Abstract
Laccases are copper-containing enzymes which oxidize phenolic substrates and transfer the electrons to oxygen. Many filamentous fungi contain several laccase-encoding genes, but their biological roles are mostly not well understood. The main interest in laccases in biotechnology is their potential to be used to detoxify phenolic substances. We report here on a novel application of laccases as a reporter system in fungi. We purified a laccase enzyme from the ligno-cellulolytic ascomycete Stachybotrys chartarum. It oxidized the artificial substrate 2,2′-azino-di-(3-ethylbenzthiazolinsulfonate) (ABTS). The corresponding gene was isolated and expressed in Aspergillus nidulans, Aspergillus niger, and Trichoderma reesei. Heterologously expressed laccase activity was monitored in colorimetric enzyme assays and on agar plates with ABTS as a substrate. The use of laccase as a reporter was shown in a genetic screen for the isolation of improved T. reesei cellulase production strains. In addition to the laccase from S. charatarum, we tested the application of three laccases from A. nidulans (LccB, LccC, and LccD) as reporters. Whereas LccC oxidized ABTS (Km = 0.3 mM), LccD did not react with ABTS but with DMA/ADBP (3,5-dimethylaniline/4-amino-2,6-dibromophenol). LccB reacted with DMA/ADBP and showed weak activity with ABTS. The different catalytic properties of LccC and LccD allow simultaneous use of these two laccases as reporters in one fungal strain.
Filamentous fungi play important roles in biotechnology as producers of low-molecular-weight compounds and enzymes. Strain improvement and antifungal compound screenings require different versatile assay systems. Another demand for those genetic tools comes from the continuously increasing number of genomic sequences of filamentous fungi (http://www.broad.mit.edu/annotation/) and the big challenge to assign gene functions. Important information can be gained by measuring promoter activity, subcellular localization of proteins, or protein-protein interactions. Whereas the last two issues can be nicely addressed by the use of green fluorescent protein, other variants of fluorescent proteins, or the two-hybrid system in Saccharomyces cerevisiae, the determination of promoter activities is often hindered (25, 31). Beta-galactosidase (lacZ) or glucuronidase (uidA) work in fungi but generally produce significant background because many fungi contain endogenous enzymes (4). Another reporter system used frequently in fungi is glucose oxidase (12, 16). This assay is based on a coupled enzyme assay, with peroxidase as a helper enzyme. If glucose oxdiase is secreted, the reporter system can be used on agar plates but requires that the colonies be incubated with glucose, a chromogenic substrate, and the peroxidase enzyme.
Therefore, we have established laccases as alternative reporter systems. Laccases are copper-containing enzymes whose biological roles are largely unknown (17, 23). Most fungi contain several endogenous genes (at least 8 in Coprinopsis cinerea), but their expression is generally tightly controlled (9, 17). In basidiomycetes, a role of laccases in lignin degradation was discussed previously (5). Furthermore, specific laccases appear to play a role in fruiting body development. In ascomycetes, some laccases are induced upon exposure to phenolic substances (9). In Aspergillus nidulans, YA has a well-characterized role during asexual development (2). If yA is mutated, conidiospores are not green pigmented, but pigment biosynthesis is blocked at a yellow precursor. A second laccase was identified during the sexual cycle, and a third one, TilA, was detected in vegetative cells at the hyphal tip (14, 27). However, the expression of tilA was extremely low. Although laccases are widely distributed enzymes, their expression is usually very low in vegetative hyphae and thus should not interfere much with heterologously expressed laccases. Here, we show the use of laccases as reporter systems in A. nidulans, Aspergillus niger, and Trichoderma reesei.
(Part of this work was patented [1, 20a].)
MATERIALS AND METHODS
Strains and culture conditions.
Standard laboratory Escherichia coli strains (XL1-Blue, DH5a, Top 10 F′) were used. Supplemented minimal (MM) and complete media (CM) for A. nidulans and standard strain construction procedures were used, as described previously (15). Stachybotrys chartarum was isolated from soil samples on an agar nutrient medium and selected for its ability to produce oxidase. The new strain was deposited under the provisions of the Treaty of Budapest as MUCL38898. S. chartarum was revived on potato dextrose agar plates (Difco) at 24°C for about 5 days. The laccase production medium for S. chartarum contains (in grams/liter) the following components: glucose, 15; lecithin, 1.5; t-aconitic acid, 1.73; KH2PO4, 3; MgSO4 · 7H2O, 0.8; CaCl2 · 2H2O, 0.1; ammonium tartrate, 1.2; soy peptone, 5; corn syrup (7359; A. E. Staley, Decatur, IL), 10; benzyl alcohol, 1; Tween 20, 1; nitrilotriacetic acid, 0.15; MnSO4 · 7H2O, 0.05; NaCl, 0.1; FeSO4 · 7H2O, 0.01; CoSO4, 0.01; CaCl2 · 2H2O, 0.01; ZnSO4 · 7H2O, 0.01; CuSO4, 0.001; Al2(SO4)3 · 12H2O, 0.001; H3BO3, 0.001; NaMoO4 · 2H2O, 0.001. A. niger strain 14-5 was generated by transforming the dgr246 p2 strain (33) with the laccase expression vector pGAPT-gDO104 (1). The overproduction strains were screened in CMA plates (dextrose, 20 g/liter; Difco Malt extract, 20 g/liter; Bacto peptone, 1 g/liter; Bacto agar, 20 g/liter). T. reesei RL-P37 was obtained from the American Type Culture Collection (Maryland) and grown on potato dextrose agar plates at 28°C for 5 days. T. reesei RL-P37L was created by transforming RL-P37 with a laccase from S. chartarum MUCL38898.
