Abstract
Approximately 70% of sequenced bacterial genomes contain prophage-like structures, yet little effort has been made to use this information to determine the functions of these elements. The recent genomic sequencing of the marine bacterium Silicibacter sp. strain TM1040 revealed five prophage-like elements in its genome. The genomes of these prophages (named prophages 1 to 5) are approximately 74, 30, 39, 36, and 15 kb long, respectively. To understand the function of these prophages, cultures of TM1040 were treated with mitomycin C to induce the production of viral particles. A significant increase in viral counts and a decrease in bacterial counts when treated with mitomycin C suggested that prophages were induced from TM1040. Transmission electron microscopy revealed one dominant type of siphovirus, while pulsed-field gel electrophoresis demonstrated two major DNA bands, equivalent to 35 and 75 kb, in the lysate. PCR amplification with primer sets specific to each prophage detected the presence of prophages 1, 3, and 4 in the viral lysate, suggesting that these prophages are inducible, but not necessarily to the same level, while prophages 2 and 5 are likely defective or non-mitomycin C-inducible phages. The combination of traditional phage assays and modern microbial genomics provides a quick and efficient way to investigate the functions and inducibility of prophages, particularly for a host harboring multiple prophages with similar sizes and morphological features.
Each drop of seawater contains nearly 1 million virus-like particles (VLPs) (3, 33). It has been suggested that marine viruses can influence the biomass, population structure, and genetic diversity of the microbial community (31, 35, 38, 40). However, in addition to VLPs, there are also many hidden viruses that are not present as free particles; rather, these “invisible” viruses integrate their DNAs into host chromosomes, becoming part of the bacterial life cycle. The integrated viral DNA, referred to as a prophage, can be induced to become a free phage particle under certain circumstances (17). It is believed that well over 50% of bacteria isolated from various environments contain prophages or are lysogenized (1). In the past 50 years, extensive work has been done to understand lysogeny in medically or industrially important bacteria (7). In contrast, only limited studies have focused on the induction of prophages from marine bacteria (19, 20, 22, 27). This is surprising, as more than 40% of culturable marine bacteria contain inducible prophages (22). The traditional and most common approach to studying prophages or temperate phages is to induce lysogenic bacteria with mitomycin C treatment or UV exposure (2, 18, 34). In most cases, bacteria isolated from environmental sources are induced without prior knowledge of the presence or absence of a prophage(s). Therefore, it is difficult to know the frequency of environmental bacteria harboring prophages simply through examination of the results from blind inductions.
The rapid increase in the number of microbial genome sequences in the databases has recently made it possible to rapidly and effectively investigate the distributions of prophages in one or more bacterial genomes. A review of the literature by Canchaya et al. (7) reported that more than 70% of bacterial genomes investigated contain prophage sequences of >10 kb, and bacteria may contain multiple prophages that constitute a substantial part of their genome (8, 30). Some prophages may contain morons or lysogenic conversion genes that could change the phenotype or enhance the ecological fitness of lysogens (5, 10). Bacterial viruses represent one of three major mobile genetic elements that contribute significantly to horizontal gene transfer in bacterial genomes (4, 13). It is noteworthy that prophages identified solely by genomic examination may be inducible or defective prophages, remnants of intact prophage genomes, or even isolated viral genes (7). In addition, existing prophages may not be inducible due to the lack of the correct inducing agent. In all cases, biological experiments designed to test the nature of the prophages are necessary and are the only means to confirm the activity of a specific prophage.
We demonstrate here a new strategy for investigating the inducibility of prophages based on knowledge of the genomic sequence of the marine bacterium Silicibacter sp. strain TM1040. Strain TM1040, originally isolated from a culture of the dinoflagellate Pfiesteria piscicida, is a member of the Roseobacter clade of the alpha-proteobacteria that interacts in a symbiotic relationship with its dinoflagellate host (26). The Roseobacter group is ubiquitous and comprises 10 to 20% of coastal and oceanic bacterioplankton (6, 14-16, 28, 37). Recently, the genome of TM1040 was sequenced and annotated (28; http:/img.jgi.doe.gov). Five putative prophages have been identified in the TM1040 genome (Table 1), prompting questions about their activity and potential for being induced to form phage particles.
