Abstract
A combination of uni- and multiplex PCR assays targeting 58 virulence genes (VGs) associated with Escherichia coli strains causing intestinal and extraintestinal disease in humans and other mammals was used to analyze the VG repertoire of 23 commensal E. coli isolates from healthy pigs and 52 clinical isolates associated with porcine neonatal diarrhea (ND) and postweaning diarrhea (PWD). The relationship between the presence and absence of VGs was interrogated using three statistical methods. According to the generalized linear model, 17 of 58 VGs were found to be significant (P < 0.05) in distinguishing between commensal and clinical isolates. Nine of the 17 genes represented by iha, hlyA, aidA, east1, aah, fimH, iroNE. coli, traT, and saa have not been previously identified as important VGs in clinical porcine isolates in Australia. The remaining eight VGs code for fimbriae (F4, F5, F18, and F41) and toxins (STa, STb, LT, and Stx2), normally associated with porcine enterotoxigenic E. coli. Agglomerative hierarchical algorithm analysis grouped E. coli strains into subclusters based primarily on their serogroup. Multivariate analyses of clonal relationships based on the 17 VGs were collapsed into two-dimensional space by principal coordinate analysis. PWD clones were distributed in two quadrants, separated from ND and commensal clones, which tended to cluster within one quadrant. Clonal subclusters within quadrants were highly correlated with serogroups. These methods of analysis provide different perspectives in our attempts to understand how commensal and clinical porcine enterotoxigenic E. coli strains have evolved and are engaged in the dynamic process of losing or acquiring VGs within the pig population.
The acquisition of virulence genes is believed to provide an evolutionary pathway to pathogenicity. As a genetically diverse group, most strains of Escherichia coli are harmless commensals of mammals (31, 54), but others are capable of causing either intestinal or extraintestinal disease (47). Manifestation of clinical symptomology and pathology appears to be closely associated with the possession of certain virulence gene combinations in E. coli (27, 35). For instance, diarrheagenic E. coli strains are classified on the basis of their virulence properties as enterotoxigenic (ETEC), enteropathogenic (EPEC), enterohemorrhagic (EHEC), enteroinvasive (EIEC), and enteroaggregative (EaggEC) (43). In general, these pathotypes have in common various virulence gene combinations for attachment and elaboration of hemolysins and enterotoxins, but there are considerable polymorphism and sequence variation in the molecular identities of genes that code for these virulence factors (8, 42).
Indeed, PCR analysis has revealed that even commensal E. coli isolates possess some of these virulence genes (10, 19). However, mere possession of a single or a few virulence genes does not endow a strain with pathogenic status unless that strain has acquired the appropriate virulence gene combination to cause disease in a specific host species (25). At present, it is debatable whether isolates that have one or a few virulence genes represent pathogenic clones that have lost virulence genes or are commensals in the process of acquiring them. Using a more phylogenetic approach, in 2000 Clermont (15) described a three-gene combination to differentiate between strains in the ECOR collection that are pathogens (phylogenetic groups B2 and D) and those members that are mainly commensals (phylogenetic groups A and B1). Surprisingly, these three genes alone were capable of providing a phylogenetic classification that closely mirrored similar groupings based on a more complex genetic analysis by multilocus enzyme electrophoresis. Furthermore, these relationships were established with a population of assembled clones, primarily of commensal origin, and in the absence of a panel of functionally accredited virulence genes.
Diarrhea in pigs can be caused by a number of pathogens, with transmissible gastroenteritis virus, rotavirus (RV), ETEC, Clostridium perfringens, and Isospora suis being the most common (61). Of these, ETEC strains are recognized as the most common cause of neonatal diarrhea (ND) in 0- to 4-day-old piglets with accompanying high morbidity and mortality rates (61). Strains of E. coli isolated from piglets with ND are mucoid (A-type capsule), often nonhemolytic, and usually confined to serogroups O8, O9, O20, O64, and O101 (23, 58). These strains have been classified as atypical “class 2” ETEC as they possess fimbrial adhesins belonging to F4, F5, F6, or F41 and are generally LT− (heat-labile toxin) and ST+ (heat-stable toxin) (23, 29, 57, 58). Older preweaning pigs, as well as postweaning (PW) animals up to 12 weeks of age, are affected by hemolytic ETEC (22). These strains are frequently represented by classic serogroups, including O8, O138, O139, O145, O141, O149, and O157, and are considered to be typical “class 1” ETEC (22, 58). They express F4 fimbrial adhesin in association with heat-labile enterotoxin LT alone or in combination with heat-stable enterotoxin STa and/or STb (40, 56, 65). More recently, Benz and Schmidt (5-7) have reported the detection of a class of diffusely adhering E. coli (DAEC) strains from piglets with diarrhea and its adhesin virulence factor, AIDA. Specific combinations of virulence genes encoding virulence factors such as adhesins and enterotoxins are the characteristic signature of pathogenic ETEC isolates.
Unlike commensals, extraintestinal pathogenic E. coli (ExPEC) strains do not establish long-term symbiotic relationships with their host (38, 65). Like strains that cause intestinal infections in humans, ExPEC strains possess virulence genes that have a range of functions, including attachment/invasion, toxin production, iron scavenging, and immune evasion (34, 37). ExPEC strains are divided into three major pathotypes (4): (i) uropathogenic (UPEC) strains that cause urinary infections, (ii) strains that cause neonatal meningitis (MENEC), and (iii) strains that cause septicemia (4). ExPEC strains possess virulence gene combinations that are distinctive from those found in their counterparts that cause intestinal disease. For instance, UPEC strains are more likely to possess P pili, S pili, afimbrial adhesin, and toxins such as hemolysin and cytotoxic necrotizing factor 1 (34, 37).
The main objectives in this study were to apply a wider array of virulence genes known to occur in intestinal and extraintestinal E. coli pathotypes associated with both human and animal disease and to optimize uni/multiplex combinatorial PCR assays for their detection in porcine isolates. The assays were then used to determine the presence of these genes in E. coli strains isolated from clinical cases of ND and postweaning diarrhea (PWD) to explore the possibility that clinical isolates can be identified by their virulence gene combinations. E. coli isolates from healthy pigs (commensals) were also included in the virulence gene analysis to assess whether there exists an evolutionary and phylogenetic relationship between pathogens and commensals modeled principally on their virulence gene repertoire. A mathematical model involving principal coordinate (PCO) analysis was used to visualize these relationships.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists 75 E. coli strains used in this study. These were obtained from scouring neonatal and weaner piglets as well as healthy animals. The ND and PWD strains were obtained from diarrhea samples cultured on blood agar plates following previously described protocols (20, 22). The commensal isolates represent a subset of E. coli previously isolated from the luminal contents of the duodenum, ileum, colon, and fecal samples of healthy pigs (19). Additional strains used as a reference source of VGs for PCR analysis are summarized in Table S1 in the supplemental material. The identity of all E. coli isolates was confirmed by a positive indole test, with no growth on Simmons citrate agar and growth on minimal lactose agar plates (19).
TABLE 1.