Molecular techniques and vectors.
Standard DNA transformation procedures were used for A. nidulans (3, 36), A. niger (10), and E. coli (26). Agrobacterium-mediated transformation was used for T. reesei (6). For PCR experiments, standard protocols were applied using a capillary Rapid Cycler (Idaho Technology, Idaho Falls, ID) for the reaction cycles. Genomic DNA was isolated from S. chartarum using the phenol-chloroform extraction method (18) and from A. nidulans with the DNeasy plant mini kit (QIAGEN, Hilden, Germany). The Universal GenomeWalker kit (Clontech) was used to isolate the 5′ and 3′ genomic DNA.
Purification of laccase from S. chartarum and gene sequence determination.
S. chartarum mycelium was removed from the culture broth, and the supernatant was concentrated by ultrafiltration using an Amicon ultrafiltration unit with a 10-kDa molecular mass cutoff. Five volumes of cold acetone were added to the concentrate, incubated for 2 h at 4 to 10°C, and then centrifuged for 30 min at 10,000 × g, and the pellet was resuspended in water. Crystalline ammonium sulfate was added to the suspension to 40% saturation, and the mixture was incubated at 4°C for 16 h with gentle mixing. The suspension was centrifuged for 30 min at 10,000 × g, and the pellet was then resuspended in 15 ml of water and concentrated by ultrafiltration using a Centriprep 3000 (Amicon). The enzyme was further purified by gel permeation chromatography using a column containing 850 ml of Sephacryl S400 high-resolution gel (Pharmacia), equilibrated with 50 mM KH2PO4/K2HPO4 (pH 7.0). The concentrated enzyme sample was loaded onto the column and eluted with 50 mM KH2PO4/K2HPO4 (pH 7.0) buffer at a flow rate of 1 ml/minute. The fractions containing the highest phenol-oxidizing enzyme activities were pooled. S. chartarum phenol-oxidizing enzyme was then isolated after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The isolated protein was treated with urea and iodoacetamide and digested with endoLysC. The fragments resulting from digestion were separated on a reverse-phase monobore C18 column, collected in a multititer plate, and sequenced via Edman degradation (peptide no. 1, DYYFPNYQSARLLXYHDHA; peptide no. 2, RGQVMPYESAGLK).
Cloning of the S. chartarum laccase gene.
Based on the peptide sequence, two degenerate primers were designed: 5′-TATTACTTTCCNAAYTAYCA-3′ (N represents a mixture of all four nucleotides [A, T, C, and G] and Y represents a mixture of T and C only) and 5′-TCGTATGGCATNACCTGNCC-3′. The gene corresponding to the protein coding region (1 kb) was isolated from genomic DNA by PCR. PCR fragments containing the 5′ gene and 3′ regions of the gene were then isolated and sequenced (NCBI accession number DQ005533). The two corresponding peptide sequences were identified in the derived protein sequence, suggesting that the corresponding gene was isolated.
Expression of S. chartarum laccase (pCF1) in A. nidulans.
Digestion of MP077 (contains the full laccase gene) with BamHI and XbaI yielded a 1.9-kb fragment carrying the S. chartarum laccase gene minus its terminator sequence. This fragment was cloned into pAL3 (34), which was linearized using the same enzymes downstream of the alcA promoter.
Expression of the S. chartarum laccase in A. niger (pGAPT-gD104).
The genomic DNA fragment containing the Stachybotrys laccase coding region flanked by two newly introduced restriction enzyme sites (BglII and XbaI) was isolated by PCR. The PCR fragment was cloned into the plasmid vector pCR-II for nucleic acid sequence verification. This DNA fragment was then cloned into the BglII-to-XbaI site of the vector (pGAPT) (1) to generate pGAPT-gD104. The expression plasmid contains the A. niger glucoamylase gene promoter, Stachybotrys laccase, A. niger glucoamylase gene terminator, and the A. nidulans pyrG gene in PUC18.
Mutagenesis.