TABLE 1.
Five putative prophage genomes found in the genome of Silicibacter sp. strain TM1040 and specific PCR primer sets for each prophage
| Prophage | Contig | Start position- stop position | Size (kb) | No. of genesa | Primer name, sequence (5′ to 3′) | Amplicon size (bp) |
|---|---|---|---|---|---|---|
| 1 | 56 | 41667-115296 | 73.6 | 90 | Pro1-f1, CGCCAAGGTGAAATCAGAG | 173 |
| Pro1-r1, TCGGGTATGAGTCGTGTG | ||||||
| 2 | 52 | 346886-376362 | 29.5 | 15 | Pro2-f2, CGTTGTCAAACCAGGTCCTC | 300 |
| Pro2-r2, CCAGTTCGGCTTCATCTACC | ||||||
| 3 | 56 | 413936-453119 | 39.2 | 47 | Pro3-f1, GCATCCGTTGACGAAACAG | 346 |
| Pro3-r1, CACCGAGTTGAGGAAAGCC | ||||||
| 4 | 55 | 219536-255560 | 36.0 | 55 | Pro4-f1, AGACACAAGCACAGAGGCG | 474 |
| Pro4-r1, TCAGGGGCGATGAAGTTAG | ||||||
| 5 | 55 | 522081-536660 | 14.6 | 17 | Pro5-f1, AAGGATTGGACCGACTATGC | 298 |
| Pro5-r1, CAAACTCAAAGCCAGCAC |
The estimated numbers of genes are based on open reading frames, as detected by DOE Joint Genome Institute software.
In this study, the TM1040 prophages were induced with mitomycin C, and induced viral particles were enumerated by SYBR gold staining. The presence of induced phage was examined using multiple approaches, including microscopic observation, genome analysis of free phage, and PCR detection of a specific prophage gene. The combination of traditional phage tools and the genomic sequence of the host provides a valuable strategy for determining the inducibility of prophages detected in silico.
MATERIALS AND METHODS
Induction of TM1040 with mitomycin C.
TM1040 was grown in marine broth 2216 (Difco) at 28°C with a shaking speed of 150 rpm throughout the induction experiment. Ten milliliters of exponentially growing TM1040 culture (optical density at 600 nm [OD600], ca. 1.0) was transferred to 200 ml fresh marine broth and incubated until the OD600 reached 0.4. The culture was then split equally into two flasks (100 ml in each), with one receiving mitomycin C (final concentration, 0.5 μg/ml) and the other serving as a control. After the treatment with mitomycin C for 30 min (32), cells in both the control and treatment groups were washed twice by centrifugation at 7,500 × g for 10 min (Beckman JA-14 centrifuge) and resuspended in 100 ml fresh marine broth. Both control and treatment cells continued to be incubated and monitored by the OD600. Subsamples (1.5 ml) were taken every 2 h for the first 12 h and then at 15, 27, 39, 55, and 78 h. Samples were fixed immediately with 1% paraformaldehyde and kept at 4°C in the dark.
Viral particle counts with SYBR gold.
The abundance of viral particles and bacterial cells at each sampling point was enumerated by epifluorescence microscopy according to the method described by Chen et al. (9). Briefly, 100 μl of fixed sample was filtered onto a 0.02-μm-pore-size 25-mm Anodisc membrane filter (Whatman) under vacuum pressure at ca. 10 mm Hg. The cells were stained with 2.5× SYBR gold solution for 10 min in the dark. Bacterial cells and viral particles were counted under blue excitation (485 nm) on a Zeiss Axioplan epifluorescence microscope. At least 200 bacterial cells or viral particles were counted per sample in 10 to 20 randomly chosen fields.
CsCl purification of induced phage.
