Abstract
Accurate enumeration of viruses within environmental samples is critical for investigations of the ecological role of viruses and viral infection within microbial communities. This report evaluates differences in viral and bacterial direct counts between estuarine sediment samples which were either immediately processed onboard ship or frozen at −20°C and later processed. Viral and bacterial abundances were recorded at three stations spanning the length of the Chesapeake Bay in April and June 2003 within three sediment fractions: pore water (PW), whole sediment (WS), and sediment after pore water removal (AP). No significant difference in viral abundance was apparent between extracts from fresh or frozen sediments. In contrast, bacterial abundance was significantly lower in the samples subjected to freezing. Both bacterial and viral abundance showed significant differences between sediment fractions (PW, WS, or AP) regardless of the fresh or frozen status. Although pore water viral abundance has been used in the past as a measurement of viral abundance in sediments, this fraction accounted for only ca. 5% of the total sediment viral abundance across all samples. The effect of refrigerated storage of sediment viral extracts was also examined and showed that, within the first 2 h, viral abundance decreased ca. 30% in formalin-fixed extracts and 66% in unfixed extracts. Finally, the reliability of direct viral enumeration via epifluorescence microscopy was tested by using DNase treatment of WS extractions. These tests indicated that a large fraction (>86%) of the small SYBR gold fluorescing particles are likely viruses.
Numerous studies have established that viruses are bountiful in marine waters (24, 46, 48, 55), freshwater (5, 18, 39), soils (1, 52, 53), and marine sediments (12, 16, 28, 36). Previous investigations have examined the influence of soil and sediment physical characteristics on the extraction and cultivation of enteric and pathogenic viruses (2, 23, 45). Burgeoning interest in the role of viruses within soil and sediment microbial communities demands that ever more precise methodologies for the direct enumeration of total viral particles in sediments (12, 13, 28) and soils be developed (53).
In environmental samples, the identity of specific viruses is not known, and only a minute fraction of the viral community can be detected through cultivation (57). Thus, accurate enumeration of viruses and bacteria is critical to characterizing viral and bacterial dynamics and understanding the interactions of these microbes within the biosphere (21, 25, 44, 54; S. W. Wilhelm and C. A. Suttle, Program Abstr. 8th Int. Symp. Microb. Ecol., p. 352, 2000). Due to time constraints, logistics, and material availability, storage of samples is often a necessity. Thus, it is important to constrain the effects of storage on subsequent viral and bacterial enumeration from environmental samples. Sample storage issues can impact any environmental study, e.g., soils, freshwater, etc., and yet are more pronounced for marine studies due to factors such as length of cruise, distance from port, and available laboratory conditions onboard ship. A recent study by Wen et al. (50) highlighted the importance of considering sample storage conditions through the discovery that refrigerated storage of aldehyde-fixed water samples results in the rapid loss of virus particles. In most studies examining viral abundance in sediments, samples were stored at 4°C (18, 28) to 10°C (22) for hours (20, 36) to days (28) to weeks (12). The only investigation of freezing effects on porous media samples addressed the use of deep-freezing for subsampling of sewage particulates (11). To our knowledge, no studies have investigated the impact of freezing on direct observations of microorganisms in sediment samples.
The main objective of the present study was to address the effects of sample storage and processing of viral extracts on direct counts of autochthonous bacteria and viruses in estuarine sediment samples. Three additional objectives were to (i) evaluate differences in the abundance of viruses in pore water and of viruses associated with sediment particles, (ii) determine the influence of sediment physical characteristics on viral and bacterial extractability and abundance, and (iii) estimate the contribution of nonviral DNA to estimates of viral abundance in sediment samples.
MATERIALS AND METHODS
Sampling sites.
Sediments were collected during research cruises in April and June 2003 at three locations representing a range of conditions across the salinity gradient of the Chesapeake Bay: polyhaline station 724 (37°24′N, 76°05′W), mesohaline station 804 (38°04′N, 76°13′W), and oligohaline station 908 (39°08′N, 76°20′W). A four-tube multicorer (MC-400 Hedrick/Marrs; Ocean Instruments) was used to collect undisturbed sediments at water depths of 15, 25, and 9 m for stations 724, 804, and 908, respectively. Each large core was subsampled once with a sterile cutoff 60-ml syringe. Two of the subsamples were processed immediately (fresh). The other two were processed upon return to shore after being capped with neoprene rubber stoppers, sealed with electrical tape, and stored vertically at −20°C (frozen). Frozen sediments were extracted in the same manner as fresh sediments after thawing. Conditions onboard ship did not facilitate special handling of anoxic samples; therefore, all samples were extracted in ambient air. Although the change from anoxic conditions may have had an effect on viral and bacterial analysis, it was included equally across all samples and thus should not have altered the conclusions of the study.
Sediment water content and porosity were determined by using the method of Danovaro et al. (14). The University of Delaware Soil Testing Laboratory examined sediments for total organic matter, particle size, as well as the chemical composition (C, N, P, Fe, and Al). Particle sizes of the sediment fractions were categorized as follows: sand (50 to 2,000 μm), silt (2 to 50 μm), and clay (<2 μm).
Viral and bacterial extractions.
Removal and collection of pore water (PW) was done by using a 17-by-100-mm culture tube containing whole sediment (WS), sealed with a perforated cap lined with a glass fiber filter (GF/F; Pall Gelman) and inverted inside a larger 50-ml centrifuge tube. The tube-within-a-tube arrangement was centrifuged at 1,000 × g in a swing-out rotor for 20 min to collect the majority of PW. Viruses and bacteria were extracted from WS and from sediment after PW removal (AP; after pore water) using a modified method of Hewson et al. (28). Briefly, 2 ml of the top 2 cm of surface sediment was placed inside a sterile 15-ml centrifuge tube to which 8 ml of 0.02-μm-pore-size-filtered 10 mM disodium pyrophosphate and 5 mM EDTA was added. Samples were vortexed horizontally for 20 min using a multitube vortex mixer set to high. After agitation, larger particles were removed by centrifugation at 2,000 × g for 25 min. The supernatant was filtered through a 0.45-μm-pore-size Sterivex filter, and formaldehyde was then added to a final concentration of 1%. Fresh sediment extracts containing 1% formaldehyde were stored between 7 and 12 h at 4°C prior to freezing with liquid nitrogen and final storage in −80°C prior to preparation for microscopy.