For A. niger, spores (3 × 106/ml) were mutated with a UV Stratalinker 1800 (Stratagene) for 30 min in small glass petri dishes (99% of spores killed). To generate genetic diversity for Trichoderma, RL-P37L spores were mutated using nitrosoguanidine (NTG) until a kill of 50% was obtained.
Expression of S. chartarum laccase in T. reesei.
The T. reesei wild type (1A52) and the transformant VHtr6 were grown on glucose- or lactose-containing medium with 2 mM 2,2′-azino-di-(3-ethylbenzthiazolinsulfonate) (ABTS).
Expression of the S. chartarum laccase in T. reesei (RL-P37) and screening for cellulase overproduction.
An expression cassette containing the Stachybotrys laccase expressed from the T. reesei cbh1 promoter and cbh1 terminator as the NotI fragment was cloned into the SmaI site of pPZP100 (13). T. reesei RL-P37 was transformed with this vector (RL-P37L) using methods described by de Groot et al. (6). For the primary screen, mutants derepressed in the presence of glucose were isolated using a screen incorporating laccase as a reporter system. Expression of laccase in T. reesei mutants was detected by a green color generated by the oxidation of the laccase substrate ABTS incorporated into Vogels agar plate medium containing 20% glucose (30). In a secondary screen, acid-swollen cellulose (ASC) was used as a substrate. ASC was made by preparing a slurry of 25 g of avicel (FMC Corp., Pennsylvania) in 100 ml of water. About 20 ml of 85% phosphoric acid was added. To this, 1 kg of 85% phosphoric acid was added, and the solution was mixed. Next, the mixture was slowly poured into 5 liters of ice water, resulting in precipitation of the ASC. The ASC was collected by filtering through miracloth (Calbiochem-Nova, California) and washed with 3 liters of water. Finally, the ASC was mixed in a blender until smooth. The secondary screening medium was Vogels containing 5% ASC. LC shake flask medium was used and was described by Ilmen et al. (19), except that 100 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; Calbiochem) was included to maintain the pH at 5.5. Total protein assays were done using a BCA total protein assay kit (Pierce).
Expression in A. nidulans (LccB, C, D).
The following primers were used to amplify the three laccase genes lccB, lccC, and lccD from genomic A. nidulans DNA: lccB fwd, 5′-CACCATGGCGCAACTTTGGGGATGG-3′, plus lccB rev, 5′-GAGTGGCTTAACAGAGTTGAGGACCACG-3′; lccC fwd, 5′-CACCATGCTGCGTTCTTCCTTTCTTCTC-3′, in combination with lccC rev, 5′-GGAGAGCCTACGAGTGTTCCTACTGG-3′; lccD fwd, 5′-CACCATGAAAGGTTTACCACACGGATTGAGC-3′, plus lccD rev, 5′-TTGTTTTTCGAGACATACTTCTTGGATGAACTC-3′. Genes were amplified, cloned into pENTR, and recombined into pMTOve as described previously (31). Plasmids were introduced into the yA2 (laccase) mutant RMS011.
Laccase assays.
The laccase activity of A. nidulans was induced on MM supplemented with different amounts of threonine. Activity was repressed with 1% glucose. Screening of transformants for inducible laccase activity was carried out on solid MM with a pH of 5 (instead of pH 6.5 in standard medium) supplemented with 5 mM ABTS. To quantify laccase activity, total protein was extracted from 100 ml of 48-h cultures by grinding the mycelium with liquid nitrogen and resuspending the cellular protein in 0.1 M sodium acetate (pH 5). Duplicate 100- or 200-μl samples of protein extract were combined with 1 ml of the extraction buffer and supplemented with 200 μl of 4.5 mM ABTS at time zero. Assay mixtures were incubated at 30°C for a few minutes until a moderate green color was obtained. The optical density at 420 nm of the samples was recorded. Activity was calculated as the change in optical density at 420 nm per minute per mg of total protein. The protein content of extracts was determined using the Bradford method. LccD and LccB were measured with the substrate combination DMA/ADBP as described previously (29, 35).
RESULTS AND DISCUSSION
Isolation and expression of Stachybotrys chartarum laccase in A. nidulans, A. niger, and T. reesei.
S. chartarum is an ascomycetous fungus which grows on plant debris and in soil and is often found in water-damaged homes. It has a complex lignocellulose-degrading enzyme system, and this is why it was expected to have a strong laccase activity (20). In this study, we isolated a laccase from S. chartarum culture broth (see Materials and Methods for details) (Table 1; Fig. 1), and two internal peptide sequences were determined to be DYYFPNYQSARLLXYHDHA and RGQVMPYESAGLK. The purified enzyme oxidized the artificial substrate ABTS. Starting from the peptide sequence, the full gene was isolated and characterized. The derived amino acid sequence shared 57.5% identity with a bilirubin oxidase (data not shown) and had low similarity to other fungal laccases (see below). In agreement with the presence of the protein in the culture broth, the protein harbors a potential secretion signal at the N terminus.
