Abstract
The diversity of microcystin-producing cyanobacteria in the western basin of Lake Erie was studied using sequence analysis of mcyA gene fragments. Distinct populations of potentially toxic Microcystis and Planktothrix were found in spatially isolated locations. This study highlights previously undocumented diversity of potentially toxic cyanobacteria.
The Laurentian Great Lakes contain ∼18% of Earth's potable water supply, making them a vital global resource. Lake Erie is the most productive of these lakes, and it maintains a position of regional socioeconomic importance (3, 9). Although the annual appearance of Microcystis in the western basin of Lake Erie since 1995 is well documented (2, 11, 14, 15), the presence of other microcystin producers has garnered less attention. Microcystis has been thought to be the primary toxin producer in Lake Erie, but observations from 2003 and 2004 suggested that the microcystin-producing community may not be composed entirely of Microcystis, as only 0.03 to 4.7% of these cells contained toxin biosynthetic machinery (11). Thus, our goal was to examine the diversity of microcystin producers in locations where high microcystin-LR concentrations were observed in 2003 and 2004. The identities and phylogenetic relationships within the microcystin-producing cyanobacterial community were assessed by the analysis of mcyA genes carried by microcystin-producing Anabaena, Planktothrix, and Microcystis. The mcyA gene encodes a microcystin synthetase found in these microcystin producers (5).
Water was collected at the 1-m depth from the western basin of Lake Erie in August 2003 and August 2004 (Fig. 1). Chlorophyll a and toxin (microcystin-LR) equivalents have been previously described (11). Cells for DNA extraction were collected from known volumes onto 47-mm-diameter, 0.22-μm-nominal-pore-size polycarbonate filters (Millipore) and DNA extracted as described previously (11). The primer pair mcyA-Cd 1R and mcyA-Cd 1F (5) was used to amplify 291- to 297-bp mcyA gene fragments. Reactions were carried out in EasyStart tubes (Molecular BioProducts) in a final volume of 50 μl containing 0.4 μM of each primer, 300 ng μl−1 bovine serum albumin (Sigma A-7030) (7), 2.5 U Taq polymerase (Promega), 0.1% Triton X-100 (Molecular BioProducts), 2 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mix, 1× PCR buffer, and 5 μl of undiluted or 10-fold-diluted DNA extract. The PCR was as follows: 95°C for 10 min; 40 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. Amplicons were separated by agarose gel electrophoresis (12), and DNA from bands of the correct size was extracted using a QIAquick gel extraction kit. Clone libraries were generated using a TOPO-TA cloning kit (Invitrogen). Thirty-eight clones from each sample were selected and the inserts sequenced at the Clemson University Genomics Institute by using the M13 forward primer (Invitrogen).
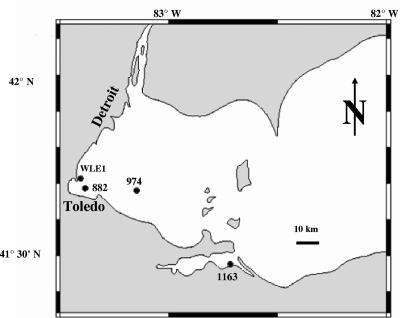
FIG. 1.
Map of station locations and corresponding station codes in the western basin of Lake Erie.
To provide information on a known isolate from Lake Erie, Microcystis aeruginosa LE-3 was grown in modified BG11 medium (6) at +25°C under continuous illumination (ca. 80 μmol of photons m−2 s−1). The cells were grown in a batch culture for 3 weeks and harvested by centrifugation. DNA was extracted using the previously described xanthogenate method (10). mcyA gene fragments were PCR amplified and cloned as described above. Sequencing was completed at the University of Tennessee Molecular Biology Resource Facility.
Sequences from natural samples and cultures were manually examined and then analyzed at both the nucleotide and amino acid levels. Individual DNA sequences from natural samples were queried against GenBank by BLAST and BLASTX searches (1). Phylogenetic relationships among unique McyA sequences were inferred through a neighbor-joining analysis using Poisson correction distance. Dendrograms were also created by using the unweighted-pair group method using average linkages and minimum-evolution approaches, which yielded comparable phylogenies (not shown). The sequences were edited, aligned, and analyzed using Bio Edit (4), Clustal W (13), and Mega 3.0 (8).