List of 75 E. coli strains from clinical cases of porcine neonatal and postweaning diarrhea and commensal isolates from healthy pigs
| Identification code | Serogroup | Origin of isolate | Hemolytic phenotypef | Phylogenetic statusg | Identification code | Serogroup | Origin of isolate | Hemolytic phenotypef | Phylogenetic statusg | |
|---|---|---|---|---|---|---|---|---|---|---|
| Neonatal | 38 | O141K85ac | Fecalb | β-h | A | |||||
| 1 | O8:F− | Fecala | Nh | A | 39 | O141K85ac | Fecalb | β-h | A | |
| 2 | O8:F41 | Fecalb | Nh | A | 40 | O141K85ac | Fecalb | β-h | A | |
| 3 | O8:K99 | Fecalb | Nh | A | 41 | O141K85ac | Fecalb | β-h | B2 | |
| 4 | O8:K88 | Fecalb | Nh | D | 42 | O141K85ac | Fecalb | β-h | A | |
| 5 | O8:K88 | Fecalb | Nh | A | 43 | O149:K88 | Fecald | β-h | A | |
| 6 | O8:K88 | Fecalb | Nh | B2 | 44 | O149K88 | Fecalb | β-h | A | |
| 7 | O8:K99 | Fecalb | Nh | B1 | 45 | O149K88 | Fecalb | β-h | A | |
| 8 | O9:F41 | Fecalb | Nh | A | 46 | O149K88 | Fecalb | β-h | A | |
| 9 | O9:F41 | Fecalb | Nh | A | 47 | O149K88 | Fecalb | β-h | A | |
| 10 | O9:F41 | Fecalb | Nh | A | 48 | O149K88 | Fecalb | β-h | A | |
| 11 | O9:987P | Fecalb | Nh | A | 49 | O149K88 | Fecalb | β-h | A | |
| 12 | O20:K99 | Fecalb | Nh | A | 50 | O149K88 | Fecalb | β-h | A | |
| 13 | O20:K99 | Fecalb | Nh | A | 51 | O149:K88 | Fecalb | β-h | A | |
| 14 | O64:K99 | Fecala | Nh | B1 | 52 | O149:K91:Hnt | Fecalc | β-h | A | |
| 15 | O99:Ksnt:H38 | Fecalc | Nh | B1 | ||||||
| 16 | O101:F41 | Fecalb | Nh | A | Commensal | |||||
| 17 | O101:K88 | Fecala | Nh | B1 | 53 | O8:H− | Ileume | β-h | A | |
| 18 | O101:Ksnt:H11 | Fecalc | Nh | A | 54 | O18a,b:H− | Ileume | nh | B1 | |
| 19 | O109:Knst:H11 | Fecalc | Nh | B1 | 55 | O40:H25 | Fecale | nh | A | |
| 20 | O157:K88 | Fecalb | β-h | A | 56 | O40:H25 | Fecale | nh | D | |
| 57 | O75:H− | Fecalc | nh | A | ||||||
| Weaner | 58 | O75:H− | Fecalc | nh | A | |||||
| 21 | O8G7 | Fecalb | β-h | A | 59 | O77:H− | Duodenume | nh | B1 | |
| 22 | O8G7 | Fecalb | β-h | D | 60 | O82:H8 | Colone | nh | A | |
| 23 | O8G7 | Fecalb | β-h | D | 61 | O106:H− | Duodenume | nh | B1 | |
| 24 | O8G7:K88 | Fecala | β-h | A | 62 | O121:H21 | Fecalc | nh | A | |
| 25 | O45 | Fecalc | β-h | B1 | 63 | O126:H− | Fecalc | nh | A | |
| 26 | O138 | Fecald | β-h | D | 64 | O130:H− | Fecalc | nh | B1 | |
| 27 | O138:K81:Hnt | Fecalc | β-h | D | 65 | O130:H11 | Facalc | nh | B1 | |
| 28 | O139 | Fecalb | β-h | D | 66 | OR:H− | Fecale | nh | A | |
| 29 | O141:K85ab | Fecald | β-h | A | 67 | OR:H38 | Fecale | nh | A | |
| 30 | O141:K85ab | Fecalb | β-h | A | 68 | Ont/R:H− | Ileume | nh | A | |
| 31 | O141ac | Fecald | β-h | A | 69 | Ont:Hnt | Ileume | nh | B1 | |
| 32 | O141:K85ac | Fecalb | β-h | A | 70 | Ont:H− | Duodenume | nh | A | |
| 33 | O141:K85ab:H4 | Fecalc | β-h | A | 71 | Ont:H− | Duodenume | nh | A | |
| 34 | O141:K85ac:H− | Fecalc | β-h | A | 72 | Ont:H− | Fecale | nh | A | |
| 35 | O141:K85ac | Fecalc | β-h | B1 | 73 | Ont:H11 | Fecalc | nh | B1 | |
| 36 | O141:K85ac | Fecalc | β-h | B1 | 74 | Ont:H27 | Ileume | nh | A | |
| 37 | O141K85ac | Fecalb | β-h | A | 75 | Ont:H45 | Duodenume | nh | B1 |
School of Veterinary Science, The University of Queensland.
Department of Primary Industries, Victoria.
Regional Veterinary Laboratory, Elizabeth Macarthur Agriculture Institute.
Chris Richards and Associates, Bendigo, Victoria.
Dixit et al. (19).
nh, nonhemolytic; β-h, beta-hemolytic.
Clermont et al. (15).
Bacterial serotyping.
Both O serotyping and H serotyping of E. coli were performed using previously reported methods (9, 12).
Hemolysis.
Hemolysis was determined by streaking E. coli isolates onto blood agar containing 10% sheep blood and incubating them at 37°C for 24 h. A clear zone around colonies where the blood cells had been utilized was characteristic of beta-hemolytic E. coli.
Maintenance of bacteria.
E. coli strains were stored at −80°C in Luria-Bertani (LB) broth containing 20% glycerol. Bacteria were recovered from frozen stocks and plated on LB agar and were never subcultured more than twice before DNA extraction.
DNA extraction.
All isolates were prepared by inoculating a single colony into 1 ml of LB broth and incubated at 37°C with shaking (∼100 rpm) overnight. DNA was extracted using a Promega DNA purification kit.
Virulence genes.
A group of 58 VGs (Table 2), reported in the literature to be associated with different E. coli pathotypes, were selected as the panel to be used in our analysis (Table 3) (3, 15, 17, 20, 28, 34, 44, 50, 52, 70, 71).
TABLE 2.
List of 58 virulence genes reported in the literature to be associated with different E. coli pathotypes associated with human and animal disease
| Virulence gene(s)/activity | E. coli pathotype | Description/function |
|---|---|---|
| Adhesins | ||
| afa/draBC | ExPEC | Central region of Dr antigen-specific fimbrial and afimbrial adhesin operons (e.g., AFA, Dr, and F1845) |
| aidA | DAEC | Adhesin involved in diffuse adherence, consisting of AIDA-I (orfB) and AIDAc (orfBc) |
| aah | DAEC | Autotransporter adhesin heptosyltransferase encoding AAH protein which modifies AIDA-I adhesin |
| bfpA | EPEC | Type IV bundle-forming pili |
| bmaE | ExPEC | M-agglutinin subunit |
| eaeA | EPEC, EHEC | Intimin |
| faeG | ETEC | F4 fimbrial adhesin |
| fanC | ETEC | F5 fimbrial adhesin |
| fasA | ETEC | F6 fimbrial adhesin |
| fedA | ETEC | F18 fimbrial adhesin |
| F41 | ETEC | Fimbrial adhesin |
| fimH | ExPEC | d-Mannose-specific adhesin, type 1 fimbriae |
| focG | ExPEC | Pilus tip molecule, F1C fimbriae (sialic acid specific) |
| iha | EHEC | Novel nonhemagglutinin adhesin (from O157:H7 and CFT073) |
| nfaE | ExPEC | Nonfimbrial adhesin I assembly and transport |
| paa | EPEC | Porcine A/E-associated gene |
| papA | ExPEC | Major structural subunit of pilus associated with pyelonephritis (P fimbriae), defines F antigen |
| papC | ExPEC | Pilus assembly, central region of pap operon |
| papEF | ExPEC | Minor tip pilins, connect PapG to shaft (PapA) |
| papG | ExPEC | Gal(1-4)Gal-specific pilus tip adhesin molecule |
| papG allele I | ExPEC | (Rare) J96-associated papG variant |
| papG allele II | ExPEC | Pyelonephritis-associated papG variant |
| papG allele III | ExPEC | Cystitis-associated (prs or pap-2) papG variant |
| papG allele l′ | ExPEC | papG variant identified in canine urine |
| saa | EHEC | STEC autoagglutinating adhesin |
| sfa/focDE | ExPEC | Central region of sfa (S fimbriae) and foc (F1C fimbriae) operons |
| sfaS | ExPEC | Pilus tip adhesin, S fimbriae (sialic acid specific) |
| Toxins | ||
| cdt | EPEC, ExPEC, NTEC | Cytolethal distending toxin |
| cdtB | ExPEC | Cytolethal distending toxin |
| cnf1 | ExPEC, NTEC | Cytotoxic necrotizing factor 1 |
| univcnf | ExPEC | Universal primer for cytotoxic necrotizing factor 1 |
| cvaC | ExPEC | Colicin V, conjugative plasmids (traT, iss, and antimicrobial resistance) |
| east1 | EaggEC | EaggEC heat-stable enterotoxin |
| exhA | EPEC, EHEC | Enterohemolysin |
| hlyA | ExPEC | α-Hemolysin |
| LT | ETEC | Heat-labile toxin |
| STa | ETEC | Heat-stable enterotoxin a |
| STb | ETEC | Heat-stable enterotoxin b |
| stx1 | EHEC | Shiga toxin I |
| stx2 | EHEC | Shiga toxin II |
| Capsule synthesis | ||
| kpsMTII | ExPEC | Group II capsular polysaccharide synthesis (e.g., K1, K5, and K12) |
| kpsMTIII | ExPEC | Group III capsular polysaccharide synthesis (e.g., K3, K10, and K54) |
| kpsMT K1 | ExPEC | Specific for K1 (group II) kpsMT |
| kpsMT “K5” | ExPEC | Specific for non-K1 and non-K2 group II kpsMT |
| rfc | ExPEC | O4 lipopolysaccharide synthesis |
| Siderophores | ||
| fyuA | ExPEC | Yersinia siderophore receptor (ferric yersiniabactin uptake) |
| ireA | ExPEC | Iron-regulated element, a siderophore receptor |
| iroNE. coli | ExPEC | Novel catecholate siderophore |
| iutA | ExPEC | Ferric aerobactin receptor (iron uptake/transport) |
| Invasins | ||
| ibeA | ExPEC | Invasion of brain endothelium |
| ipaH | EIEC | Invasion plasmid antigen |
| Additional virulence genes | ||
| chuA | EHEC | Gene required for heme transport in EHEC O157:H7 |
| iss | ExPEC | Serum survival gene |
| ompT | ExPEC | Outer membrane protein A and T (protease) |
| PAI | ExPEC | Pathogenicity-associated island, provides mechanism for coordinate horizontal transfer of VF genes between lineages |
| TSPE4C2 | Anonymous DNA fragment | |
| traT | ExPEC | Surface exclusion, serum survival |
| yjaA | Identified in E. coli K12, function currently unknown |
TABLE 3.