TABLE 1.
Purification of the S. chartarum laccasea
| Fraction | Vol (ml) | No. of units/ml | Total no. of units | Protein (mg/ml) | Total protein (mg) | Sp act (U/mg) |
|---|---|---|---|---|---|---|
| UFb | 500 | 18 | 9,015 | 13 | 6,495 | 1.4 |
| Acetone precipitation | 53 | 36 | 1,929 | 7 | 372 | 5.2 |
| Ammonium sulfate | ||||||
| 40% | 50 | 36 | 1,789 | 5 | 249 | 7.2 |
| 80% | 5 | 255 | 1,276 | 26 | 131 | 9.7 |
| Gel filtration (S200HR) | 400 | 2.5 | 1,016 | 0.16 | 63.2 | 16 |
Volumes have been corrected for the original starting material volume, and total units have been recalculated based on the starting volumes based on the specific activity of 22.6 U/mg.
UF, ultrafiltration concentrate.
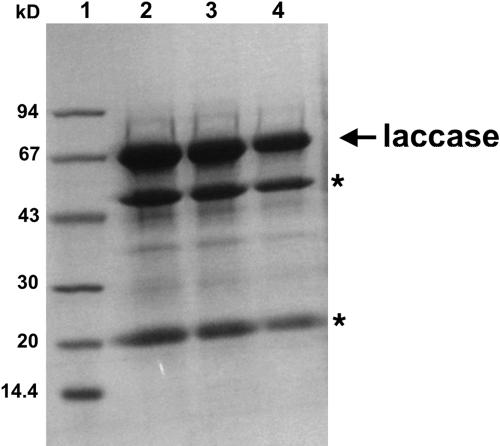
FIG. 1.
Sodium dodecyl sulfate gel of purified S. chartarum laccase. Lane 1 is the molecular mass marker. Lanes 2, 3, and 4 were loaded with 50 ng, 25 ng, and 10 ng of purified laccase, respectively. Proteolytic fragments of the laccase are indicated by an asterisk.
To test the possibility of heterologous expression of the S. chartarum laccase, the gene was cloned into different expression vectors (see Materials and Methods) and introduced into A. nidulans (G191), A. niger (dgr246p2), and T. reesei 1A52 (Fig. 2). In A. nidulans, the construct was under the control of the A. nidulans alcA promoter and thus was repressed in the presence of glucose, derepressed when the fungus grows in the presence of glycerol, and induced with ethanol or threonine (8). Strains which expressed the laccase gene were identified with their ABTS-oxidizing activity. The agar around the colonies was stained bright blue. Although glucose represses the alcA promoter, colonies showed a small blue halo. This can be explained because mature A. nidulans colonies are glucose limited, and thus, the alcA promoter is at least derepressed and, thus, the expression is leaky. Wild-type A. nidulans colonies did not turn blue, despite the fact that the laccase YA is highly expressed in conidiospores (1a, 2). Five pCF1 transformants (G1, G2, G5, G6, and G7) were selected for quantification of the ABTS-oxidizing activity, an example of which is shown in Fig. 2B. The values represent the averages of two measurements. The experiment was repeated one or two times with independent cultures, and essentially the same results were obtained. The pattern of induction follows that expected for the alcA promoter, based on data previously obtained using the beta-galactosidase reporter gene (21). For example, strain no. 6 showed 66-fold induction of laccase activity on inducing (threonine) compared to repressing (glucose) medium. Intermediate activities were obtained in the presence of 2.2 mM glucose and 100 mM glycerol. In similar experiments, the S. chartarum laccase was introduced into A. niger and T. reesei, where gene expression was controlled through the glucoamylase promoter glaA and the cbh1 promoter (1), respectively (Fig. 2C and D). The glaA promoter was induced with xylose, and the corresponding A. niger strain (14-5) converted ABTS to the blue product. With maltose as a carbon source, no laccase expression was observed. In comparison, the cbh1 promoter was induced with lactose and repressed with glucose. These results suggested that laccase may be used as a reporter system. In comparison to beta-galactosidase and glucose oxidase as a reporter, laccase has the advantage of being more suitable for screening on solid media (Fig. 3A). The use of beta-galactosidase assays in plates has proven to be problematic due to the acid conditions (pH 5 to 6) used to culture fungi. At this pH, the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) indicator does not give a strong color reaction. In contrast, both the laccase enzyme and the ABTS indicator function well at pH 5. In addition, many fungi contain endogenous beta-galactosidase activities, whereas the expression of laccases is linked to certain developmental stages (e.g., yA in A. nidulans) or they are induced in the presence of phenolic substrates (9). Another advantage of laccases as reporters is that many fungi contain those enzymes and, if heterologous expression of one of the laccases described here would not work, it would be possible to generate a reporter system using an endogenous gene instead.