To determine viral extraction efficiency, multiple extractions were performed for sediment samples at each station. A 2-ml surface sediment sample was collected and subjected to extraction as previously mentioned. The resulting supernatant was decanted and stored as described above. The remaining sediment pellet was resuspended in the manner described above and subjected to repeated extractions three more times.
Viral and bacterial enumeration.
For microscopic enumeration, all extracts were prepared using a modified method of Chen et al. (9). For total viral abundance, sediment viral extract was filtered through a 25-mm, 0.02-μm-pore-size filter (Anodisc; Whatman International, Ltd.), backed with a 0.20-μm-pore-size filter (Supor; Pall Gelman) and an extra thick glass fiber filter (GF/F; Pall Gelman, catalog no. 66075) at approximately 20 kPa on a vacuum manifold. Filters were stained for 15 min in the dark with a final concentration of 2.5× SYBR Gold (Molecular Probes), which is supplied from the manufacturer as a 10,000× concentrate (37). Filters were air dried, topped with an antifade solution (37), and pressed with a weighted glass pane to flatten the filter under the coverslip, removing any wrinkles or bubbles. Ten microscope fields of view and at least 200 virus and bacteria particles were imaged by using a Zeiss Axioscope-2 epifluorescence microscope (Carl Zeiss, Inc.) with a ×100 Plan Neofluar oil objective lens under fluorescein isothiocyanate excitation. Each field was digitally captured by using an ORCA-ER camera (Hamamatsu Corp.), taken in a 12-bit Tiff format, and enumerated by using the FoVea Pro (Reindeer Software) plug-in with Adobe Photoshop. Viruses were identified as small green fluorescent spheres and could be differentiated from bacteria based on the relative size and fluorescence intensity. Abundances were calculated based on the method of Noble and Fuhrman (37), and statistical calculations were determined by using Minitab (release 14.12.0; Minitab, Inc.) and SPSS (version 11.0.2; SPSS, Inc.).
Virus enumeration accuracy.
Since SYBR gold stains all double-stranded DNA (as well as other nucleic acids), it is possible that some small fluorescent particles within the virus-sized fraction were not viruses. To determine the accuracy of viral counts, WS extracts from stations 908 and 804 were treated with RNase-free DNase (origin, bovine pancreas; Promega), with heat (98°C for 15 min), and with heat followed by DNase. For the DNase treatments, 1 U of DNase was added to a total volume of 100 μl of extractant, followed by incubation at room temperature for 30 min. Next, 1 μl of stop solution (20 mM EGTA [pH 8.0]) was used to inactivate the DNase. All samples were then prepared and enumerated as previously described.
Effects of refrigerated storage on sediment viral extracts.
Because the negative effects of aldehyde fixates and storage at 4°C on viral direct counts from water and sediment samples are known (8, 12, 50), an experiment was undertaken to determine whether and to what extent these storage conditions would have affected viral direct counts in sediment extracts. The top 2 cm of sediment in each multicorer tube was collected (ca. 100 g of sediment) and mixed thoroughly to ensure a homogenous blend. Sediments were extracted in the same manner as previously described. After extraction, the pooled supernatant was divided into aliquots in four 50-ml conical centrifuge tubes. Two of these aliquots were fixed with a final concentration of 1% formaldehyde (fixed replicates), and the other two were not fixed (unfixed replicates). All replicates were stored at 4°C for 21 d. At each time point, a 1.5-ml subsample from each 50-ml replicate was collected, flash frozen in liquid nitrogen, and stored at −80°C until slide preparation. Slides were prepared as described above for each time point. A curve fit based on r2 values was estimated for viral decay over short-term (<10 h) and long-term (10 h to 18 days) storage by using SPSS (version 11.0.2; SPSS, Inc.).
RESULTS
Freezing effects.
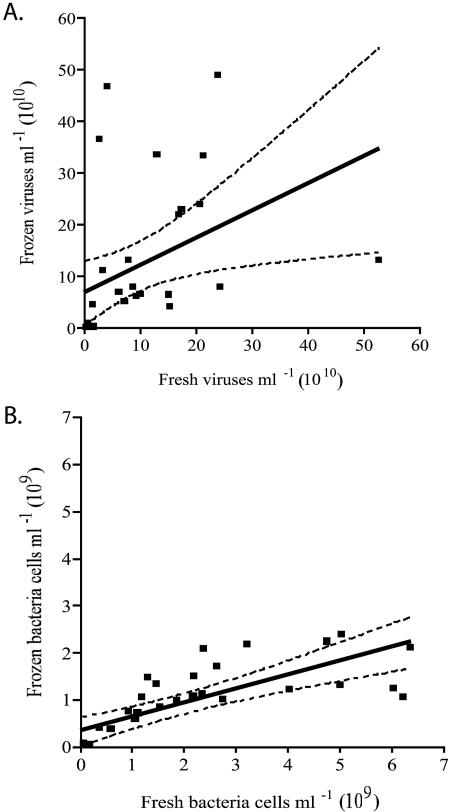
Paired two-tailed t tests showed that the difference in viral abundance between freshly extracted sediment and sediments frozen prior to extraction was not significant at a 95% confidence interval (CI; P > 0.05), regardless of sediment fraction (PW, WS, or AP). The slope of a linear regression applied to paired samples of frozen versus fresh viral counts was 0.6 (P = 0.02, r2 = 0.2) (Fig. 1A). The scatter plot did indicate two unusual datum points (not included on the graph due to distance from the regression data) in which the ratio of fresh to frozen viral counts was greater than 3. These outliers were removed and not included in the regression. In contrast, regression analysis of the bacterial densities in freshly extracted sediments showed that they were significantly higher (P < 0.01) than those found in frozen sediments (slope = 0.3, r2 = 0.6) (Fig. 1B), and paired two-tailed t tests showed that the difference in bacterial abundance between freshly extracted sediment and those frozen prior to extraction was significant at a 95% CI (P = 0.01).