One liter of induced viral lysate was centrifuged at 10,000 × g in a Beckman J2-21 centrifuge (Beckman Coulter, Inc., Fullerton, CA). The supernatant was filtered through a 0.45-μm-pore-size filter (type HA; Millipore) to remove host cells and cellular debris. Phage particles in the filtrate were treated with polyethylene glycol 8000 (final concentration, 100 g liter−1) overnight at 4°C. The phage particles were precipitated by centrifugation at 30,000 × g in a Beckman JA-21 rotor for 1 h. The pellet was resuspended with 6 ml SM buffer (10 mM NaCl, 50 mM Tris, 10 mM MgSO4, and 0.1% gelatin) and incubated overnight at 4°C. The phage suspension was added to CsCl to a final concentration of 0.5 g ml−1 and centrifuged for 24 h at 200,000 × g, using a T-8100 rotor in a Sorvall Discovery 100S centrifuge. The visible viral band was extracted with a 22-gauge syringe needle and then dialyzed twice in SM buffer overnight at 4°C. The CsCl-purified phage lysate was stored at 4°C until further analysis.
Extraction of phage DNA.
CsCl-purified phage were first treated with a proteinase K cocktail (100 μg ml−1 proteinase KI, 50 mM Tris, 25 mM EDTA, and 1% [final concentration] sodium dodecyl sulfate) at 55°C for 3 h. The phage DNA was extracted using phenol and chloroform (23), and the DNA pellet was dissolved in TE buffer (10 mM Tris, 1 mM EDTA) and stored at 4°C.
Transmission electron microscopy (TEM).
One drop of CsCl-purified phage lysate was left on a 200-mesh Formvar/carbon-coated copper grid for 15 min. The phage adsorbed on the grid were stained with 0.5% aqueous uranyl acetate for ca. 30 s and examined with a Zeiss CEM902 transmission electron microscope operated at 80 kV (University of Delaware, Newark). Images were taken using a Megaview II digital camera (Soft Imaging System Corp., Lakewood, CO).
Plaque assay.
Plaque assays were done according to a protocol described elsewhere (36).
PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis, including gel plug preparation and proteinase K treatment, followed the method described by Wommack et al. (41). PFGE was performed using a CHEF DR-III clamped homogeneous electric field system (Bio-Rad, Richmond, Calif.) with a 1% agarose gel, a 1- to 25-s pulse ramp, an electrophoresis rate of 6.0 V/cm with an included angle of 120° at a constant temperature of 14°C, and a run time of 24 h. Gels were stained with SYBR gold (Molecular Probes, Eugene, OR) and visualized with a Kodak EDAS 290 gel documentation system (Eastman Kodak Company, New Haven, CT).
PCR amplification of prophage DNA.
Five different PCR primer sets corresponding to each of the five prophage genomes were designed based on the unique genes found in each prophage (Table 1). Two primer sets, one (338f and 907r) based on conserved regions of the bacterial 16S rRNA gene (29) and the other based on the virD4 gene of Silicibacter sp. strain TM1040, were also included to detect the presence of host genomic DNA. PCR amplification was performed in a 25-μl volume containing 1× reaction buffer (Promega, Madison, WI) with 1.5 mM MgCl2, a 100 μM concentration of each deoxynucleoside triphosphate, 10 pmol of each primer, 1 U Taq DNA polymerase (Promega), and 5 to 10 ng phage DNA as the template. The PCR program for all reactions included an initial denaturing step at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, annealing at 50°C for 30 s, and 72°C for 1 min. In addition, a multiplex PCR that contained all five prophage primer sets (equally mixed) was also conducted under the PCR conditions described above.
Identification of prophages in silico.
The complete Silicibacter sp. strain TM1040 genome sequence is available under GenBank accession number NZ_AAFG00000000. Functional assignments were made to open reading frames (ORFs) that had BLAST E values of e−35 or less. The beginning and end of a specific prophage genome were determined by the following two criteria: homology of the specific ORF to known phage genes and the likelihood that the ORF was part of an operon containing other ORFs with homology to known phage genes.