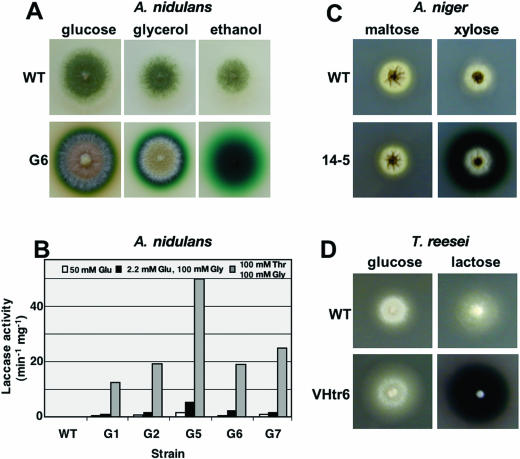
FIG. 2.
Expression of S. chartarum laccase in A. nidulans (A, B), A. niger (C), and T. reesei (D). (A) The A. nidulans wild type (WT) (FGSC4) and one positive transformant (G6) were grown on glucose-, glycerol-, and ethanol-containing media supplemented with 2 mM ABTS. (B) Quantification of laccase activity of five transformants with pCF1. Strains were grown on 50 mM glucose (Glu), 2.2 mM glucose plus 100 mM glycerol (Gly), or 100 mM threonine (Thr) plus 100 mM glycerol as indicated. Activity was determined in the supernatant of the culture extract and is given as μmol of ABTS per min and mg of dry mycelium. The average of two measurements is shown. (C) The A. niger wild type (dgr246p2) and transformant 14-5 were grown on maltose- and xylose-containing media with 2 mM ABTS. (D) The T. reesei wild type (1A52) and transformant VHtr6 were grown on glucose- and lactose-containing media with 2 mM ABTS.
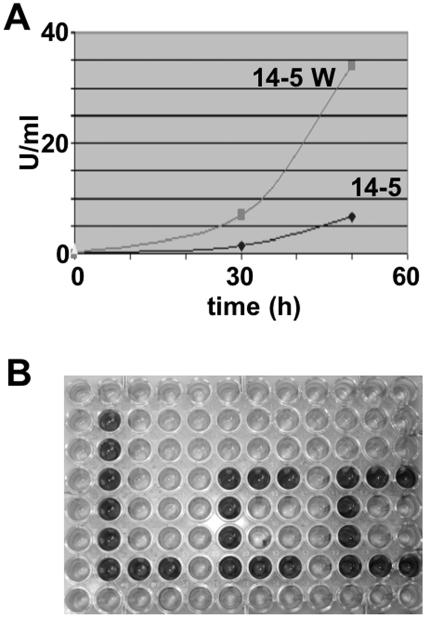
FIG. 3.
Application of laccase for strain improvement. (A) A. niger strain improvement for laccase production. Strain 14-5 was mutagenized, and strains were selected which produced bigger ABTS halos on agar plates. Strain 14-5W was selected, and laccase production was compared to that of the wild type in liquid culture. Activities are given in units per ml of culture supernatant. (B) LccC-expressing A. nidulans and wild-type A. nidulans were inoculated in liquid culture (threonine medium supplemented with 1 mM ABTS) in microtiter plates.
Laccase as a reporter for genetic screens.
Biotechnological applications of laccase require high amounts of enzyme. Therefore, S. chartarum laccase was expressed in A. niger under the control of the very strong glucoamylase promoter (see Materials and Methods). The expression plasmid pGAPT-gD104 was introduced into the dgr246 p2 (pyrG-negative) strain of A. niger (33) by standard polyethylene glycol methods (10). Forty transformants were isolated and grown on agar plates and then in shake flasks containing CSL medium (containing glucose, maltose, and fructose) (7). The transformants grown in CSL for 2 days were then transferred to CSM medium (containing only maltose) (10). ABTS activity assays were performed (data not shown). The best producer was transformant 14-5 (results not shown). In the next step we wanted to improve laccase expression of strain 14-5 further and generated a large strain collection by UV mutagenesis and NTG for ABTS plate screening (Materials and Methods). We isolated several strains with increased diameters of the green-blue halo. Laccase activity was subsequently tested in liquid medium. Strain 14-5W showed six- and fivefold increases in laccase activity at 30 and 50 h, respectively, compared to strain 14-5 (Fig. 3A). The activity was lower at 65 h (results not shown). These results show that laccase activity can be used to screen for strains with different laccase activity in a visual screen on agar plates. Similarly, visual screening of blue-green color in ABTS-containing liquid medium was possible in microtiter plates (Fig. 3B). This demonstrates the value of laccase for high-throughput screenings.