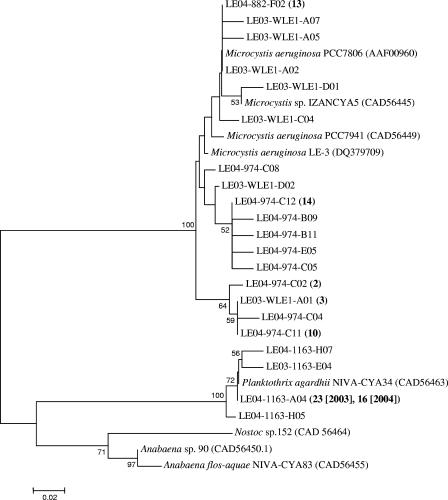
Overall, the results from BLAST and BLASTX searches confirmed that the cloned sequences originated from the mcyA gene of the microcystin synthetase complex. Relative to known sequences, the natural samples clustered into two distinct groups. Phylogenetic analyses confirmed that 100% of the sequences from station 1163 clustered with Planktothrix aghardii, whereas 100% of the sequences from stations WLE1, 882, and 974 clustered with Microcystis isolates (Fig. 2), although sequences within the Microcystis clade were remarkably diverse. All Microcystis McyA sequences from GenBank as well as most sequences from natural samples were 76 amino acids in length, whereas a subset of Microcystis-like sequences (20 sequences from station 974 and 1 sequence from station WLE1) appeared to have two extra amino acids. Sequence alignment indicated that these two additional amino acids occurred at the same positions where similar residues are located in Planktothrix, Anabaena, and Nostoc (Fig. 3). Due to the systematic location of the extra amino acids, we consider it unlikely that these residues in the Microcystis-like sequences are PCR artifacts. Despite the extra amino acids, the Lake Erie sequences were most similar to Microcystis (93 to 97% identity at the nucleotide level and 93 to 96% identity at the amino acid level), although no exact matches were found.
FIG. 2.
Neighbor-joining tree displaying the relationship between partial McyA sequences (76 to 78 amino acids). The coding system for the clones is as follows: LE03-1163, Lake Erie, year 2003, station 1163; LE03-WLEI, Lake Erie, year 2003, station WLEI; LE04-1163, Lake Erie, year 2004, station 1163; LE04-974, Lake Erie, year 2004, station 974; LE04-882, Lake Erie, year 2004, station 882. The numbers in parentheses after the codes for the clones indicate the number of identical sequences represented by these sequences. Accession numbers are in parentheses after the names for reference sequences obtained from GenBank. Bootstrap values of >50% (for 2,000 iterations) are displayed at the branch nodes. The scale bar represents substitutions per site.
FIG. 3.
Multiple alignment of McyA amino acid sequences of cultured isolates of Microcystis aeruginosa LE-3 (translated from the mcyA gene sequence under accession no. DQ379709), Microcystis aeruginosa PCC7806 (accession no. AAF00960), Planktothrix aghardii NIVA-CYA34 (CAD56463), Anabaena flos-aquae NIVA-CYA83 (CAD56455), and Nostoc sp. strain 152 (CAD56464) with novel Microcystis-like McyA sequences from natural samples that contain two extra amino acids, unlike Microcystis sequences from cultured isolates.
Microcystis, Planktothrix, and Anabaena are the most common microcystin-producing cyanobacteria, and their potentially toxic genotypes can be detected and identified by the unique sequences within the mcyA amplicons (5). In the context of this study, we have examined this conserved genetic element to determine which organism(s) within a population may be responsible for microcystin production in spatially separated regions of a lake. This analysis has identified a genetic element not associated with any currently known toxic Microcystis organism, suggesting the existence of previously unknown sequence diversity within the mcyA genes of this genus. The effect of the additional two residues on microcystin production is also unknown. Analysis of McyA sequences from natural samples indicates a spatial separation of these toxigenic populations within Lake Erie. Although the data presented here indicate no mixing between Planktothrix and Microcystis populations, this may not be entirely the case. It is possible that Planktothrix mcyA fragments were preferentially amplified from samples collected at station 1163 due to a high abundance of toxic P. aghardii relative to toxic Microcystis. Regardless, these data reveal a new diversity of microcystin-producing cyanobacteria in Lake Erie, although the conditions supporting and separating these populations are not yet understood. Overall, we couch the observations within the framework of developing a better understanding of the factors that regulate cyanobacterial blooms in Lake Erie and other environments.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported here are DQ379674 to DQ379709.