RESULTS
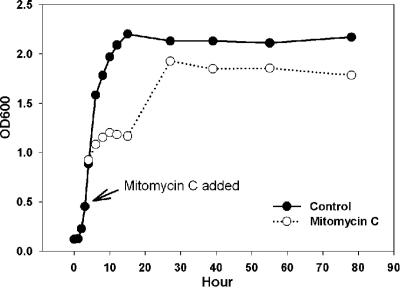
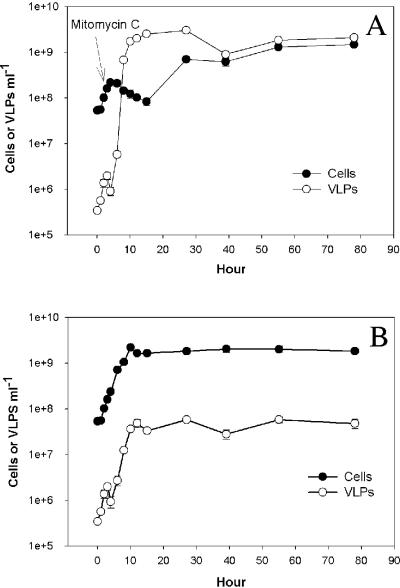
Exponentially growing cultures of Silicibacter sp. strain TM1040 were briefly exposed to the prophage-inducing chemical mitomycin C. The growth of Silicibacter sp. strain TM1040 was inhibited in the first 12 h after the addition of mitomycin C and then increased exponentially to a level close to the control level (Fig. 1). Dramatic increases in the number of VLPs (from 106 to 109/ml) and the virus-to-bacterium ratio (from 0.004 to 30) were evident within 8 h of mitomycin treatment (Fig. 2A). It was noticed that the volume of TM1040 cells increased three- to fourfold during this period (data not shown). Autoinduction of VLPs in the control appeared to be parallel to the density of host cells and remained below 108 VLPs/ml (Fig. 2B).
FIG. 1.
Effect of mitomycin C treatment on growth of Silicibacter sp. strain TM1040. Cell densities were determined by measuring the optical densities at 600 nm in a culture treated with 0.5 μg/ml mitomycin C for 30 min and in a control culture without treatment.
FIG. 2.
Viral particle yield following mitomycin C induction of Silicibacter sp. strain TM1040. Microscopic counts of TM1040 cells and viral-like particles were done with (A) a mitomycin C-treated culture and (B) a control culture without mitomycin C.
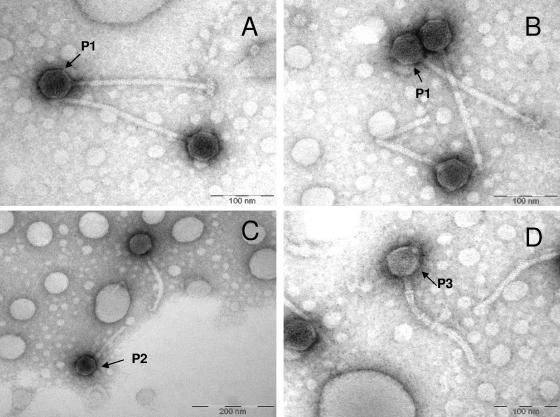
TEM examination indicated that the viral lysate was dominated by one type of siphovirus (P1) (Fig. 3A and B). The head size and tail length of phage P1 were 55 ± 1.5 and 211 ± 17 nm (n = 10), respectively. Occasionally, phages with slightly different sizes and shapes (i.e., P2 and P3) were observed (Fig. 3C and D). It was difficult to identify different types of induced phages by TEM due to the lack of distinct phage head and tail morphology. The lysates were also used in a plaque assay with TM1040 but did not yield visible plaques. Therefore, linking the phages observed by TEM to the multiple prophages found by genomic analysis was not possible by these means.