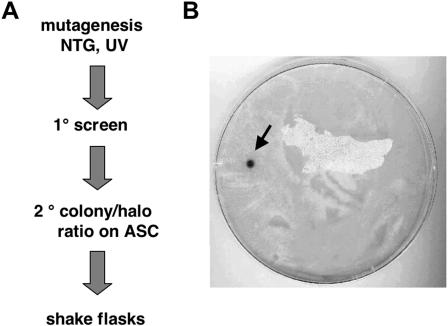
To further prove the potential of laccase as a reporter system for genetic screens, we wanted to improve cellulase production in T. reesei. To this end, we constructed a transgenic strain with a laccase expression plasmid where laccase is under the control of the T. reesei cellulase promoter (cbh1) and introduced it by Agrobacterium-mediated plasmid transfer into a uridine auxotrophic T. reesei strain (RL-P37). The strategy was to isolate mutants with increased laccase production due to mutagenesis of trans-acting factors which interact with the cbh1 promoter, e.g., factors which mediate carbon catabolite repression. These factors should also act on the endogenous cellulase promoters, and mutagenesis should increase the yield of cellulase as it was shown before (24). The corresponding T. reesei strain (RL-P37L) was mutagenized, and strains with increased laccase activity were isolated (see Materials and Methods). In a primary screen from about 1 million NTG-mutated T. reesei spores, strains were isolated that produced laccase under catabolite repression conditions (Fig. 4). About 150 mutants with strong laccase activity were subsequently screened for improved cellulase production on medium containing ASC. Mutant strains formed clearing zones in the ASC medium and the halo-to-colony size ratio was measured. Higher ratios were thought to be indicative of improved cellulase production. Twelve strains were selected for further study in shake flasks. Two mutants, LC5 and LC36, were found to produce 15 and 20% more total secreted protein than the control, respectively (results not shown). These two strains produced 25 and 30% higher cellulase activities than RL-P-37, as determined in a filter paper assay (24). These results show that laccases can be used for different applications and are powerful tools for strain improvement and strain characterization.
FIG. 4.
T. reesei strain improvement for cellulase production. (A) Scheme of mutagenesis and strain selection. (B) Agar plate with a lawn of mutated strains on ABTS-containing plates. The arrow points to a colony with derepressed laccase expression. For details, see the text.
A. nidulans LccB, LccC, and LccD show different substrate specificities.
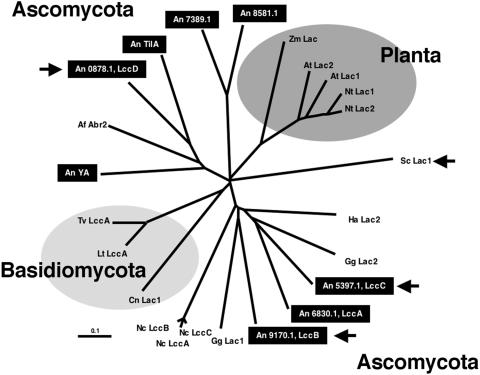
The S. chartarum laccase oxidized ABTS as a substrate. To test whether there exist laccases with different substrate spectra, useful as genetic reporters, we analyzed the genome of A. nidulans for laccase-encoding genes by searching the A. nidulans genome database (http://www.broad.mit.edu/annotation/fungi/aspergillus/) (11) with the sequence of A. nidulans YA (2) and TilA (27) as well as with Abr2 from Aspergillus fumigatus (32). We detected six new putative laccase genes, two of which were more closely related to ascorbate oxidases (Fig. 5). In this study, we worked with the remaining four laccases, lccA to lccD. For further analysis we used the automatically translated protein sequence from the Whitehead sequencing center (11). However, alignment of the putative proteins revealed that neglect of a predicted intron in lccA and addition of 2 bp restored the L1 region typical for laccases (22). Therefore, this modified LccA protein sequence was used in the alignment. Experimental determination of the exon-intron border was impossible because we were unable to detect any transcript in RNA derived from different developmental stages in Northern blots or amplify cDNA by reverse transcription-PCR and subsequent use of nested primers. These experiments demonstrated the extremely low (if any) expression of lccA to lccD under the conditions tested. All six A. nidulans laccase proteins are comprised of 485 (LccA) to 665 amino acids. Analysis of the proteins for signal peptides (http://www.cbs.dtu.dk/services/SignalP/) identified this motif for protein secretion in LccB to LccD, YA, and TilA but not in LccA. Therefore, all but LccA are likely to be extracellular proteins. The fact that the activities of LccB, LccC, and LccD were detected on agar plates without lysis of the hyphae demonstrates that these three laccases are indeed secreted (Fig. 6).
FIG. 5.