Summary of 12 multiplex (I to XII) and 6 uniplex (XIII to XVIII) primer sets for the amplification of the 58 virulence genes in this studya
| Primer set (reference[s]) and virulence genea | Primer name | DNA sequence (5′→3′) | Amplified product (bp) | Primer concn |
|---|---|---|---|---|
| I (34) | ||||
| PAI | RPAi-F | GGACATCCTGTTACAGCGCGCA | 930 | 0.6 μM |
| RPAi-R | TCGCCACCAATCACAGCCGAAC | |||
| papAH | PapA-F | ATGGCAGTGGTGTCTTTTGGTG | 720 | 0.6 μM |
| PapA-R | CGTCCCACCATACGTGCTCTTC | |||
| fimH | FimH-F | TGCAGAACGGATAAGCCGTGG | 508 | 0.6 μM |
| FimH-R | GCAGTCACCTGCCCTCCGGTA | |||
| kpsMTIII | kpsII-F | GCGCATTTGCTGATACTGTTG | 392 | 0.6 μM |
| kpsII-R | CATCCAGACGATAAGCATGAGCA | |||
| papEF | kpsII-F | GCGCATTTGCTGATACTGTTG | 336 | 0.6 μM |
| kpsII-R | CATCCAGACGATAAGCATGAGCA | |||
| ibeA | ibe10-F | AGGCAGGTGTGCGCCGCGTAC | 170 | 0.6 μM |
| ibe10-R | TGGTGCTCCGGCAAACCATGC | |||
| II (34) | ||||
| fyuA | FyuA-F | TGATTAACCCCGCGACGGGAA | 880 | 0.6 μM |
| FyuA-R | CGCAGTAGGCACGATGTTGTA | |||
| bmaE | bmaE-F | ATGGCGCTAACTTGCCATGCTG | 507 | 0.6 μM |
| bmaE-R | AGGGGGACATATAGCCCCCTTC | |||
| sfa/focDE | sfa 1-F | CTCCGGAGAACTGGGTGCATCTTAC | 410 | 0.6 μM |
| sfa 2-R | CGGAGGAGTAATTACAAACCTGGCA | |||
| iutA | AerJ-F | GGCTGGACATCATGGGAACTGG | 300 | 0.6 μM |
| AerJ-R | CGTCGGGAACGGGTAGAATCG | |||
| papG allele III | Allele III-F | GGCCTGCAATGGATTTACCTGG | 258 | 0.6 μM |
| Allele III-R | CCACCAAATGACCATGCCAGAC | |||
| kpsMT K1 | K1-fc-F | TAGCAAACGTTCTATATTGGTGC | 153 | 0.6 μM |
| KpsII-R | CATCCAGACGATAAGCATGAGCA | |||
| III (34) | ||||
| hlyA | hly-F | AACAAGGATAAGCACTGTTCTGGCT | 1,177 | 0.6 μM |
| hly-R | ACCATATAAGCGGTCATTCCCGTCA | |||
| rfc | rfc-F | ATCCATCAGGAGGGGACTGGA | 788 | 0.6 μM |
| rfc-R | AACCATACCAACCAATGCGAG | |||
| nfaE | nfaE-F | GCTTACTGATTCTGGGATGGA | 559 | 0.3 μM |
| nfaE-R | CGGTGGCCGAGTCATATGCCA | |||
| papG allele I | Allele I-F | TCGTCTCAGGTCCGGAATTT | 461 | 0.3 μM |
| Allele I-R | TGGCATCCCCCAACATTATCG | |||
| kpsMTII | kpsII-F | GCGCATTTGCTGATACTGTTG | 272 | 0.3 μM |
| kpsII-R | CATCCAGACGATAAGCATGAGC | |||
| papC | PapC-F | GTGGCAGTATGAGTAATGACCGTTA | 200 | 0.3 μM |
| PapC-R | ATATCCTTTCTGCAGGGATGCAATA | |||
| IV (34) | ||||
| cvaC | ColV-C-F | CACACACAAACGGGAGCTGTT | 680 | 0.6 μM |
| ColV-C-R | CTTCCCGCAGCATAGTTCCAT | |||
| cdtB | cdt-a1-F | AAATCACCAAGAATCATCCAGTTA | 430 | 0.6 μM |
| cdt-a2-R | AAATCTCCTGCAATCATCCAGTTTA | |||
| cdt-s1-F | GAAAGTAAATGGAATATAAATGTCCG | |||
| cdt-s2-R | GAAAATAAATGGAACACACATGTCCG | |||
| focG | FocG-F | CAGCACGGCAGTGGATACGA | 360 | 0.6 μM |
| FocG-R | GAATGTCGCCTGCCCATTGCT | |||
| traT | TraT-F | GGTGTGGTGCGATGAGCACAG | 290 | 0.6 μM |
| TraT-R | CACGGTTCAGCCATCCCTGAG | |||
| papG allele II | Allele II-F | GGGATGAGCGGGCCTTTGAT | 190 | 0.6 μM |
| Allele II-R | CGGGCCCCCAAGTAACTCG | |||
| V (34) | ||||
| papG allele I | pG-F | CTGTAATTACGGAAGTGATTTCTG | 1,190 | 0.6 μM |
| pG1"-R | TCCAGAAATAGCTCATGTAACCCG | |||
| papG alleles II and III | pG-F | CTGTAATTACGGAAGTGATTTCTG | 1,070 | 0.6 μM |
| pG-R | ACTATCCGGCTCCGGATAAACCAT | |||
| afa/draBC | Afa-F | GGCAGAGGGCCGGCAACAGGC | 559 | 0.3 μM |
| Afa-R | CCCGTAACGCGCCAGCATCTC | |||
| cnf1 | cnf1-F | AAGATGGAGTTTCCTATGCAGGAG | 498 | 0.3 μM |
| cnf2-R | CATTCAGAGTCCTGCCCTCATTATT | |||
| sfaS | SfaS-F | GTGGATACGACGATTACTGTG | 240 | 0.3 μM |
| SfaS-R | CCGCCAGCATTCCCTGTATTC | |||
| kpsMT K5 | K5-F | CAGTATCAGCAATCGTTCTGTA | 159 | 0.6 μM |
| kpsII-R | CATCCAGACGATAAGCATGAGCA | |||
| VI (34, 70) | ||||
| univcnf | CONCNF-F | ATCTTATACTGGATGGGATCATCTTGG | 1,105 | 0.6 μM |
| CONCNF-R | GCAGAACGACGTTCTTCATAAGTATC | |||
| iha | IHA-F | CTGGCGGAGGCTCTGAGATCA | 827 | 0.6 μM |
| IHA-R | TCCTTAAGCTCCCGCGGCTGA | |||
| iroNE. coli | IRONEC-F | AAGTCAAAGCAGGGGTTGCCCG | 665 | 0.6 μM |
| IRONEC-R | GACGCCGACATTAAGACGCAG | |||
| ompT | OMPT-F | ATCTAGCCGAAGAAGGAGGC | 559 | 0.6 μM |
| OMPT-R | CCCGGGTCATAGTGTTCATC | |||
| Allele I′ | ALLELE I′-F | CTACTATGTTCATGCTCAGGTC | 479 | 0.6 μM |
| ALLELE I′-R | CCTGCATCCTCCACCATTATCGA | |||
| iss | ISS-F | CAGCAACCCGAACCACTTGATG | 323 | 0.6 μM |
| ISS-R | AGCATTGCCAGAGCGGCAGAA | |||
| ireA | IRE-F | GATGACTCAGCCACGGGTAA | 254 | 0.6 μM |
| IRE-R | CCAGGACTCACCTCACGAAT | |||
| VII (51) | ||||
| ehxA | ehxA-F | GCATCATCAAGCGTACGTTCC | 534 | 20 pmol |
| ehxA-R | AATGAGCCAAGCTGGTTAAGCT | |||
| eaeA | eaeA-F | GACCCGGCACAAGCATAAGC | 384 | 20 pmol |
| eaeA-R | CCACCTGCAGCAACAAGAGG | |||
| stx2 | stx2-F | GGCACTGTCTGAAACTGCTCC | 255 | 20 pmol |
| stx2-R | TCGCCAGTTATCTGACATTCTG | |||
| stx1 | stx1-F | ATAAATCGCCTATCGTTGACTAC | 180 | 20 pmol |
| stx1-R | AGAACGCCCACTGAGATCATC | |||
| VIII (20) | ||||
| eltA | LTA-1 | GGCGACAGATTATACCGTGC | 696 | 3.2 pmol |
| LTA-2 | CCGAATTCTGTTATATATGTC | |||
| fasA | F6-Fw | TCTGCTCTTAAAGCTACTGG | 333 | 3.2 pmol |
| F6-Rv | AACTCCACCGTTTGTATCAG | |||
| estII | STb-1 | ATCGCATTTCTTCTTGCATC | 172 | 3.2 pmol |
| STb-2 | GGGCGCCAAAGCATGCTCC | |||
| IX (20) | ||||
| faeG | F4-Fw | GGTGATTTCAATGGTTCG | 764 | 3.2 pmol |
| F4-Rv | ATTGCTACGTTCAGCGGAGCG | |||
| fanC | F5-Fw | TGGGACTACCAATGCTTCTG | 450 | 3.2 pmol |
| F5-Rv | TATCCACCATTAGACGGAGC | |||
| estI | STa1 | TCTTTCCCCTCTTTTAGTCAG | 166 | 3.2 pmol |
| STa2 | ACAGGCAGGATTACAACAAAG | |||
| X (20) | ||||
| fedA | FedA-1 | GTGAAAAGACTAGTTTATTTC | 510 | 3.2 pmol |
| FedA-2 | CTTGTAAGTAACCGCGTAAGC | |||
| F41 | F41-Fw | GAGGGACTTTCATCTTTTAG | 431 | 3.2 pmol |
| F41-Rv | AGTCCATTCCATTTATAGGC | |||
| XI (44) | ||||
| aah (orfA) | UN19 | CTGGGTGACATTATTGCTTGG | 370 | 10 pmol |
| UN20 | TTTGCTTGTGCGGTAGACTG | |||
| aidA AIDA-I (orfB) | UN21 | TGCAAACATTAAGGGCTCG | 450 | 10 pmol |
| UN22 | CCGGAAACATTGACCATACC | |||
| aidA AIDAc (orfB) | UN23 | CAGTTTATCAATCAGCTCGGG | 543 | 10 pmol |
| UN24 | CCACCGTTCCGTTATCCTC | |||
| XII (15) | ||||
| chuA | ChuA.1 | GACGAACCAACGGTCAGGAT | 279 | 20 pmol |
| ChuA.2 | TGCCGCCAGTACCAAAGACA | |||
| yjaA | YjaA.1 | TGAAGTGTCAGGAGACGCTG | 211 | 20 pmol |
| YjaA.2 | ATGGAGAATGCGTTCCTCAAC | |||
| TSPE4.C2 | TspE4C2.1 | GAGTAATGTCGGGGCATTCA | 152 | 20 pmol |
| TspE4C2.2 | CGCGCCAACAAAGTATTACG | |||
| XIII (71) | ||||
| east1 | east 11a | CCATCAACACAGTATATCCGA | 111 | 50 pmol |