FIG. 1.
Regression analysis of virus (slope = 0.6, r2 = 0.2, P = 0.02) (A) and bacterium (slope = 0.3, r2 = 0.6, P < 0.0001) (B) abundances comparing fresh to frozen storage conditions. The CI values are based on original abundances and are shown at a 99% CI.
Viral and bacterial abundance.
Enumeration of certain sediment sample extracts via epifluorescence microscopy was initially problematic. Sediments from stations with a higher clay and humic content frequently required serial dilution (1:1,000) to reduce the high background fluorescence which initially interfered with enumerations, as seen in other studies (27). Once diluted, samples were more easily enumerated.
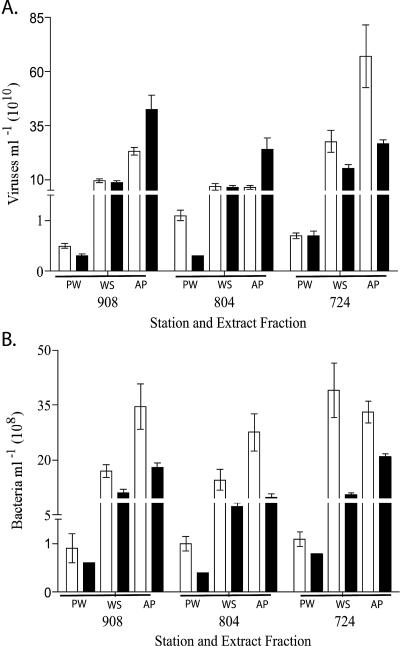
Viral abundances between April 2003 and June 2003 sampling dates at each station were not significantly different from one another. The average viral abundance for freshly extracted PW samples ranged from 4.6 × 109 viruses ml−1 (station 908) to 1.1 × 1010 viruses ml−1 (station 804). Frozen viral extracts showed average abundances in PW of from 3.0 × 109 viruses ml−1 (station 804) to 6.7 × 109 viruses ml−1 (station 724) (Fig. 2A). In addition, PW viral abundance, regardless of storage condition, was significantly lower (P < 0.05) than viral abundances within either WS or AP extracts, contributing ca. 5% to the total viral abundance seen in WS extracts. For freshly extracted WS, average viral abundance ranged from 6.7 × 1010 viruses ml−1 (station 804) to 2.8 × 1011 viruses ml−1 (station 724), whereas average viral abundances from frozen WS extracts ranged from 6.4 × 1010 viruses ml−1 (station 804) to 1.5 × 1011 viruses ml−1 (station 724) (Fig. 2A). Surprisingly, average viral abundances for AP extracts from fresh and frozen sediments were frequently ca. 20% higher than WS viral abundances.
FIG. 2.
Mean viral (A) and bacterial (B) abundances according to sediment fraction and storage condition. Bars: □, freshly extracted samples; ▪, samples frozen prior to extraction.
Bacterial abundance within fresh PW samples averaged from 9.3 × 107 bacteria ml−1 (station 908) to 1.1 × 108 bacteria ml−1 (station 724), whereas bacterial abundance within pore water from frozen sediments ranged from 4.3 × 107 bacteria ml−1 (station 804) to 7.5 × 107 bacteria ml−1 (station 724) (Fig. 2B). Average bacterial abundances within freshly extracted WS ranged from 1.5 × 109 bacteria ml−1 (station 804) to 3.9 × 109 bacteria ml−1 (station 724). In WS extracts from frozen cores, the bacterial abundance ranged from 7.4 × 108 bacteria ml−1 (station 804) to 1.1 × 109 bacteria ml−1 (station 908) (Fig. 2B). Bacterial counts showed significant differences between sediment fractions (PW, WS, and AP) and storage conditions (P < 0.01). As with viruses, the contribution of free bacteria in sediment PW to total bacterial abundance was significantly less than WS or AP fractions (P < 0.05, one-way analysis of variance [ANOVA]). Virus-to-bacteria ratios (VBR) for freshly extracted cores averaged 70 for WS samples, 80 for PW samples, and 153 for AP samples. Conversely, due to the negative impact of freezing on bacterial counts, frozen extract average VBRs were much higher for WS and AP fractions (116 and 243, respectively), and PW ratios were similar to fresh extracts, with a VBR of 70.
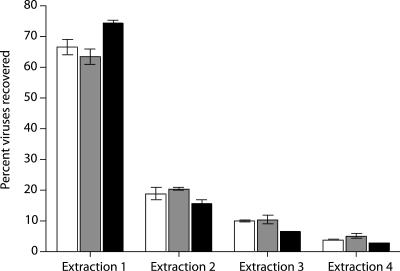
The efficiencies of viral extraction using sodium pyrophosphate and EDTA with vortexing were 67, 64, and 75% for stations 908, 804, and 724, respectively, for the first extraction (Fig. 3). Additional extractions eluted ca. 20% for stations 908 and 804 and 15% for station 724, whereas a third extraction eluted 7% (station 724) to 10% (stations 908 and 804). A fourth and final extraction resulted in <5% recoverable viruses for each station.
FIG. 3.
Effect of multiple extractions on viral recovery from Chesapeake Bay sediment samples. The percentages of viruses recovered are based on summed viral abundances from all four extractions. Extraction results from stations 908 (□), 804 (░⃞), and 724 (▪) are shown.