Phylogenetic tree of different plant and fungal laccases and ascorbate oxidases. The tree was generated with the ClustalW algorithm from the ClustalX 1.8 program with standard parameters. The gene names and accession numbers are indicated as follows: A. fumigatus, Af Abr2 (AF104823); A. nidulans, An TilA (AJ305224), An YA (X52552), An0878.1, An5397.1, An6830.1, An7389.1, An8581.1, An9170.1; Arabidopsis thaliana, At Lac1 (NP_565881), At Lac2 (NP_195739); Cryptococcus neoformans, Cn Lac1 (A36962); Gaeumannomyces graminis var. graminis, Gg Lac1 (AJ437319), Gg Lac2 (AJ437320); Hortaea acidophila, Ha Lac1 (AAY33971); Lentinus tigrinus, Lt Lac1 (AAX07469); Neurospora crassa, Nc LccA (AAA33591), Nc LccB (AAA33592), Nc LccC (AAA33590); Nicotiana tabacum, Nt Lac1 (AAC49538), Nt Lac2 (AAC49536); Trametes versicolor, Tv LacA (A35883); Zea mays, Zm Lac1 (AAX83113); S. chartarum, Sc Lac1 (AAY23005). The dendrogram was visualized by TreeView. The scale is about 10% calculated sequence divergence. The arrows indicate the four laccases studied in this paper.
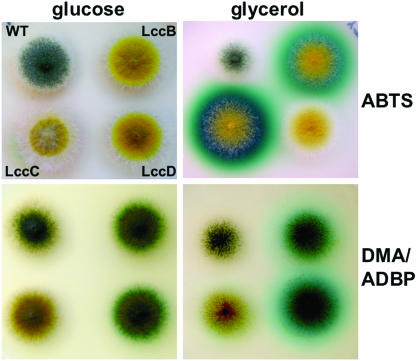
FIG. 6.
Substrate specificity of LccB, LccC, and LccD of A. nidulans. Strains were grown on glucose medium (upper panel) or glycerol medium (middle panel) supplemented with ABTS. The ABTS staining of the middle panel was documented, and subsequently, the colonies overlaid with ADBP/DMA (14, 28). The conversion of the substrates is displayed in the lower panel. ABTS staining of the middle colony is lost upon incubation with ADBP/DMA. WT, wild type.
To test the activity of the extracellular laccases, we expressed lccB, lccC, and lccD under the control of the alcA promoter in A. nidulans and used ABTS as a substrate in agar plates (Fig. 6). We used A. nidulans RMS011 as a recipient for the plasmids because this strain has a mutation in the conidiospore-specific laccase gene yA. We found that only LccC reacted strongly with ABTS (Km = 0.3 mM), whereas LccD did not convert ABTS. LccB showed only weak activity. LccB and LccD converted the substrate combination DMA/ADBP (28). Hence, LccC and LccD can be used simultaneously for the analyses of two promoters, similar to the use of beta-galactosidase and beta-glucuronidase (4). Laccases thus appear to be a versatile reporter system in fungi.
Acknowledgments
We thank H. Körner (University of Karlsruhe) for help with the enzyme assays.
The work was funded by the German Science foundation (DFG), the Fonds of the Chemical Industry, and the Max-Planck-Institute for Terrestrial Microbiology, and the special program “Lebensmittel und Gesundheit” from the ministry of Baden-Württemberg. C.F. was supported by a BBSRC CASE Studentship.
REFERENCES
- 1.Amory, A., et al. September1999. International patent WO9949020A2.
- 1a.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aramayo, R., and W. E. Timberlake. 1990. Sequence and molecular structure of the Aspergillus nidulans yA (laccase I) gene. Nucleic Acids Res. 18:3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balance, D. J., F. P. Buxton, and G. Turner. 1983. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene. Biochem. Biophys. Res. Commun. 112:284-289. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage, A. A., and J. Van den Brulle. 1995. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J. Bacteriol. 177:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crestini, C., L. Jurasek, and D. S. Argyropoulos. 2003. On the mechanism of the laccase-mediator system in the oxidation of lignin. Chemistry 9:5371-5378. [DOI] [PubMed] [Google Scholar]
- 6.de Groot, M. J., P. Bundock, P. J. Hooykaas, and A. G. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 7.Dunn-Coleman, N. S., P. Bloebaum, R. M. Berka, E. Bodie, N. Robinson, G. Armstrong, M. Ward, M. Przetak, G. L. Carter, R. LaCost, et al. 1991. Commercial levels of chymosin production by Aspergillus. Biotechnology 9:976-981. [DOI] [PubMed] [Google Scholar]
- 8.Felenbok, B., M. Flipphi, and I. Nikolaev. 2001. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 69:149-204. [DOI] [PubMed] [Google Scholar]
- 9.Fernández-Larrea, J., and U. Stahl. 1996. Isolation and characterization of a laccase gene from Podospora anserina. Mol. Genet. Genomics 252:539-551. [DOI] [PubMed] [Google Scholar]
- 10.Fowler, T., R. M. Berka, and M. Ward. 1990. Regulation of the glaA gene of Aspergillus niger. Curr. Genet. 18:537-545. [DOI] [PubMed] [Google Scholar]