| east 11b | GGTCGCGAGTGACGGCTTTGT | |||
| XIV (17) | ||||
| cdt | CDT3A | GAGTTATTCCTTCCCCAGGC | 108 | 50 pmol |
| CDT3B | CAAAGGCATCAACAGCAGAA | |||
| XV (3) | ||||
| paa | M155-F1 | ATGAGGAAACATAATGGCAGG | 350 | 50 pmol |
| M155-R1 | TCTGGTCAGGTCGTCAATAC | |||
| XVI (50) | ||||
| saa | SAADF | CGTGATGAACAGGCTATTGC | 119 | 50 pmol |
| SAADR | ATGGACATGCCTGTGGCAAC | |||
| XVII (52) | ||||
| ipah | ipaHIII | GTTCCTTGACCGCCTTTCCGATACCGTC | 600 | 50 pmol |
| ipaHIV | GCCGGTCAGCCACCCTCTGAGATAC | |||
| XVIII (28) | ||||
| bfpA | EP1 | AATGGTGCTTGCGCTTCGTGC | 326 | 50 pmol |
| EP2 | GCCGCTTTATCCAACCTGGTA |
The forward and reverse primer sequences for each virulence gene, together with the size of the exception amplicon products, are also shown.
PCR analysis.
A series of 12 multiplex PCR sets (Table 3, sets I to XII) was adapted from published protocols and optimized for the amplification of 52 VGs. The remaining six VGs were individual PCR amplifications represented by sets XIII to XVIII (Table 3). PCR conditions were as described in Table S2 in the supplemental material. PCRs were conducted using a PC960 air-cooled thermal cycler (Corbett Research) with program cycles listed in Table S3 in the supplemental material. Amplicons were visualized by electrophoresis (80 V, 500 mA for 2.5 h for sets I to VI and 1.5 h for sets VII to XVIII) in 2% agarose gels prepared in 0.5× Tris-borate-EDTA (TBE) buffer (45 mM Tris base, 45 mM boric acid, 10 mM EDTA, pH 8) containing 4 μl of 5 μg/ml ethidium bromide. Amplicons were sized with corresponding 100-bp DNA markers (New England Biolabs) and processed in a Gel Doc system (Bio-Rad).
ECOR assignment.
E. coli isolates were assigned to one of the four main groups identified in the ECOR collection (32, 45) by the method of Clermont (15). Any strains that failed to yield amplicons for the three Clermont genes—chuA, yjaA, and TSPE4.C2—by PCR were further identified using the BBL Crystal enteric/nonfermenter identification system (Becton Dickinson) according to the manufacturer's protocol. When confirmed to be E. coli, these isolates were then classified as ECOR group A strains (26).
GLM.
The generalized linear model (GLM) was used to assess the significance of differences between groups of isolates (ND, PWD, and commensals) for each gene and to rank the relative importance of these genes based on the deviance value contributed to by group differences. Consider a model that can relate a dependent variable, xjr (value 0 = absent or 1 = present), to the group parameters (β) according to a logistic function
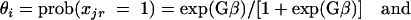 |
(1) |
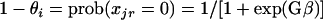 |
(2) |
where G is an N × t design matrix (values = 0 or 1), with N as the number of observations and t as the number of E. coli groups. A logit transformation was used to linearize the above relationship to the following form:
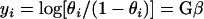 |
(3) |
The group parameters (β) were then estimated using the maximum likelihood estimation, and the deviance value (chi square) was determined using a likelihood ratio test. Details of this method were described by Cox in 1970 (16). The comparisons between group means were determined by forming factors of group contrasts, and chi-square values associated with these contrasts were calculated.
Cluster analysis.
An agglomerative hierarchical algorithm was used to establish cluster relationships between E. coli strains essentially as described by Kaufman and Rousseeuw in 1990 (34a). Let x be an n × v data matrix with elements xir having values of 0 = absent or 1 = present where n is the number of isolates and v is the number of genes. A simple matching coefficient was calculated as follows: m12 = 1 − d2ij/v and the sum of squared distances between all pairs of isolates to form a dissimilarity matrix, Q, with the element given by qij = −1/2 d2ij, where d2ij = Σ(xir − xjr)2.
The between-cluster dissimilarity, d(K1,K2), was then determined by the complete linkage method (the furthest neighbor method): that is
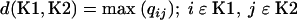 |
(4) |
Once the longest distance was established for the first cluster, the same calculations were performed to compute the second longest cluster, and this was repeated until all cluster combinations were set within the framework defined by the first cluster.
PCO analysis.
The technique of PCO analysis was used to convert the data matrix of 58 VGs (or dimensions) into 2 or 3 major coordinates (reduced number of dimensions). The conversion of 58 dimensional data (v1 … v58) to the principal coordinates (PCO1 … PCOk) is represented by multiple linear equations as follows:
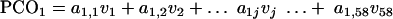 |
(5) |
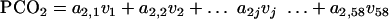 |
(6) |
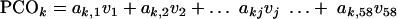 |
(7) |
The coefficients (a1,j, etc.) are latent vectors and are derived from an association matrix of “simple matching” coefficients and calculated via the principal component analysis technique. Gower (26a) in 1966 introduced the principal coordinate analysis by implementing the conventional principal component analysis on the squared distance matrix Q, which has latent root λ = [λ1 λ2 λ3 … λn]T and latent vector a, such that Qa = λa.
The methodology described in reference 42 was used to derive the latent root λ and the latent vector a. All computations were carried out using GenStat release 7.1 (64).
RESULTS
Bacterial serogroups and phenotypes.
The 75 porcine isolates were characterized for serogroup, hemolytic phenotype, and phylogenetic status as shown in Table 1. Three main serogroups represented by O8, O9, and O101 predominated in ND isolates, while PWD isolates were represented primarily by O8G7, O141, and O149.
All the commensal isolates belonged to serotypes that did not match either ND or PWD strains. Almost half of these were not typeable by currently available protocols. All ND and commensal isolates, with one exception each, were nonhemolytic on blood agar. In contrast, PWD isolates were all beta-hemolytic.
Phylogenetic status based on the Clermont virulence gene combination.