Acknowledgments
We thank the captain and crew of the CCGS Limnos as well as Bob Hess, Greg Boyer, George Bullerjahn, Gary LeCleir, Tom Bridgeman, and two anonymous reviewers for assistance and helpful discussions. We thank Wayne Carmichael for providing the M. aeruginosa LE-3 isolate.
This article includes research supported by funds from the National Oceanic and Atmospheric Administration Coastal Ocean Program (MERHAB-LGL, no. NA 160 P2788), an Oceans and Human Health external grant (NA05NOS4781251), the National Science Foundation (grant DEB-0129118), and Helsingin Sanomat Centennial Foundation.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brittain, S. M., J. Wang, L. Babcock-Jackson, W. W. Carmichael, K. L. Rinehart, and D. A. Culver. 2000. Isolation and characterization of microcystins, cyclic heptapeptide hepatotoxins from a Lake Erie strain of Microcystis aeruginosa. J. Great Lakes Res. 26:241-249. [Google Scholar]
- 3.Fuller, K., H. Sherar, and J. Wittig (ed.). 1995. The Great Lakes, an environmental atlas and resource book, 3rd ed. Government of Canada, Toronto, Ontario, Canada, and U.S. Environmental Protection Agency, Great Lakes National Program Office, Chicago, Ill.
- 4.Hall, T. A. 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 5.Hisbergues, M., G. Christiansen, L. Rouhiainen, K. Sivonen, and T. Borner. 2003. PCR-based identification of microcystin-producing genotypes of different cyanobacterial genera. Arch. Microbiol. 180:402-410. [DOI] [PubMed] [Google Scholar]
- 6.Kerry, A., D. E. Laudenbach, and C. G. Trick. 1988. Influence of iron limitation and nitrogen-source on growth and siderophore production by cyanobacteria. J. Phycol. 24:566-571. [Google Scholar]
- 7.Kirchman, D. L., L. Y. Yu, B. M. Fuchs, and R. Amann. 2001. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26:13-22. [Google Scholar]
- 8.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformat. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 9.Munawar, M., I. F. Munawar, H. Dermott, H. Niblock, and S. Carou. 2002. Is Lake Erie a resilient ecosystem? Aquat. Ecosyst. Health Management 5:79-93. [Google Scholar]
- 10.Ouellette, A. J. A., S. M. Handy, and S. W. Wilhelm. 2006. Toxic Microcystis is widespread in Lake Erie: PCR detection of toxin genes and molecular characterization of associated cyanobacterial communities. Microb. Ecol. 51:154-165. [DOI] [PubMed] [Google Scholar]
- 11.Rinta-Kanto, J. M., A. J. A. Ouellette, G. L. Boyer, M. R. Twiss, T. B. Bridgeman, and S. W. Wilhelm. 2005. Quantification of toxic Microcystis spp. during the 2003 and 2004 blooms in western Lake Erie using quantitative real-time PCR. Environ. Sci. Technol. 39:4198-4205. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanderploeg, H. A., J. R. Liebig, W. W. Carmichael, M. A. Agy, T. H. Johengen, G. L. Fahnenstiel, and T. F. Nalepa. 2001. Zebra mussel (Dreissena polymorpha) selective filtration promoted toxic Microcystis blooms in Saginaw Bay (Lake Huron) and Lake Erie. Can. J. Fish. Aquat. Sci. 58:1208-1221. [Google Scholar]
- 15.Vincent, R. K., X. M. Qin, R. M. L. McKay, J. Miner, K. Czajkowski, J. Savino, and T. Bridgeman. 2004. Phycocyanin detection from LANDSAT TM data for mapping cyanobacterial blooms in Lake Erie. Remote Sensing Environ. 89:381-392. [Google Scholar]