- 11.Galagan, J. E., S. E. Calvo, C. Cuomo, L.-J. Ma, J. R. Wortman, S. Batzoglou, S.-I. Lee, M. Bastürkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. d'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Peñalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 12.Geisen, R. 1995. Expression of the Aspergillus niger glucose oxidase gene in Penicillium nalgiovense. World J. Microbiol. Biotechnol. 11:322-325. [DOI] [PubMed] [Google Scholar]
- 13.Hajdukiewiez, P., Z. Svab, and P. Maliga. 1994. The small, versatile pPZP family of Agrobacerium binary vectors for plant transformation. Plant Mol. Biol. 25:989-994. [DOI] [PubMed] [Google Scholar]
- 14.Hermann, T. E., M. B. Kurtz, and S. P. Champe. 1983. Laccase localized in Hulle cells and cleistothecial primordia of Aspergillus nidulans. J. Bacteriol. 154:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill, T. W., and E. Käfer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48:20-21. [Google Scholar]
- 16.Hodgkins, M., D. Mead, D. J. Ballance, A. Goodey, and P. Sudbery. 1993. Expression of the glucose oxidase gene from Aspergillus niger in Hansenula polymorpha and its use as a reporter gene to isolate regulatory mutations. Yeast 9:625-635. [DOI] [PubMed] [Google Scholar]
- 17.Hoegger, P. J., M. Navarro-Gonzalez, S. Kilaru, M. Hoffmann, E. D. Westbrook, and U. Kues. 2004. The laccase gene family in Coprinopsis cinerea (Coprinus cinereus). Curr. Genet. 45:9-18. [DOI] [PubMed] [Google Scholar]
- 18.Hynes, M. J., C. M. Corrick, and J. A. King. 1983. Isolation of genomic clones containing the amdS gene of Aspergillus nidulans and their use in the analysis of structural and regulatory mutations. Mol. Cell. Biol. 3:1430-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilmen, M., A. Saloheimo, M. L. Onnela, and M. E. Penttila. 1997. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 63:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis, B. B. 2002. Chemistry and toxicology of molds isolated from water-damaged buildings. Adv. Exp. Med. Biol. 504:43-52. [DOI] [PubMed] [Google Scholar]
- 20a.Karos, M., et al. April2006. International patent WO2006/034811A2.
- 21.Kennedy, J., and G. Turner. 1996. d-(L-a-Aminoadipyl)-L-cysteinyl-D-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol. Genet. Genomics 253:189-197. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, S., P. Phale, S. Durani, and P. Wangikar. 2003. Combined sequence and structure analysis of the fungal laccase family. Biotechnol. Bioeng. 83:386-394. [DOI] [PubMed] [Google Scholar]
- 23.Mayer, A. M., and R. C. Staples. 2002. Laccase: new functions for an old enzyme. Phytochemistry 60:551-565. [DOI] [PubMed] [Google Scholar]
- 24.Montenecourt, B. S., and D. E. Eveleigh. 1977. Preparation of mutants of Trichoderma reesei with enhanced cellulase production. Appl. Environ. Microbiol. 34:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osmani, A. H., J. Davies, C. E. Oakley, B. R. Oakley, and S. A. Osmani. 2003. TINA interacts with the NIMA kinase in Aspergillus nidulans and negatively regulates astral microtubules during metaphase arrest. Mol. Biol. Cell 14:3169-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russel. 1999. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Scherer, M., and R. Fischer. 2001. Molecular characterization of a blue-copper laccase, TILA, of Aspergillus nidulans. FEMS Microbiol. Lett. 199:207-213. [DOI] [PubMed] [Google Scholar]
- 28.Scherer, M., and R. Fischer. 1998. Purification and characterization of laccase II of Aspergillus nidulans. Arch. Microbiol. 170:78-84. [DOI] [PubMed] [Google Scholar]
- 29.Scherer, M., H. Wei, R. Liese, and R. Fischer. 2002. Aspergillus nidulans catalase-peroxidase gene (cpeA) is transcriptionally induced during sexual development through the transcription factor StuA. Eukaryot. Cell 1:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor, H., and C. W. Tabor. 1970. Metabolism of amino acids and amines, vol. XVII. Academic Press, New York, N.Y.
- 31.Toews, M. W., J. Warmbold, S. Konzack, P. E. Rischitor, D. Veith, K. Vienken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in Escherichia coli (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, H. F., M. H. Wheeler, Y. C. Chang, and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward, M., L. J. Wilson, and K. H. Kodama. 1993. Use of Aspergillus overproducing mutants, cured for integrated plasmid, to overproduce heterologous proteins. Appl. Microbiol. Biotechnol. 39:738-743. [DOI] [PubMed] [Google Scholar]
- 34.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 35.Wolfenden, B. S., and R. L. Willson. 1982. Radical-cations as reference chromogens in kinetic studies of one-electron transfer reactions. J. Chem. Soc. Perkin Trans. II:805-812. [Google Scholar]
- 36.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]