The phylogenetic groupings of porcine ETEC were established using Clermont's three-gene PCR for chuA, yjaA, and TSPE4.C2 (Table 1). On this basis, 90% and 81% of ND and PWD isolates, respectively, were classified as belonging to phylogenetic groups A (chuA yjaA TSPE4.C2 mutant or chuA yjaA+ TSPE4.C2 mutant) and B1 (chuA yjaA TSPE4.C2+) together with almost all the commensal strains. The phylogenetic group B2 was represented by only one isolate each in the ND and PWD collection. There was only one group D representative in ND (isolate 4) and commensal (isolate 56) isolates compared to five in the PWD assemblage (isolates 22, 23, and 26 to 28).
Distribution of virulence genes.
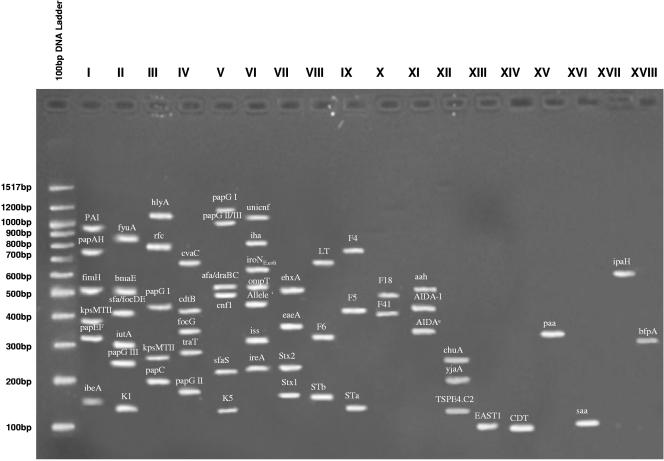
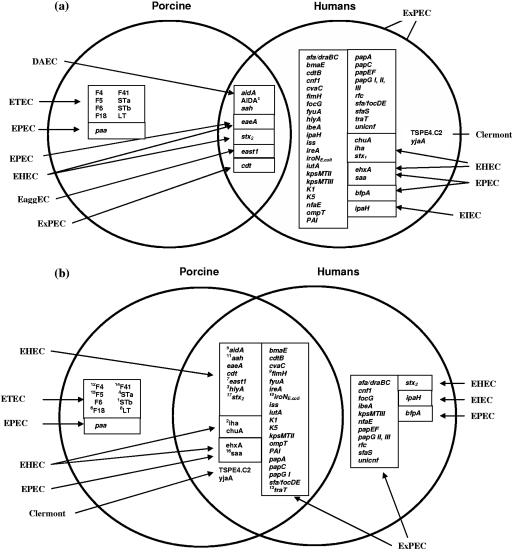
The identification of 58 VGs by PCR in 18 uni/multiplex sets is depicted in Fig. 1. Prior to this study, E. coli strains isolated from clinical cases of ND and PWD were analyzed for the presence of F4, F5, F6, F18, F41, STa, STb, and LT (Fig. 2a). Other VGs, including AIDA (aah, aidA, and AIDAc), eaeA, stx2, east1, and cdt, are found in both porcine and human isolates. As a consequence of this investigation, an additional 25 VGs can now be used to classify porcine clinical E. coli (Fig. 2b). Of the 25 VGs identified in porcine isolates, 19 of these VGs are primarily associated with ExPEC isolates from humans (34), with 4 associated with EHEC (iha, chuA, saa, and ehxA) and 1 associated with EPEC (saa).
FIG. 1.
Ethidium bromide-stained 2% agarose gel showing amplified bands corresponding to the 58 virulence genes associated with the representative pathotypes of E. coli known to cause intestinal and extraintestinal disease in humans and animals. Note that multiplex V contains an internal control and is identified as papG II/III.
FIG. 2.
The 58 virulence genes analyzed in this study that had previously been reported in porcine ETEC infections and human clinical isolates (a). Of the 58 virulence genes analyzed, 25 additional genes have now been identified in the porcine ETECs in this study (b). The superscript numbers represent the chi-square significance ranking.
Prevalence of significant virulence genes in commensal, ND and PWD isolates.
In this analysis, 17 out of 58 VGs were found to be significantly different (P < 0.05) for isolates from the three different sources of origin (Table 4). The chi-square ranking varied from 48.3 for STb to 7.5 for stx2. Nine of these 17 genes represented by iha, hlyA, aidA, east1, aah, fimH, iroNE. coli, traT, and saa have not been previously identified as important virulence genes in the identification of porcine diarrheagenic isolates. Not surprisingly, the prevalence of virulence genes encoding STb, STa, F18, LT, Stx2, F4, F5, and F41 was considered to be significant in differentiating between clinical and commensal isolates.
TABLE 4.
Prevalence of statistically significant genes (P < 0.05) by chi-square analysis of virulence genes in E. coli from scouring neonatal and weaner pigs and healthy pigs
| Virulence gene | % of E. coli strains with gene
|
Chi-square value (P)a | ||
|---|---|---|---|---|
| Neonatal | Weaner | Commensal | ||
| STb | 25 | 81.3 | 0 | 48.33 |
| iha | 15 | 81.3 | 4.3 | 44.93 |
| hlyA | 10 | 71.9 | 4.3 | 37.55 |
| STa | 35 | 68.8 | 0 | 34.43 |
| aidA (orfB) | 10 | 56.3 | 0 | 30.12 |
| F18 | 10 | 46.9 | 0 | 23.04 |
| east1 | 10 | 40.6 | 17.4 | 18.82 |
| LT | 15 | 37.5 | 0 | 15.81 |
| fimH | 100 | 62.5 | 69.6 | 14.28 |
| iroNE. coli | 25 | 0 | 0 | 14.25 |
| aah (orfA) | 20 | 56.3 | 13 | 13.79 |
| F4 | 5 | 37.5 | 4.3 | 11.25 |
| traT | 95 | 65.6 | 56.5 | 10.15 |
| F41 | 0 | 0 | 17.4 | 9.98 |
| F5 | 25 | 6.3 | 0 | 9.07 |
| saa | 40 | 9.4 | 13 | 7.56 |
| stx2 | 5 | 34.4 | 13 | 7.47 |
A total of 17 of the 58 virulence genes were able to differentiate between the three groups based on chi-square analysis.
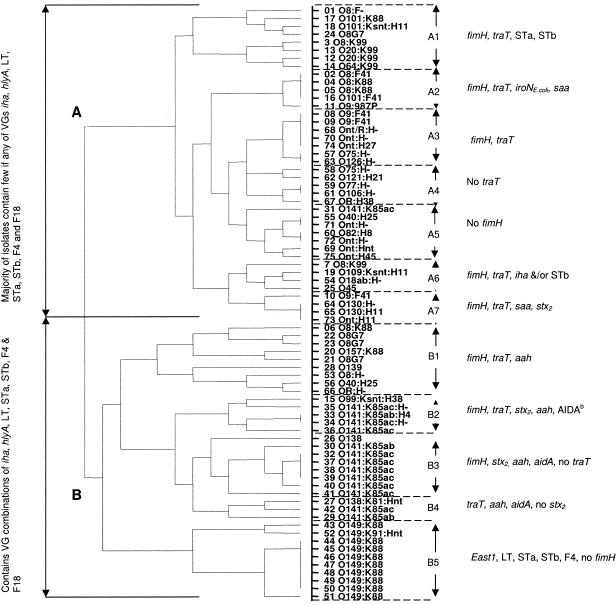
Cluster analysis—an association between virulence gene signatures and serogroups.
An agglomerative hierarchical algorithm was employed to form a gradual nesting of individual isolates into larger clusters. On this basis, all 75 isolates were distributed into one of two main branches (Fig. 3), A and B, which effectively partitioned neonatal diarrhea/commensal isolates from postweaning diarrhea strains, respectively. However, three ND isolates (no. 6 [O8:K88], 20 [O157:K88], and 15 [O99:Ksnt:H38]) and three commensal isolates (no. 53 [O8:H−], 56 [O40:H25], 66 [OR:H−]) were located in the B branch, while two PWD isolates (no. 24 [O8G7] and 25 [O45]) were in the A branch. Within the B branch, PWD isolates in B1, B2 to B4, and B5 were highly associated with their respective serogroup clusters: O8:G7, O141:K85, and O149:K88.
FIG. 3.
An agglomerative hierarchical algorithim was used to derive a cluster analysis dendrogram to establish the relationship between individual E. coli strains isolated from clinical cases of porcine neonatal and postweaning diarrhea and commensal isolates from healthy pigs based on the possession or nonpossession of the 17 significant virulence genes as determined by chi-square analysis.
Ownership of the 17 significant VGs by each strain is shown in Table S4 in the supplemental material. Two genes, fimH and traT, were for the most part present in both A and B branches. The traT gene was missing in almost all B3 members, while fimH was not detected in B5 strains. Unlike branch B strains, branch A strains were missing VGs corresponding to the STb, iha, STa, aidA, F18, east1, and LT genes that were signature genes for PWD strains. Virulence genes such as saa and iroNE. coli were detected in some of the ND strains but were absent from PWD isolates (all members of branch B). A number of other ExPEC VGs that were not statistically significant were also detected occasionally in ND isolates. These genes include the bmaE, cdtB, cvaC, iutA, papA, papG allele I, papC, and K1 genes, which were never identified in PWD or commensal isolates.