Station characteristics.
The salinity of the benthic boundary water at the three stations varied from oligohaline (station 908, 4.8 to 8.8 parts per thousand [ppt]) to mesohaline (station 804, 17.1 to 21.1 ppt) to polyhaline (station 724, 21.4 to 26.6 ppt). Temperatures of sediment samples at time of collection ranged from 5 to 17°C. Sediment samples from stations 908 and 804 contained similarly high proportions of silt and clay, whereas sediments from station 724 contained more sand with less clay content (Table 1) . Station 804 sediment samples contained the highest percentage of organic matter (OM), whereas the lowest percentage of OM was found at the lower bay station 724 (Table 1). Bivariate analysis of fresh sample viral abundances and sediment characteristics determined by Spearman's rank correlation showed a negative relationship between viral abundance and clay content (r2 = −1.0, P < 0.01) and a positive correlation between viral abundance and sand content (r2 = 1.0, P < 0.01). In addition, viral abundance was negatively correlated to porosity and water content (r2 = −0.886, P < 0.05). Chemical analyses, including percent carbon, nitrogen, and trace elements, were completed for each station as well (data not shown). WS viral abundances were significantly correlated (r2 = −0.900, P < 0.05) to K, Mg, and B; however, WS bacterial abundances showed a significant correlation (r2 = −0.900, P < 0.05) only to the total percent S content.
TABLE 1.
Sediment characteristics in surface sediments sampled to a depth of 2 cm
| Station | Mean (SE) |
||||||
|---|---|---|---|---|---|---|---|
| WCa (%) | Porosity | Sand (%) | Silt (%) | Clay (%) | pH | OMb (%) | |
| 908 | 53 (0.02) | 0.7 (0.02) | 12 (8.5) | 47 (6.6) | 41 (6.3) | 7.0 (0.18) | 5.3 (0.20) |
| 804 | 60 (0.03) | 0.8 (0.02) | 22 (7.7) | 44 (5.9) | 34 (3.5) | 7.0 (0.20) | 7.0 (0.20) |
| 724 | 42 (0.02) | 0.6 (0.02) | 43 (5.9) | 39 (5.1) | 18 (1.9) | 7.4 (0.09) | 1.3 (0.11) |
WC, water content.
As determined by loss of weight on ignition.
Viral enumeration accuracy.
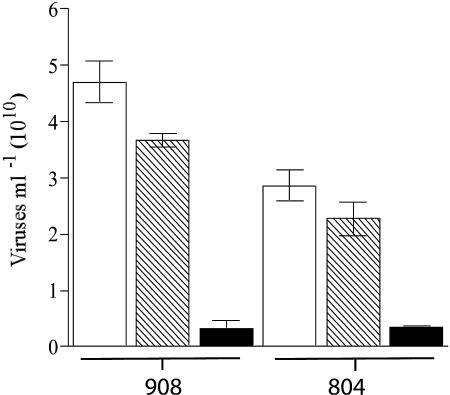
No significant differences were found between the number of viruses in untreated extracts and viruses in DNase-treated extracts (P < 0.05). Viral estimates for the heat-plus-DNase-treated viral extract were significantly lower than for the DNase-only treatment and the untreated control (P < 0.01). The fractions of virus-like particles that remained visible after heat-plus-DNase treatment were 7.7% (station 908) and 13.9% (station 804) of the WS total counts (Fig. 4). Because these particles survived the heat-DNase treatment, they cannot be operationally defined as viruses.
FIG. 4.
DNase test on WS samples: untreated viral extracts (□), DNase-treated viral extracts (▧), and heated and DNase-treated viral extracts (▪).
Effects of refrigerated storage on sediment viral extracts.
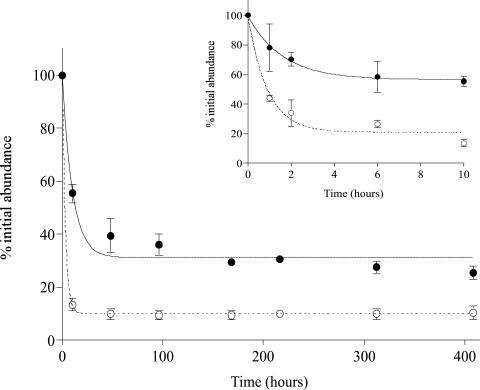
For both fixed and unfixed sediment extracts stored at 4°C, viral abundances decreased considerably during the 21-day incubation period. Within 2 h, viral abundance in the fixed sample decreased by 29% ± 7%), whereas the abundance in the unfixed sample decreased by 66% ± 12% (Fig. 5). After 10 h, the viral abundance was only 56% ± 5% and 13% ± 3% of the original abundance for fixed and unfixed extracts, respectively. By day 17, these percentages of original abundance declined to 25 and 10%, respectively (Fig. 5). Changes in viral abundance within fixed extracts were calculated for <10 h by using the equation γ = 0.7322 + [−0.0626 × ln(h)] (r2 = 0.72) and for unfixed extracts by using the equation γ = 0.4393 + [−0.1216 × ln(h)] (r2 = 0.97), where γ is the percent remaining viruses and h is the time in hours. For times greater than 10 h, the decay of viruses for fixed extracts was modeled using the equation γ = 0.7656 + [−0.0873 × ln(h)] (r2 = 0.80). For unfixed extracts, no accurate model was attainable for samples stored at 4°C longer than 10 h. Estimated abundances in the present study were corrected by using these viral decay models.
FIG. 5.
Effects of refrigerated storage and formalin fixation loss of viral particles from sediment extracts for the first 10 h (A) and from 0 to 408 h (B). Symbols: •, fixed viral extracts; ○, unfixed viral extract samples.
DISCUSSION
Fresh or frozen.