FIG. 3.
TEM images of siphoviruses found in the lysate obtained from Silicibacter sp. strain TM1040 induced with 0.5 μg/ml mitomycin C. A P1-like siphovirus dominated the viral lysate (panels A and B), while other similar siphoviruses, such as P2 (panel C) and P3 (panel D), were found in the lysate at a low frequency.
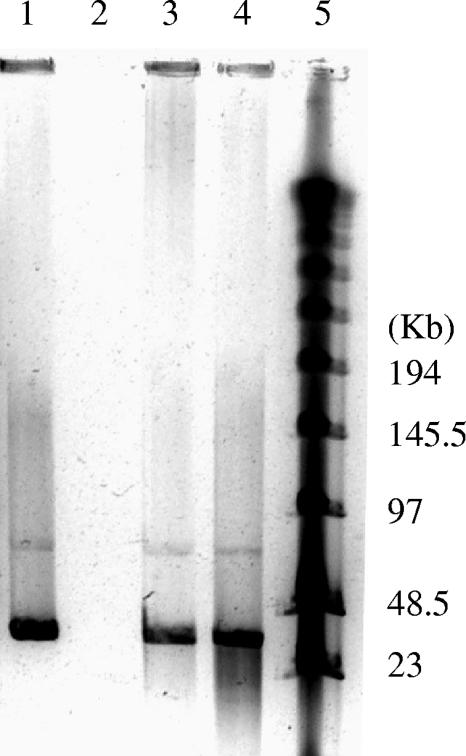
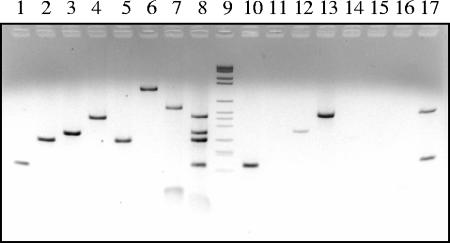
PFGE analysis revealed that two DNA bands (ca. 75 and 35 kb) came from double-stranded DNA phages (Fig. 4). The 75-kb band is weak and likely represents prophage 1. The 35-kb band may contain more than one inducible phage because the predicted genome sizes of prophages 3 and 4 are close to this range (Table 1). PCR amplification of mitomycin C-treated samples yielded three bands corresponding to the expected sizes of prophages 1, 3, and 4 (Fig. 5, lanes 10, 12, and 13). The relative yield of prophage 3 was much less than that of prophages 1 and 4 (Fig. 5), i.e., the intensity of the DNA band corresponding to prophage 3 was weaker. The nucleotide sequences of these PCR amplicons were identical to those of prophages 1, 3, and 4, indicating that all three prophages are present in the induced viral lysate. PCR using eubacterial primers or TM1040-specific primers produced a product only when DNA from TM1040 was used as a template (Fig. 5, lanes 6 and 7), not when CsCl-purified viral lysate served as a template (Fig. 5, lanes 15 and 16), suggesting that the DNAs of CsCl-purified phages were clean and free of host DNA. Multiplex PCR with combined prophage primers was able to detect at least four prophages in the host DNA (Fig. 5, lane 8) and two prophages (prophages 1 and 4) in the viral lysate (Fig. 5, lane 17). It is noteworthy that the PCR products from prophages 2 and 5 were both about 300 bp and therefore likely to have formed a single band in the multiplex PCR.
FIG. 4.
PFGE analysis of viral genomic DNAs isolated from the induced viral lysate. From left to right, the lanes contain the following: 1, DNA obtained from the induced phage particles; 2, phage DNA with DNase treatment; 3, phage DNA with RNase treatment; 4, phage DNA with S1 nuclease treatment; and 5, size markers (New England Biolabs).
FIG. 5.
