Muller, N. Pande, Z. Shang, N. Yu, and R. R. Gutell. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 5.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dollhopf, S. L., S. A. Hashsham, and J. M. Tiedje. 2001. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb. Ecol. 42:495-505. [DOI] [PubMed] [Google Scholar]
- 7.Ellis, L. B., D. Roe, and L. P. Wackett. 2006. The University of Minnesota Biocatalysis/Biodegradation Database: the first decade. Nucleic Acids Res. 34:D517-D521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 2001. PHYLIP Phylogeny Inference Package, version 3.6. Department of Genetics, University of Washington, Seattle, Wash.
- 11.Frickey, T., and A. N. Lupas. 2004. PhyloGenie: automated phylome generation and analysis. Nucleic Acids Res. 32:5231-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galtier, N., and J. R. Lobry. 1997. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 44:632-636. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 14.Holmquist, M. 2000. Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr. Protein Pept. Sci. 1:209-235. [DOI] [PubMed] [Google Scholar]
- 15.Holoman, T. R., M. A. Elberson, L. A. Cutter, H. D. May, and K. R. Sowers. 1998. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl. Environ. Microbiol. 64:3359-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, H., and S. Larter. 2005. Biodegradation of petroleum in subsurface geological reservoirs, p. 91-121. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, D.C.
- 17.Huber, R., and M. Hannig. 2005. Thermotogales. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., vol. 3.14. Springer-Verlag, New York, N.Y.
- 18.Huber, R., T. Langworthy, H. Konig, M. Thomm, C. Woese, U. Sleytr, and K. Stetter. 1986. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 C. Arch. Microbiol. 144:324-333. [Google Scholar]
- 19.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, M. R., C. I. Montero, S. B. Conners, K. R. Shockley, S. L. Bridger, and R. M. Kelly. 2005. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 55:664-674. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 22.Konings, D. A., and R. R. Gutell. 1995. A comparison of thermodynamic foldings with comparatively derived structures of 16S and 16S-like rRNAs. RNA 1:559-574. [PMC free article] [PubMed] [Google Scholar]
- 23.Korber, B. 2001. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), Computational and evolutionary analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 24.Lien, T., M. Madsen, F. A. Rainey, and N. K. Birkeland. 1998. Petrotoga mobilis sp. nov., from a North Sea oil-production well. Int. J. Syst. Bacteriol. 48:1007-1013. [DOI] [PubMed] [Google Scholar]
- 25.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchant, R., I. M. Banat, T. J. Rahman, and M. Berzano. 2002. The frequency and characteristics of highly thermophilic bacteria in cool soil environments. Environ. Microbiol. 4:595-602. [DOI] [PubMed] [Google Scholar]
- 27.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 28.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, C. M. Fraser, et al. 1999. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 29.Nesbø, C. L., M. Dlutek, and W. F. Doolittle. 2006. Recombination in Thermotoga: implications for species concepts and biogeography. Genetics 172:759-769. [DOI] [PMC free article] [PubMed]
- 30.Nesbø, C. L., and W. F. Doolittle. 2003. Active self-splicing group I introns in the 23S rRNA genes of hyperthermophilic bacteria, derived from introns in eukaryotic organelles. Proc. Natl. Acad. Sci. USA 100:10806-10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ollivier, B., and J. Cayol. 2005. Fermentive, iron-reducing, and nitrogen-reducing microorganisms, p. 71-88. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, D.C.
- 32.Omelchenko, M. V., Y. I. Wolf, E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, M. J. Daly, E. V. Koonin, and K. S. Makarova. 2005. Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planckaert, M. 2000. Microbiology of oil reservoirs. Antonie Leeuwenhoek 77:3-20. [Google Scholar]
- 34.Rahman, T. J., R. Marchant, and I. M. Banat. 2004. Distribution and molecular investigation of highly thermophilic bacteria associated with cool soil environments. Biochem. Soc. Trans. 32:209-213. [DOI] [PubMed] [Google Scholar]
- 35.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 36.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 38.Suhre, K., and J. M. Claverie. 2003. Genomic correlates of hyperthermostability, an update. J. Biol. Chem. 278:17198-17202. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 2001. PAUP*. Phylogenetic Analysis Using Parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 40.Veerassamy, S., A. Smith, and E. R. Tillier. 2003. A transition probability model for amino acid substitutions from blocks. J. Comput. Biol. 10:997-1010. [DOI] [PubMed] [Google Scholar]
- 41.Watts, J. E., Q. Wu, S. B. Schreier, H. D. May, and K. R. Sowers. 2001. Comparative analysis of polychlorinated biphenyl-dechlorinating communities in enrichment cultures using three different molecular screening techniques. Environ. Microbiol. 3:710-719. [DOI] [PubMed] [Google Scholar]
- 42.Whelan, S., and N. Goldman. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691-699. [DOI] [PubMed] [Google Scholar]
- 43.Wu, Q., K. R. Sowers, and H. D. May. 2000. Establishment of a polychlorinated biphenyl-dechlorinating microbial consortium, specific for doubly flanked chlorines, in a defined, sediment-free medium. Appl. Environ. Microbiol. 66:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan, T., T. M. Lapara, and P. J. Novak. 2006. The reductive dechlorination of 2,3,4,5-tetrachlorobiphenyl in three different sediment cultures: evidence for the involvement of phylogenetically similar Dehalococcoides-like bacterial populations. FEMS Microbiol. Ecol. 55:248-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida, N., N. Takahashi, and A. Hiraishi. 2005. Phylogenetic characterization of a polychlorinated-dioxin-dechlorinating microbial community by use of microcosm studies. Appl. Environ. Microbiol. 71:4325-4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.