Of the four objectives in this study, the foremost was to evaluate the effects of freezing sediment samples on subsequent estimation of the viral and bacterial abundance. The data presented here demonstrate that viral direct counts within estuarine sediment samples were not significantly affected by frozen storage; however, the overall trend did favor fresh extraction of viruses from sediment samples (Fig. 1A). In contrast and not unexpectedly, bacterial direct counts were significantly reduced in the frozen samples. Freezing bacterium samples without a cryoprotectant is known to destroy some bacteria by ice crystal disruption of cellular membranes (43). Previous studies of Swiss agricultural soils showed that freezing greatly reduced bacterial direct counts by up to 22% over nonfrozen samples (40). Not surprisingly, the VBR was higher in frozen sediment extracts than in freshly extracted sediments; this was probably due to the reduced number of bacteria in the frozen sediment samples. Thus, accurate estimation of the VBR requires the immediate processing of sediments. These results indicate that frozen storage irrevocably alters sediment bacterial abundance and is not recommended when conducting bacterial analyses of sediment, especially for cultivation studies. The use of frozen storage methods should be approached with caution when bacterial enumerations are of vital importance.
Viral abundance in estuarine sediments.
Pore water has been collected and analyzed for viral abundance within sediments of the Chesapeake Bay (16) and the Chukchi and Bering Seas (46), as well as for the presence of lytic agents in coastal sediments in British Columbia (32). The data from these previous investigations revealed that viral abundance spanned from a low of 2.7 × 107 viruses ml−1 in the Arctic (46) to a high of 4.4 × 108 viruses ml−1 in the Chesapeake (16). Viral abundances from sediment pore water in the present study were more than 10-fold higher than in previous reports (Table 2). This difference could be attributed to several factors, including location, type of stain used, and environmental conditions. For the present study, the nucleic acid stain SYBR gold was used for epifluorescence enumeration, whereas Yo-Pro-1 was used by Drake et al. (16) and transmission electron microscopy (TEM) was used by Steward et al. (46). It is well known that TEM typically yields the most conservative direct viral counts, whereas Yo-Pro 1 yields the highest estimates (6, 9, 26, 49). The low viral counts from arctic sediment samples probably reflect real differences in the abundance of viruses within this cold environment and the typical underestimation of viral abundance by TEM (49). Another important difference between the present study and the only other report of sediment viral abundance in the Chesapeake Bay (16) was sample storage. In the present study, the samples were collected, fixed with 1% formaldehyde, and snap-frozen in liquid nitrogen within 12 h of sampling prior to actual filter preparation, whereas Drake et al. (16) did not report any fixation of samples, and there is no clear explanation of the temperature or length of sample storage. In light of the data on viral decay in fixed and unfixed sediment extracts shown in Fig. 5, as well as other reports of viral loss in water (8, 50) and sediment samples (12, 17), it is likely that the PW viral abundances previously reported for Chesapeake Bay sediments were artifactually low.
TABLE 2.
Comparison of values of PW and WS extracted viruses
| Location | PW viral abundance (viruses ml−1) | WS viral abundance (viruses ml−1 and cm−3) | VBR | Counting method | Source or reference |
|---|---|---|---|---|---|
| Chesapeake Bay, United States | 2.6 × 109 to 1.4 × 1010 | 2.3 × 1010 to 3.8 × 1011 | 37-107 (PW) | SYBR gold | Present study |
| 36-102 (WS) | |||||
| Chesapeake Bay, United States | 3.6 × 109 to 5.4 × 109 | 5.14 × 1010 to 2.3 × 1011 | TEM | Present study (data not shown) | |
| California coast, United States | 2.0 × 108 to 2.45 × 109 | 10-98 | SYBR green I | 27 | |
| Kühwörter Wasser, Austria | 4.3 × 109 to 7.2 × 109 | 0.9-3.2 | SYBR gold | 20 | |
| Nivå Bay, Denmark | 4.9 × 107 to 7.5 × 108 | 1-10 | SYBR green I | 36 | |
| Alabama wetland, United States | 2.8 × 104 to 4.5 × 107 | 0.03-0.7 | SYBR green I | 18 | |
| Brisbane River, Australia | 2.4 × 108 to 2.2 × 1011 | SYBR green I | 28 | ||
| Mediterranean deep sea | 1.0 × 109 to 2.0 × 109 | 2-5 | SYBR green I | 15 | |
| Chesapeake Bay, United States | 2.2 × 108 to 4.4 × 108 | 29-85 | Yo-Pro-1 | 16 | |
| Bering and Chukchi Seas | 2.7 × 107 | TEM | 46 | ||
| Key Largo, Fla. | 1.4 × 108 to 5.3 × 108 | TEM | 38 |
Among the limited reports of viral abundance in marine sediments (Table 2), the present study is the first to assess the relative contribution of viruses within PW and attached to sediment particles. In every case, viral abundances within sediment PW comprised ca. 5% of total viral abundance in whole sediments. Since viruses within PW represent only a small fraction of the total viral community, reporting only PW viral abundance grossly underestimates sediment viral abundance and is not a particularly relevant parameter for characterizing sediment microbial communities.
According to our definitions for sediment fractions, previous sediment studies reported only viral and bacterial abundance within whole sediment (15, 28). In the present study three sediment fractions were examined and, interestingly, bacterial and viral direct counts from the freshly extracted AP fractions ranged from only 2% of the WS direct counts (station 724, April 2003) to 85% greater than the WS abundance estimations (station 724, June 2003). For frozen AP fractions, the range was similar, albeit for different stations. Although there was no statistically significant relationship of AP and WS counts, in most cases (four of six for fresh extracts, and five of six for frozen) AP viral counts were higher than WS viral counts. A possible reason for this trend of increased viral abundance in the AP fraction could stem from the removal of the pore water itself. Pore water removal may have altered the physical and chemical conditions of the sediment sample, in particular the ionic strength of the subsequent extraction solution. Sediment that contained PW and extractant solution would have had a higher ionic strength, thus promoting virus adsorption to sediment particles. PW removal would have lowered the ionic strength of the extraction, thus reducing the electrostatic interaction between viruses and sediment particles and weakening viral adsorption. To test this hypothesis, ionic strength was measured in sediment samples taken on a recent cruise. Indeed, ionic strength showed a significant (P < 0.05) decrease in AP samples versus WS samples at each station and was significantly correlated to sediment fraction (WS or AP) (P = 0.001) (data not shown). Although ionic strength seems to explain the increase in the efficiency of viral extraction after the removal of pore water, there are probably other factors involved since samples from station 724 (sandy, least clay) did not show this trend.