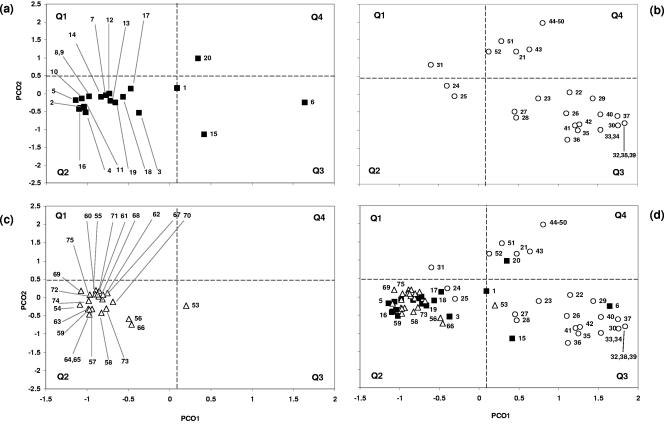
Genetic relationships between commensal and clinical E. coli clones from scouring neonatal piglets and weaners.
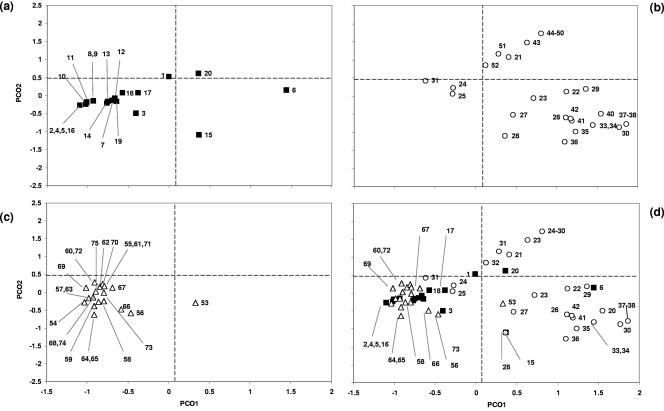
PCO analysis was applied using the 17 defined chi-square-significant genes (Fig. 4) and the full panel of 58 VGs (Fig. 5). In both cases, 16 out of 20 ND isolates were clustered in quadrant 2 (Q2). Similarly, 22 out of 23 commensal isolates were also located in quadrant 2. Eighteen of the 32 PWD isolates were located in Q3, and 11 were located in Q4. Analyzing the ND, PWD, and commensal isolates with PCO using the 17 significant VGs resulted in a tighter cluster between ND and commensal isolates which resulted in isolate 1 moving from borderline Q2 to Q3 in the 58-VG analysis to Q1 in the 17-VG analysis. The analysis of the 17 VGs of the PWD isolates resulted in isolate 31 moving from Q1 to Q2. PWD isolates clustered in Q3 and Q4 (Fig. 4b) were also grouped coordinately with their serogroup.
FIG. 4.
Principal coordinate analysis using the 17 significant virulence genes (chi-square analysis, P < 0.05) to establish a spatial relationship between the 75 E. coli clones and their clinical origin. The coordinate values of scouring neonates (a [n = 1 to 20]), weaners (b [n = 21 to 52]), and commensals (c [n = 53 to 75]) are shown as a composite in panel d.
FIG. 5.
Principal coordinate analysis using all 58 virulence genes to establish a spatial relationship between the 75 E. coli clones and their clinical origin. The coordinate values of scouring neonates (a [n = 1 to 20]), weaners (b [n = 21 to 52]), and commensals (c [n = 53 to 75]) are shown as a composite in panel d.
DISCUSSION
Intestinal or extraintestinal strains of clinical and commensal E. coli isolates from different animal species, with or without clinical disease, have been found to be extremely diverse in their genetic makeup. Such diversity has been demonstrated by different analytical methods, including multilocus enzyme electrophoresis, multilocus sequencing typing (36, 62, 67, and 69), and the presence of virulence genes by PCR. Over time, genetic diversity has been exploited by selection and adaptation so that pathogenic strains have tended to become host specific, with strains identified in scouring pigs being phenotypically and serotypically different from those that cause diarrhea in humans (30). Within each host species, genetic differences can still be found in different pathogenic isolates. For example, E. coli strains associated with intestinal disease are genotypically different from those that cause extraintestinal disease. In pigs, a similar difference can be found between isolates responsible for ND and PWD. A still unanswered question is how do virulence genes continue to be acquired or lost in each “adapted” individual's evolutionary pathway? Commensal E. coli isolates on the other hand have attracted very little attention because they are not overtly involved in causing disease. These silent autochthonous bacteria, while nonpathogenic, could potentially also harbor virulence genes but are incapable of causing disease because they lack the appropriate virulence gene combinations.
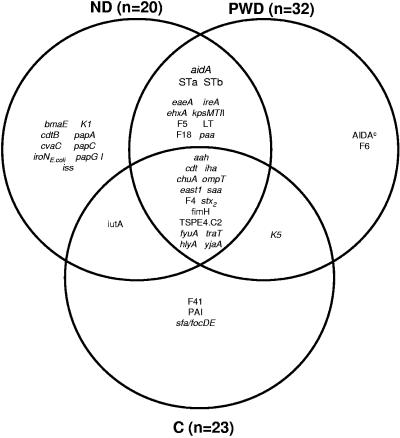
To obtain a better understanding about the role of virulence genes in the pathogenicity of porcine isolates, a total of 58 virulence genes were assembled and their presence or absence in commensal, ND, and PWD isolates was analyzed. As shown in Fig. 6, 15 genes out of 58 were shared in common between clinical and commensal isolates; 11 genes were common between ND and PWD isolates. Nine ND, two PWD, and three commensal isolates also carried specific virulence genes that were not shared. Exclusion or inclusion of virulence genes as depicted in the Venn diagram can be biased in favor of minority genes because even a single gene that has occurred only once in a single isolate can be partitioned and included in the diagram. Notwithstanding this limitation, it is clear that a number of commensal isolates have acquired one or more virulence genes. For example, isolates 64 and 65, which belong to serogroup O130, may not possess pathogenic functionality because they have a nonpathogenic serogroup or lack the appropriate combination of virulence genes. However, it is conceivable that over time, further virulence gene acquisitions, particularly entire pathogenicity islands, can reequip such a commensal with the potential to develop into a pathogen.
FIG. 6.
Distribution of additional virulence genes identified in the ND, PWD, and commensal E. coli isolates.
According to the GLM, nine virulence genes represented by iha, hlyA, aah, aidA, east1, fimH, iroNE. coli, traT, and saa, in addition to eight normally associated with pathogenic ETEC (coding for STb, STa, F18, LT, Stx2, F4, F5, and F41), were found to be significant in distinguishing between commensal, ND, and PWD clones. Iha has been described as a nonhemagglutinin adhesin found in O157:H7 and CFT073 (63). AIDA (adhesin-involved-in-diffuse-adherence) consists of the autotransporter adhesin heptosyltransferase (aah, formally orfA) gene which encodes the 44.8-kDa protein AAH, which modifies AIDA-I (orfB) by 19 heptose residues. AIDA-I remains covalently associated with the bacterial surfaces and is responsible for diffuse adherence (DA) patterns when E. coli cells attach to HeLa and HEp-2 cell culture lines (5-7). saa codes for an STEC autoagglutinating adhesin (50). fimH codes for d-mannose-specific adhesin or type 1 fimbriae (59, 60). hlyA α-hemolysin is a member of the RTX family of cytotoxins which is phenotypically observed on sheep blood agar (washed and unwashed) (11). east1 codes for an enteroaggregative E. coli heat-stable enterotoxin (53). iroNE. coli codes for a novel catecholate siderophore. traT is one of the F factor genes involved in encoding the outer membrane protein; the TraT lipoprotein is responsible for surface exclusion activity (1, 2, 13).
One of the main limitations inherent with Venn diagrams is the inability to evaluate relationships between strains based on virulence gene combinations. An agglomerative hierarchical algorithm was used to establish cluster relationships between isolates based on mutual possession or nonpossession of all possible virulence gene combinations out of the 17 significant virulence genes examined in this study. As shown in Fig. 3, isolates clustered on the basis of virulence genes were grouped according to their serogroups and this in turn enabled PWD isolates to be distinguished from ND and commensal strains. A similar cluster analysis utilizing all 58 virulence genes yielded a clustering pattern that was essentially similar to that obtained with the panel of 17 significant virulence genes. The pattern of ownership of these 17 virulence genes provided an opportunity to identify subclusters that were closely associated with the serogroup of member strains (see Table S4 in the supplemental material). Thus, the B1, B2 to B4, and B5 strains subclustered in accordance with serogroups O8, O141, and O149, respectively. It is interesting to note in terms of gene acquisitions that ND and commensal isolates carried primarily ExPEC virulence genes, while the sources of PWD virulence genes were from a range of pathotypes including ETEC, ExPEC, EHEC/EPEC, EaggEC, and DAEC.