PCR detection of the five prophages. Silicibacter sp. strain TM1040 genomic DNA and DNAs obtained from the induced viral lysate were each used as templates for PCR amplification using five primer sets specific for the respective prophages (Table 1). Lanes 1 to 5 show the PCR products of prophages 1 to 5, respectively, amplified from the TM1040 genomic DNA, while lanes 10 to 14 show the results of PCR for prophages 1 to 5, respectively, amplified from CsCl-purified viral lysate. Positive PCR control reactions for host DNA contained Silicibacter sp. strain TM1040 genomic DNA as a template and PCR primer sets designed to amplify virD4 (lane 6) and the 16S rRNA gene (lane 7), respectively. Control reactions to ensure that viral lysates were free of host genomic DNA contamination contained viral lysate DNA as templates and either virD4 (lane 15) or 16S rRNA gene (lane 16) primers. Lanes 8 and 17 show the results of multiplex PCRs using all five prophage primer sets with host DNA and viral lysate, respectively. Lane 9 contains DNA size markers (2,178, 1,766, 1,230, 1,033, 653, 517, 453, 394, 298, 234/220, and 154 bp, from top to bottom of gel).
DISCUSSION
We took advantage of the knowledge of the TM1040 genome sequence to determine if the prophage elements observed could be induced to produce phage particles. The results clearly show that a large amount of phage particles was released from TM1040 in the first 8 h after treatment with mitomycin C. The yield of VLPs with treatment was about 30- to 50-fold higher than the control yield. Although three of the five putative prophages were detected by PCR in the induced lysate, it is not known whether these prophages are induced at different levels. PCR also detected the presence of prophage 1 in the control at 8 h (data not shown), suggesting that prophage 1 is spontaneously induced to form viral particles at low levels without mitomycin C treatment.
It is often difficult to differentiate between multiple species of phages with similar genome sizes and morphologies by visual inspection using TEM. In our case, the morphologies of the majority of phage particles indicated that they were siphoviruses (often the case for temperate phages), with only slight differences in head and tail dimensions. Therefore, TEM observations are unlikely to provide information about which five species of prophage are present in the viral lysate. The combination of traditional phage assays with molecular genetic tools, however, does provide a new strategy for studying the role of prophages in microbial genomes. Many environmental factors (e.g., pollutants, sunlight, nutrients, etc.) may act to trigger the induction of prophages in marine ecosystems (11, 12, 21, 24, 25, 39). Low-level induction or autoinduction of prophages under natural environmental conditions has not been well studied. PCR and quantitative PCR with prophage-specific primers are particularly useful for exploring the impact of environmental factors on the production of induced phages.
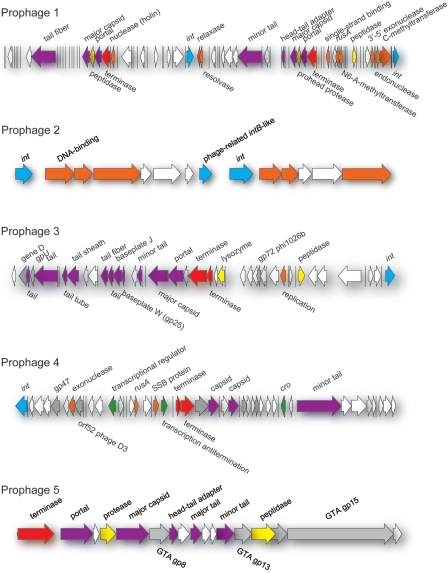
Our genomic comparison suggests that the TM1040 prophages share very limited homology (Fig. 6). They are diverse in terms of the sizes of their genomes, the numbers and sizes of their ORFs, and the amino acid sequences of their ORFs. Among the genes contained within each prophage genome, only the integrase (int) gene, the terminase gene, and a major capsid protein gene are found in all three of the inducible prophages. However, these ORFs share no homology with one another in either their nucleotide or amino acid sequence. Close examination of the genome of each prophage suggests that the inducible phages contain more ORFs than the noninducible ones (Table 1). Prophage 2 contains three int genes and several genes involved in DNA metabolism. Prophage 5 contains phage genes responsible for termination and lysis and several phage structural genes, including one phage capsid protein gene, but does not contain an int gene. Judging by the compositions and sizes of their genomes, it is likely that prophages 2 and 5 are remnants of prophages and are not inducible by mitomycin C. Therefore, induction assays will remain an important component of studies aimed at understanding the functional genomics of prophages identified by in silico genomic analyses.