Viral abundances in marine sediments are typically significantly higher than those found in the overlying water column (15-17, 19, 28, 38, 46). As results of the present study show, viruses extracted from the sediments of the Chesapeake Bay are more abundant than any previously studied marine sediment environment (Table 2) such as Key Largo, Fla. (38), the California coast (27), and the Mediterranean deep sea (15). As with PW abundance, methodological differences between studies likely contributed to these observed differences. In each study, a different means of extraction was used to remove viruses from sediments, including potassium citrate with minimal shaking time (38), sodium pyrophosphate with an increased shaking time (27), and pyrophosphate with sonication (15). The present study used conditions similar to those used for viral extraction from Brisbane River estuary sediments (27) but with twice the shaking time, which may have provided for increased desorption of viruses from sediment particles. In addition, each study used a different means of enumeration, including TEM (38) and SYBR green I (15, 27). Viral abundance in the present study was assessed by using SYBR gold, which is known to have a higher quantum efficiency than SYBR green I (9) and thus may have increased our chances of detecting viruses that otherwise might have been missed.
Aside from the techniques, there are substantive differences in sediment site location and types across the limited data set of viral direct counts in aquatic sediments (Table 2). For aquatic environments, viral abundance and activity are thought to positively correspond to increasingly eutrophic conditions (47). Recent studies have found that bacterial abundance and activity (7, 10, 54) are the primary factors positively correlating to viral abundance in aquatic environments. These trends can likely be extended to benthic microbial communities. Thus, not surprisingly, a previous study of a similarly eutrophic site located in the Brisbane River, Australia (29), found correspondingly high sediment viral abundances (2.2 × 1011 viruses cm−3). Given the analogous eutrophic conditions of this location and the Chesapeake Bay, it is only reasonable that high viral abundances were seen within these similar environments.
Physical conditions of sediments.
It is often more difficult to measure viral and bacterial abundance in porous media such as soils and sediments than in water samples due to the necessity of removing viruses and bacteria from the sediment matrix. Although a priori studies have shown the effects of clay (33, 35), pH (3, 31), and organic matter (42, 56) on viral mobility and transport, none have addressed extractability. Correlation analyses between various sediment characteristics and viral and bacterial abundance may show that the physical attributes of sediments, i.e., porosity, textural qualities, pH, and organic matter composition, may contribute to observed differences in virus extractability or abundance between sediment samples.
For Chesapeake Bay sediments, correlation analysis showed that the sand and clay composition had the most significant correlations to viral abundances (P < 0.01), with lesser contributions from porosity and water content (P < 0.05). The influence of sand and clay on sediment viral abundance could be attributed to the pore volume and surface area available for viral adsorption. Sandier sediments have a smaller total pore volume and total surface area than clayey sediments, thus affecting adsorption site availability. In addition, because of its high sorptive properties, clay prevents the movement of viruses and bacteria, as well as some pollutants, and can influence the persistence of viruses within sediments and soils (4, 33). Because of these strong ionic interactions, the recovery of viruses from sediments with a higher clay content will be less efficient (30, 45).
Sediments at stations 908 and 804 had higher clay contents and lower viral and bacterial abundances than at station 724 for both fresh and frozen WS extractions. Interestingly, frozen storage of sediments from each station had different impacts on viral abundance in the WS and AP fractions. Station 724, which had the lowest clay and the highest sand content, showed 51 and 60% decreases in average viral counts within frozen samples from the WS and AP fractions, respectively. In contrast, stations 908 and 804, which had much higher clay contents, showed 70 and 46% increases, respectively, in average viral abundances of frozen AP samples but remained relatively unchanged for WS samples (Fig. 2A). Previous studies have shown that for soils, freeze/thaw cycling affects clay by disrupting its physical properties and releasing ions from otherwise nonexchangeable interactions (34, 41). Because of the high clay content at stations 908 and 804, it is possible that in the lower ionic strength in AP samples, along with freezing, disrupted clay structures and permitted more exhaustive viral extraction.
Viral enumeration accuracy.
Multiple extractions of sediment samples from each station were performed to determine the efficacy of the extraction technique. As found for other sediment (12) and soil (52) extraction procedures, the majority (ca. 70%, Fig. 3) of extractable estuarine sediment viruses were removed in the initial extraction. Subsequent extractions removed ca. 20, 10, and <5% of the extractable viruses in the second, third, and fourth extractions, respectively (Fig. 3). Similar reductions in extraction efficiency were shown in coastal and deep-sea sediments (12) and agricultural soils (52). Thus, the viral abundance estimates for Chesapeake Bay sediments reported here are conservative and likely represent ca. 70% the total extractable viral community.
Similarly, as found in Fischer et al. (19), DNase testing here indicated that epifluorescence microscopic enumerations are valid estimates of virus abundance in sediments. This type of examination has been previously reported for water (26) and soils (53). In this test, viruses are defined as SYBR-positive particles resistant to DNase digestion but susceptible to the combined effects of heating to 95°C followed by DNase digestion. Unlike soils where all SYBR-positive particles were eliminated by the heat-DNase treatment, sediment extracts contained an insignificant number (<13% of total particles) of fluorescent particles (either autofluorescent or SYBR stained) after this treatment. Because these particles are in such low abundance, they do not significantly contribute to overestimation of viral abundance within sediment extracts. The nature of these DNase and heat-resistant particles is currently unknown.