Principal coordinate analysis provided another method of examining relationships between individual isolates by reducing 58 virulence genes or dimensions to three principal coordinates, or three dimensions, that can be plotted along the x, y, and z axes. While the three-dimensional plots can provide a better spatial perspective, the two-axis plots shown in Fig. 4 and 5 provide valuable insight into the relationship between individual clones. The spatial relationship supports the previous analytical methods that ND, PWD, and commensal isolates can be segregated on the basis of their virulence gene signatures. Unlike the dendrogram analysis, PCO plots have positioned the commensal isolates within the ND quadrant showing that there is greater similarity between these because they have fewer virulence genes in common compared to PWD isolates. Two-dimensional PCO plots depict Q2 primarily containing ND and commensal isolates, while Q3 and Q4 are dominated by PWD isolates. This clustering of ND and commensal isolates together and the distinct separation of the PWD isolates were apparent in the PCO analysis using only 17 significant genes. A similar computation using the full panel of 58 virulence genes did not significantly alter the clustering pattern.
Until this study, only eight virulence genes (coding for F4, F5, F6, F18, F41, STa, STb, and LT) were recognized and used routinely to characterize porcine ETEC (20, 39, 48, 49). More recently, additional virulence genes have been identified in porcine ETEC, including aidA and aah (44), cdt (17), and east1 (14). This study has revealed a further 25 virulence genes from other pathotypes that have not previously been identified in porcine isolates. Of these, 19 are closely associated with ExPECs, and four members of the virulence genes in this group represented by fimH, hlyA, iroNE. coli, and traT play a significant role in distinguishing between porcine commensal, ND, and PWD isolates (Table 4). The deployment of GLM and PCO methods of analysis was mandated by an accumulation of relatively large numbers of virulence genes acquired in the course of this study. In the past, when only eight virulence genes were used to characterize porcine ETEC, differences or similarities were recorded as percentages of occurrence of each gene in an assembly of isolates (18, 24, 46, 48, and 68). Such data essentially failed to demonstrate relationships between isolates and also did not relate virulence genes to serogroups and pathotypes. It is important to acknowledge that ownership of a virulence gene is not equivalent to its expression. Genes that get turned on to facilitate infection are usually dependent upon environmental queues emanating from community microorganisms (competition stress) or the host (immune stress) (33, 55).
A conclusion that can be drawn from this study is that virulence genes cannot fully define each pathotype. Instead, the virulence genes associated with each pathotype contribute to its functionality in causing characteristic symptomatology that typifies the disease syndrome. For instance, ND strains probably have a survival advantage over PWD strains because they do not require fimbriae to colonize the intestinal wall of the neonate with its immature and underdeveloped immune system. Under a different set of selection pressures, clones that possess disadvantageous virulence genes could be eliminated and replaced with other clones. Such changes can be associated with the increasing use of E. coli immunization of sows to generate protective antibodies in the colostrums (69). This would change the intestinal environment and profoundly alter the capacity of both commensals and pathogens to colonize the intestinal environment. The presence of virulence genes from human isolates in porcine E. coli strains should not be interpreted as a process of active virulence gene acquisition. The data suggests that both porcine commensal and clinical strains have over the years acquired and maintained these genes as part of a survival mechanism to engender greater diversity and hence increase their survival capability in the host animal.
Although the analytical techniques adopted in this study have focused on a panel of virulence genes representing different pathotypes of intestinal and extraintestinal E. coli, they can be applied to other gene panels, including those of genes that encode antibiotic resistance. As additional gene panels become available, the data matrix can be expanded and processed utilizing GLM, clustering, and PCO as the sequence format for biometric interrogation. The tedium of data acquisition using multiple gene analysis by PCR can be offset to some extent by multiplexing. Nevertheless, compared to DNA microarrays, PCR is relatively cost-effective (labor and capital) and allows a higher throughput for multiple clonal analyses. Furthermore, unlike DNA hybridization, the level of sensitivity and specificity of multiplex PCR is affected to a lesser extent by DNA concentrations and plasmid copy numbers. Unfortunately, virulence gene detection by PCR or microarray will still lack the level of definition provided by multilocus sequence polymorphism and cannot be used to detect subtle point mutations.
Supplementary Material
Acknowledgments
We thank David Gordon at the Australian National University for his continued advice and logistical support in the use of virulence genes as an epidemiological tool. Commensal strains were obtained from isolations and characterizations carried out by S. Dixit with assistance from R. Bugler. We thank A. Kusevski for technical assistance in serotyping the strains of E. coli.
T.A.C. was supported by an Australian Postgraduate Award (Industry) in conjunction with Bunge Meat Industries (now QAF). This work was funded in part by a Research and Development Start Grant to International Animal Health Products and NSW Department of Primary Industries.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtman, M., N. Kennedy, and R. Skurray. 1977. Cell-cell interactions in conjugating Escherichia coli: role of TraT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74:5104-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agüero, M. E., L. Aron, A. G. DeLuca, K. N. Timmis, and F. C. Cabello. 1984. A plasmid-encoded outer membrane protein, TraT, enhances resistance of Escherichia coli to phagocytosis. Infect. Immun. 46:740-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batisson, I., M. P. Guimond, F. Girard, H. An, C. Zhu, E. Oswald, J. M. Fairbrother, M. Jacques, and J. Harel. 2003. Characterization of the novel factor Paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 71:4516-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekal, S., R. Brousseau, L. Masson, G. Prefontaine, J. Fairbrother, and J. Harel. 2003. Rapid identification of Escherichia coli pathotypes by virulence gene detection with DNA microarrays. J. Clin. Microbiol. 41:2113-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benz, I., and M. A. Schmidt. 1992. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect. Immun. 60:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 8.Bertin, Y., K. Boukhors, N. Pradel, V. Livrelli, and C. Martin. 2001. Stx2 subtyping of Shiga toxin-producing Escherichia coli isolated from cattle in France: detection of a new Stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39:3060-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bettleheim, K. A., and C. J. Thompson. 1987. New method of serotyping Escherichia coli: implementation and verification. J. Clin. Microbiol. 25:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beutin, L., O. Marchés, K. A. Bettelheim, K. Gleier, S. Zimmermann, H. Schmidt, and E. Oswald. 2003. HEp-2 cell adherence, actin aggregation, and intimin types of attaching and effacing Escherichia coli strains isolated from healthy infants in Germany and Australia. Infect. Immun. 71:3995-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beutin, L., M. A. Montenegro, I. Ørskov, F. Ørskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandler, M. E., and K. A. Bettelheim. 1974. A rapid method of identifying Escherichia coli “H” antigens. Zentbl. Bakteriol. Mikrobiol. Hyg. 1 Abt. Orig. A 129:74-79. [PubMed] [Google Scholar]
- 13.Chang, H., S. Y. Sheu, and S. J. Lo. 1999. Expression of foreign antigens on the surface of Escherichia coli by fusion to the outer membrane protein TraT. J. Biomed. Sci. 6:64-70. [DOI] [PubMed] [Google Scholar]
- 14.Choi, C., W. Cho, H. Chung, T. Jung, J. Kim, and C. Chae. 2001. Prevalence of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) gene in isolates in weaned pigs with diarrhoea and/or edema disease. Vet. Microbiol. 81:65-71. [DOI] [PubMed] [Google Scholar]
- 15.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox, D. R. 1970. Analysis of binary data. Chapman and Hall Ltd., London, United Kingdom.
- 17.da Silva, A. S., and D. da Silva Leite. 2002. Investigation of putative CDT gene in Escherichia coli isolates from pigs with diarrhea. Vet. Microbiol. 89:195-199. [DOI] [PubMed] [Google Scholar]
- 18.DebRoy, C., and C. W. Maddox. 2001. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 1:129-140. [PubMed] [Google Scholar]
- 19.Dixit, S. M., D. M. Gordon, X. Y. Wu, T. Chapman, K. Kailasaphathy, and J. C. Chin. 2004. Diversity analysis of commensal porcine Escherichia coli—associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150:1735-1740. [DOI] [PubMed] [Google Scholar]
- 20.Do, T., C. Stephens, K. Townsend, K. Wu, T. Chapman, J. Chin, M. Bara, and D. J. Trott. 2003. Enterotoxigenic Escherichia coli pathotypes in pigs with diarrhea in South East Queensland. Aust. Vet. J. 83:293-299. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Fahy, V. A., I. D. Connaughton, S. J. Driesen, and E. M. Spicer. 1987. Postweaning colibacillosis, 189-201. In J. L. Barnett, E. S. Batterham, G. M. Cronin, C., Hansen, P. H. Hemsworth, D. P. Hennessy, P. E. Hughes, N. E. Johnston, and R. H. King (ed.), Manipulating pig production. Australasian Pig Science Association, Werribee, Victoria, Australia.