FIG. 6.
Maps of the five Silicibacter sp. strain TM1040 prophage genomes. The relative sizes and directions of transcription of the individual ORFs in each prophage genome are indicated by the arrows. The following colors are used to represent the potential functions of ORFs, as defined by BLASTP homology E values of e−35 or less: blue, integrase; orange, DNA metabolism; green, regulation; red, terminase; yellow, lytic functions; and purple, structural components (head, tail, tail fibers, etc.). ORFs with E values of >e−35 with homology to known phage genes in the database are indicated by gray arrows. White arrows indicate ORFs encoding hypothetical or conserved proteins without known functions. Vertical bars within the genome map symbolize small ORFs of unknown function. The genetic neighborhood surrounding each prophage genome contains the following flanking ORFs. The left side of prophage 1 is flanked by a hypothetical ORF separated from the prophage genome by 1.48 kb. The right side is flanked by three ORFs associated with purine metabolism (TPR protein gene, putR, and putA). The left side of prophage 2 is flanked by a hypothetical zinc-binding protein gene, while the right side is flanked by an ORF coding for a hypothetical protein. Prophage 3 is flanked on its left by an operon carrying genes involved in branched-chain amino acid ABC transport, and its right side lies adjacent to an ORF encoding 4-hydroxybenzoyl-coenzyme A thioesterase. Prophage 4 is flanked by a lon ortholog on its left side and an ortholog of the transcriptional regulatory protein gene tcsR on its right. A conserved hypothetical protein gene flanks the left side of prophage 5, while an ortholog of the gene for cold shock-like protein (CspE) flanks its right side. The prophage genomic maps were generated from the genomic sequences by using Vector NTI (Invitrogen) and Adobe Illustrator software.
Currently, no uniform criteria have been established for identifying prophages in bacterial genomic sequences (7). In general, searching for int genes and phage-like genes in the vicinity of the int gene is a practical way to detect a prophage. However, the actual biological activity of putative prophages still needs to be proved based on phage induction experiments because the majority of prophages in bacterial genomes may be defective and apparently in a dynamic process of gradual genetic decay (7). A defective prophage may be mutated or truncated prophage DNA which is physically linked on a bacterial genome.
Microbial genome sequencing projects are producing a wealth of new information on the potential roles of mobile genetic elements, such as plasmids, phages, and transposable elements, in prokaryotes (4, 13). The TM1040 prophage genomes account for about 5% of the host genome. The role of these prophages in horizontal gene transfer (HGT) has not yet been estimated. It will be interesting to know whether the HGT frequency via viral transduction is higher in a host that carries multiple prophages than in a host with one prophage. In aquatic environments, where a large portion of bacterioplankton is free-living, HGT by transduction could be more significant to the survival of the cells than other HGT mechanisms, such as conjugation or transformation, which usually require cell-to-cell contact. Much can be learned from Silicibacter sp. strain TM1040 and its prophages with respect to the roles that phage induction and viral-mediated HGT play in microbial diversification in the marine ecosystem.
Acknowledgments
We gratefully acknowledge support for this research from the National Science Foundation (MCB-0132070 and MCB-0238515 to F.C. and MCB-0537041 to R.B.).
We thank M. A. Moran and K. Williamson for reading and editing the manuscript and for insightful discussions. We thank Alla Lapidus and Paul Richardson of DOE JGI for nucleotide sequencing and automated annotation of Silicibacter sp. strain TM1040.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. Viruses of prokaryotes: general properties of bacteriophages. CRC Press, Inc., Boca Raton, Fla.