Based on these findings, the following procedures should be followed when processing aquatic sediments for direct counts of viruses. First, sediment samples should be extracted soon after collection since this will yield the most accurate estimates for both bacterial and viral abundance. Second, immediate snap-freezing in liquid nitrogen of fixed or unfixed sediment viral extracts results in the best preservation of virus particles for subsequent microscopic and molecular genetic assays (8, 50). Finally, the least-variable estimates of total viral abundance in aquatic sediments from the Chesapeake Bay come through the extraction of whole sediments without prior extraction of PW.
Acknowledgments
This research was supported in part by an NSF Microbial Observatories grant (MCB-0132070) awarded to K.E.W.
We thank D. M. Winget for technical assistance, Y. Jin for interpretation of ionic strength data, K. M. Ritalahti and K. E. Williamson for helpful comments, K. J. Czymmek and D. H. Powell at the Delaware Biotechnology Institute Bio-imaging Center for microscopy assistance, Old Dominion University for use of the multicorer, and the excellent support of the captain and crew of the R/V Cape Henlopen.
REFERENCES
- 1.Ashelford, K. E., M. J. Day, and J. C. Fry. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl. Environ. Microbiol. 69:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bales, R. C., S. M. Li, K. M. Maguire, M. T. Yahya, and C. P. Gerba. 1993. Ms-2 and poliovirus transport in porous media: hydrophobic effects and chemical perturbations. Water Resources Res. 29:957-963. [Google Scholar]
- 3.Bales, R. C., S. M. Li, K. M. Maguire, M. T. Yahya, C. P. Gerba, and R. W. Harvey. 1995. Virus and bacteria transport in a sandy aquifer, Cape-Cod, MA. Ground Water 33:653-661. [Google Scholar]
- 4.Beiras, R., E. His, and M. N. L. Seaman. 1998. Effects of storage temperature and duration on toxicity of sediments assessed by Crassostrea gigas oyster embryo bioassay. Environ. Toxicol. Chem. 17:2100-2105. [Google Scholar]
- 5.Bettarel, Y., T. Sime-Ngando, C. Amblard, J. F. Carrias, and C. Portelli. 2003. Virioplankton and microbial communities in aquatic systems: a seasonal study in two lakes of differing trophy. Freshwat. Biol. 48:810-822. [Google Scholar]
- 6.Bettarel, Y., T. Sime-Ngando, C. Amblard, and H. Laveran. 2000. A comparison of methods for counting viruses in aquatic systems. Appl. Environ. Microbiol. 66:2283-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bongiorni, L., M. Magagnini, M. Armeni, R. Noble, and R. Danovaro. 2005. Viral production, decay rates, and life strategies along a trophic gradient in the north Adriatic Sea. Appl. Environ. Microbiol. 71:6644-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brussaard, C. P. 2004. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 70:1506-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, F., J. R. Lu, B. J. Binder, Y. C. Liu, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corinaldesi, C., E. Crevatin, P. Del Negro, M. Marini, A. Russo, S. Fonda-Umani, and R. Danovaro. 2003. Large-scale spatial distribution of virioplankton in the Adriatic Sea: testing the trophic state control hypothesis. Appl. Environ. Microbiol. 69:2664-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabrowski, W., R. I. Mackie, and M. Zielina. 2002. Does freezing affect sediment sampling results? Water Air Soil Pollut. 140:367-370. [Google Scholar]
- 12.Danovaro, R., A. Dell'Anno, A. Trucco, M. Serresi, and S. Vanucci. 2001. Determination of virus abundance in marine sediments. Appl. Environ. Microbiol. 67:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danovaro, R., E. Manini, and A. Dell'Anno. 2002. Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl. Environ. Microbiol. 68:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danovaro, R., D. Marrale, N. Della Croce, P. Parodi, and M. Fabiano. 1999. Biochemical composition of sedimentary organic matter and bacterial distribution in the Aegean Sea: trophic state and pelagic-benthic coupling. J. Sea Res. 42:117-129. [Google Scholar]
- 15.Danovaro, R., and M. Serresi. 2000. Viral density and virus-to-bacterium ratio in deep-sea sediments of the Eastern Mediterranean. Appl. Environ. Microbiol. 66:1857-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake, L. A., K.-H. Choi, A. G. E. Haskell, and F. C. Dobbs. 1998. Vertical profiles of virus-like particles and bacteria in the water column and sediments of Chesapeake Bay, USA. Aquat. Microb. Ecol. 16:17-25. [Google Scholar]
- 17.Duhamel, S., and S. Jacquet. 2006. Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. J. Microbiol. Methods 64:316-332. [DOI] [PubMed] [Google Scholar]
- 18.Farnell-Jackson, E. A., and A. K. Ward. 2003. Seasonal patterns of viruses, bacteria and dissolved organic carbon in a riverine wetland. Freshwat. Biol. 48:841-851. [Google Scholar]
- 19.Fischer, U. R., A. K. T. Kirschner, and B. Velimirov. 2005. Optimization of extraction and estimation of viruses in silty freshwater sediments. Aquat. Microb. Ecol. 40:207-216. [Google Scholar]
- 20.Fischer, U. R., C. Wieltschnig, A. K. Kirschner, and B. Velimirov. 2003. Does virus-induced lysis contribute significantly to bacterial mortality in the oxygenated sediment layer of shallow oxbow lakes? Appl. Environ. Microbiol. 69:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 22.Glud, R. N., and M. Middelboe. 2004. Virus and bacteria dynamics of a coastal sediment: implication for benthic carbon cycling. Limnol. Oceanogr. 49:2073-2081. [Google Scholar]