- 23.Fairbrother, J. M., S. Lariviere, and W. M. Johnson. 1988. Prevalence of fimbrial antigens and enterotoxins in nonclassical serogroups of Escherichia coli isolated from newborn pigs with diarrhea. Am. J. Vet. Res. 49:1325-1328. [PubMed] [Google Scholar]
- 24.Frydendahl, K. 2002. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhea and edema disease in pigs and comparison of diagnostic approaches. Vet. Microbiol. 85:169-182. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore, M. S., and J. J. Ferretti. 2003. The thin line between gut commensal and pathogen. Science 299:1999-2002. [DOI] [PubMed] [Google Scholar]
- 26.Gordon, D. M., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575-3586. [DOI] [PubMed] [Google Scholar]
- 26a.Gower, J. C. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325-338. [Google Scholar]
- 27.Grauke, L. J., I. T. Kudva, J. W. Yoon, C. W. Hunt, C. J. Williams, and C. L. Hovde. 2002. Gastrointestinal tract location of Escherichia coli O157:H7 in ruminants. Appl. Environ. Microbiol. 68:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunzburg, S. T., N. G. Tornieporth, and L. W. Riley. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J. Clin. Microbiol. 33:1375-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harel, J., H. Lapointe, A. Fallara, L. A. Lortie, M. Bigras-Poulin, S. Larivière, and J. M. Fairbrother. 1991. Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J. Clin. Microbiol. 29:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hart, C. A., R. M. Batt, and J. R. Saunders. 1993. Diarrhea caused by Escherichia coli. Ann. Trop. Paediatr. 13:121-131. [DOI] [PubMed] [Google Scholar]
- 31.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 32.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoey, E. D. E., L. Sharp, C. Currie, C. A. Lingwood, D. L. Gally, and D. G. E. Smith. 2003. Verotoxin 1 binding to intestinal crypt epithelial cells results in localization of lysosomes and abrogation of toxicity. Cell Microbiol. 5:85-97. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 34a.Kaufman, L., and P. J. Rousseeuw. 1990. Finding groups in data. An introduction to cluster analysis. John Wiley & Sons, Inc., New York, N.Y.
- 35.Law, D. 2000. Virulence factors of Escherichia coli O157 and other shiga toxin-producing E. coli. J. Appl. Microbiol. 88:729-745. [DOI] [PubMed] [Google Scholar]
- 36.Lemee, L., A. Dhalluin, M. Pestel-Caron, J.-F. Lemeland, and J.-L. Pons. 2004. Multilocus sequence typing analysis of human and animal Clostridium difficile isolates of various toxigenic types. J. Clin. Microbiol. 42:2609-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrs, C. F., L. Zhang, P. Tallman, et al. 2002. Escherichia coli virulence gene distributions show interesting difference between urinary tract infection, fecal and periurethral isolate collections. J. Med. Microbiol. 51:138-142.11863265 [Google Scholar]
- 38.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melin, L., M. Katouli, A. Lindberg, C. Fossum, and P. Wallgren. 2000. Weaning of piglets. Effects of an exposure to a pathogenic strain of Escherichia coli. J. Vet. Med. 47:663-675. [DOI] [PubMed] [Google Scholar]
- 40.Moon, H., and S. C. Whipp.1970. Development of resistance with age by swine intestine to effects of enteropathogenic Escherichia coli. J. Infect. Dis. 122:220-223. [DOI] [PubMed] [Google Scholar]
- 41.Morrison, D. F. 1976. Multivariate statistical methods, 2nd ed. McGraw-Hill, New York, N.Y.
- 42.Nagy, B., and P. Z. Fekete. 1999. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet. Res. 30:259-284. [PubMed] [Google Scholar]
- 43.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niewerth, U., A. Frey, T. Voss, C. Le Bouguénec, G. Baljer, S. Franke, and M. A. Schmidt. 2001. The AIDA autotransporter system is associated with F18 and Stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ojeniyi, B., P. Ahrens, and A. Meyling. 1994. Detection of fimbrial and toxin genes in Escherichia coli and their prevalence in piglets with diarrhea. The application of colony hybridization assays, polymerase chain reaction and phenotypic assays. J. Vet. Med. 41:49-59. [DOI] [PubMed] [Google Scholar]
- 47.Ørskov, F., and I. Ørskov.1992. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38:699-704. [PubMed] [Google Scholar]
- 48.Osek, J. 1999. Prevalence of virulence factors of Escherichia coli strains isolated from diarrheic and healthy piglets after weaning. Vet. Microbiol. 68:209-217. [DOI] [PubMed] [Google Scholar]
- 49.Osek, J. 2000. Virulence factors and genetic relatedness of Escherichia coli strains isolated from pigs with post-weaning diarrhea. Vet. Microbiol. 71:211-222. [DOI] [PubMed] [Google Scholar]
- 50.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paton, A. W., and J. C. Paton. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rich, C., A. Alfidja, J. Sirot, B. Joly, and C. Forestier. 2001. Identification of human enterovirulent Escherichia coli strains by multiplex PCR. J. Clin. Lab. Anal. 15:100-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savarion, S. J., A. Fasano, J. Watson, B. M. Martin, M. M. Levine, S. Guandalini, and P. Guerry. 1993. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat stable toxin. Proc. Natl. Acad. Sci. USA 90:3093-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selander, R. K., D. A. Caugant, and T. S. Whittam. 1987. Genetic structure and variation in natural populations of Escherichia coli, p. 1625-1648. In F. C. Neidhardt, J. L. Ingraham, K.B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 55.Sermon, J., M.-R. P. Wevers, L. Janse, P. De Spiegeler, K. Vanoirbeek, A. Aertsen, and C. W. Michiels. 2005. CorA affects tolerance of Escherichia coli and Salmonella enterica serovar Typhimurium to the lactoperoxidase enzyme system but not to other forms of oxidative stress. Appl. Environ. Microbiol. 71:6515-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, H. W., and C. L. Gyles. 1970. The relationship between two apparently different enterotoxins produced by enteropathogenic strains of Escherichia coli of porcine origin. J. Med. Microbiol. 3:387-401. [DOI] [PubMed] [Google Scholar]
- 57.Söderlind, O., B. Thafvelin, and R. Möllby. 1988. Virulence factors in Escherichia coli strains isolated from Swedish piglets with diarrhea. J. Clin. Microbiol. 26:879-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sojka, W. J. 1971. Enteric diseases in new-born piglets, calves and lambs due to Escherichia coli infection. Vet. Bull. 41:509-522. [Google Scholar]
- 59.Sokurenko, E. V., H. S. Courtney, D. E. Ohman, P. Klemm, and D. L. Hasty. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J. Bacteriol. 176:748-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sokurenko, E. V., V. Chesnokova, D. E. Dykhuizen, I. Ofek, X. Wu, K. A. Krogfelt, C. Struve, M. A. Schembri, and D. L. Hasty. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. USA 95:8922-8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Straw, B. E., D. E. Dewey, and M. R. Wilson. 1999. Differential diagnosis of swine diseases, p. 41-59. In B. E. Straw, S. D’Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames.
- 62.Tarr, C. L., T. M. Large, C. L. Moeller, D. W. Lacher, P. I. Tarr, D. W. Acheson, and T. S. Whittam. 2002. Molecular characterization of a serotype O121:H19 clone, a distinct Shiga toxin-producing clone of pathogenic Escherichia coli. Infect. Immun. 70:6853-6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R.Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tortora, G. J. 1995. Microbiology: an introduction, p. 273-279, 654-698. In B. Roesch (ed.), Principles of human anatomy. Harper Collins College Publishers, New York, N.Y.
- 65.VSN International. s2003. GenStat Release, ed. 7.1. VSN International, Hemel Hempstead, United Kingdom.
- 66.Whipp, S. C., H. W. Moon, and R. A. Argenzio. 1981. Comparison of enterotoxigenic activities of heat-stable enterotoxins from class 1 and class 2 Escherichia coli of swine origin. Infect. Immun. 31:245-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whittam, T. S., M. L. Wolfe, K. Wachsmuth, F. Ørskov, I. Ørskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wittig, W., and C. Fabricius.1992. Escherichia coli types isolated from porcine E. coli infections in Saxony from 1963-1990. Zentbl. Bakteriol. 277:389-402. [DOI] [PubMed] [Google Scholar]
- 69.Woodward, J. M., I. D. Connaughton, V. A. Fahy, A. J. Lymbery, and D. J. Hampson. 1993. Clonal analysis of Escherichia coli of serogroups O9, O20, and O101 isolated from Australian pigs with neonatal diarrhea. J. Clin. Microbiol. 31:1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, X. Y., T. Chapman, D. M. Gordon, D. N. Thuy, S. Driesen, M. J. Walker, and J. Chin. 2003. Molecular virulence gene typing of clinical E.coli isolates from pigs with post weaning diarrhea, p. 59. In J. E. Paterson, Manipulating pig production. Australasian Pig Science Association, Werribee, Victoria, Australia.
- 71.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.