- 2.Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 3.Bergh, O., K. Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 4.Bordenstein, S. R., and W. S. Reznikoff. 2005. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3:688-698. [DOI] [PubMed] [Google Scholar]
- 5.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchan, A., J. M. González, and M. A. Moran. 2005. An overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67:238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49:277-300. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F., J. R. Lu, B. Binder, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chibani-Chennoufi, S., A. Bruttin, M. L. Dillmann, and H. Brussow. 2004. Phage-host interaction: an ecological perspective. J. Bacteriol. 186:3677-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran, P. K., C. A. Kellog, and J. H. Paul. 1998. Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar. Ecol. Prog. Ser. 164:125. [Google Scholar]
- 12.Cochran, P. K., and J. H. Paul. 1998. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microbiol. 64:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost, L. S., R. Leplae, A. O. Summers, and A. Toussaint. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722-732. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni, S. J., and M. S. Rappé. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley and Sons, New York, N.Y.
- 15.González, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman, M. E., and R. A. Weisberg. 1971. Prophage insertion and excision, p. 113-138. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Hershey, A. D., and W. Dove. 1983. Introduction to lambda, p. 3-20. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Jiang, S. C., C. A. Kellogg, and J. H. Paul. 1998. Characterization of marine temperate phage-host systems isolated from Mamala Bay, Oahu, Hawaii. Appl. Environ. Microbiol. 64:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang, S. C., and J. H. Paul. 1996. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 142:27-38. [Google Scholar]
- 21.Jiang, S. C., and J. H. Paul. 1994. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 104:163-172. [Google Scholar]
- 22.Jiang, S. C., and J. H. Paul. 1998. Significance of lysogeny in the marine environment: studies with isolates and a model of lysogenic phage production. Microb. Ecol. 35:235-243. [DOI] [PubMed] [Google Scholar]
- 23.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 25.McDaniel, L., and J. H. Paul. 2005. Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine Synechococcus species. Appl. Environ. Microbiol. 71:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moebus, K. 1983. Lytic and inhibition responses to bacteriophages among marine bacteria, with special reference to the origin of phage-host systems. Helgoländer Meeresuntersuch. 36:375-391. [Google Scholar]
- 28.Moran, M. A., A. Buchan, J. M. González, J. F. Heidelberg, W. B. Whitman, R. P. Kiene, J. R. Henriksen, G. M. King, R. Belas, C. Fuqua, L. Brinkac, M. Lewis, S. Johri, B. Weaver, G. Pai, J. A. Eisen, E. Rahe, W. M. Sheldon, W. Ye, T. R. Miller, J. Carlton, D. A. Rasko, I. T. Paulsen, Q. Ren, S. C. Daugherty, R. T. Deboy, R. J. Dodson, A. S. Durkin, R. Madupu, W. C. Nelson, S. A. Sullivan, M. J. Rosovitz, D. H. Haft, J. Selengut, and N. Ward. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910-913. [DOI] [PubMed] [Google Scholar]
- 29.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 31.Paul, J. H. 1982. Use of Hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl. Environ. Microbiol. 43:939-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prigent, M., M. Leroy, F. Confalonieri, M. Dutertre, and M. S. DuBow. 2005. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara desert. Extremophiles 9:289-296. [DOI] [PubMed] [Google Scholar]
- 33.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 34.Roberts, J. W., and R. Devoret. 1983. Lysogenic induction, p. 123-144. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Suttle, C. A. 2005. Viruses in the sea. Nature 437:356-361. [DOI] [PubMed] [Google Scholar]
- 36.Suttle, C. A., and F. Chen. 1992. Mechanisms and rates of decay of marine viruses in seawater. Appl. Environ. Microbiol. 58:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki, M. T., C. M. Preston, F. P. Chavez, and E. F. DeLong. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in the Monterey Bay. Aquat. Microb. Ecol. 24:117-127. [Google Scholar]
- 38.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 39.Williamson, S. J., L. A. Houchin, L. McDaniel, and J. H. Paul. 2002. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total community analysis using pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]