- 23.Goyal, S. M., and C. P. Gerba. 1979. Comparative adsorption of human enteroviruses, simian rotavirus, and selected bacteriophages to soils. Appl. Environ. Microbiol. 38:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara, S., I. Koike, K. Terauchi, H. Kamiya, and E. Tanoue. 1996. Abundance of viruses in deep oceanic waters. Mar. Ecol. Prog. Ser. 145:269-277. [Google Scholar]
- 25.Heldal, M., and G. Bratbak. 1991. Production and decay of viruses in aquatic environments. Mar. Ecol. Prog. Ser. 72:205-212. [Google Scholar]
- 26.Hennes, K. P., and C. A. Suttle. 1995. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr. 40:1050-1055. [Google Scholar]
- 27.Hewson, I., and J. A. Fuhrman. 2003. Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic waters. Microb. Ecol. 46:337-347. [DOI] [PubMed] [Google Scholar]
- 28.Hewson, I., J. M. O'Neil, J. A. Fuhrman, and W. C. Dennison. 2001. Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol. Oceanogr. 46:1734-1746. [Google Scholar]
- 29.Hewson, I., J. M. O'Neil, C. A. Heil, G. Bratbak, and W. C. Dennison. 2001. Effects of concentrated viral communities on photosynthesis and community composition of co-occurring benthic microalgae and phytoplankton. Aquat. Microb. Ecol. 25:1-10. [Google Scholar]
- 30.Johnson, R. A., R. D. Ellender, and S. C. Tsai. 1984. Elution of enteric viruses from Mississippi estuarine sediments with lecithin-supplemented eluents. Appl. Environ. Microbiol. 48:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinoshita, T., R. C. Bales, K. M. Maguire, and C. P. Gerba. 1993. Effect of pH on bacteriophage transport through sandy soils. J. Contam. Hydrol. 14:55-70. [Google Scholar]
- 32.Lawrence, J. E., A. M. Chan, and C. A. Suttle. 2002. Viruses causing lysis of the toxic bloom-forming alga Heterosigma akashiwo (Raphidophyceae) are widespread in coastal sediments of British Columbia, Canada. Limnol. Oceanogr. 47:545-550. [Google Scholar]
- 33.Lipson, S. M., and G. Stotzky. 1983. Adsorption of reovirus to clay-minerals: effects of cation-exchange capacity, cation saturation, and surface-area. Appl. Environ. Microbiol. 46:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marion, G. M. 1995. Freeze-thaw processes and soil chemistry. Special report 95-12. U.S. Army Corps of Engineers Cold Regions Research Engineering Lab, Washington, D.C.
- 35.Marshall, K. C. 1975. Clay mineralogy in relation to survival of soil bacteria. Annu. Rev. Phytopathol. 13:357-373. [Google Scholar]
- 36.Middelboe, M., R. N. Glud, and K. Finster. 2003. Distribution of viruses and bacteria in relation to diagenetic activity in an estuarine sediment. Limnol. Oceanogr. 48:1447-1456. [Google Scholar]
- 37.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 38.Paul, J. H., J. B. Rose, S. C. Jiang, C. A. Kellogg, and L. Dickson. 1993. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl. Environ. Microbiol. 59:718-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peduzzi, P., and F. Schiemer. 2004. Bacteria and viruses in the water column of tropical freshwater reservoirs. Environ. Microbiol. 6:707-715. [DOI] [PubMed] [Google Scholar]
- 40.Pesaro, M., F. Widmer, G. Nicollier, and J. Zeyer. 2003. Effects of freeze-thaw stress during soil storage on microbial communities and methidathion degradation. Soil Biol. Biochem. 35:1049-1061. [Google Scholar]
- 41.Polubesova, T. A., L. T. Shirshova, M. Lefevre, and V. A. Romanenkov. 1996. Effect of freezing and thawing on the surface chemical properties of soils and clays. Eurasian Soil Sci. 28:104-114. [Google Scholar]
- 42.Powelson, D. K., J. R. Simpson, and C. P. Gerba. 1991. Effects of organic matter on virus transport in unsaturated flow. Appl. Environ. Microbiol. 57:2192-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prescott, L. M., D. A. Klein, and J. P. Harley. 2001. Microbiology, 5th ed. McGraw-Hill Companies, New York, N.Y.
- 44.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 45.Quignon, F., M. Sardin, L. Kiene, and L. Schwartzbrod. 1997. Poliovirus-1 inactivation and interaction with biofilm: a pilot-scale study. Appl. Environ. Microbiol. 63:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steward, G. F., D. C. Smith, and F. Azam. 1996. Abundance and production of bacteria and viruses in the Bering and Chukchi seas. Mar. Ecol. Prog. Ser. 131:287-300. [Google Scholar]
- 47.Weinbauer, M. G., D. Fuks, and P. Peuzzi. 1993. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl. Environ. Microbiol. 59:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weinbauer, M. G., and M. G. Hofle. 1998. Distribution and life strategies of two bacterial populations in a eutrophic lake. Appl. Environ. Microbiol. 64:3776-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weinbauer, M. G., and C. A. Suttle. 1997. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat. Microb. Ecol. 13:225-232. [Google Scholar]
- 50.Wen, K., A. C. Ortmann, and C. A. Suttle. 2004. Accurate estimation of viral abundance by epifluorescence microscopy. Appl. Environ. Microbiol. 70:3862-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reference deleted.
- 52.Williamson, K. E., M. Radosevich, and K. E. Wommack. 2005. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71:3119-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williamson, K. E., K. E. Wommack, and M. Radosevich. 2003. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 69:6628-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wommack, K. E., R. T. Hill, M. Kessel, E. Russek-Cohen, and R. R. Colwell. 1992. Distribution of viruses in the Chesapeake Bay. Appl. Environ. Microbiol. 58:2965-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhuang, J., and Y. Jin. 2003. Virus retention and transport as influenced by different forms of soil organic matter. J. Environ. Qual. 32:816-823. [DOI] [PubMed] [Google Scholar]
- 57.Zobell, C. E. 1946. Marine microbiology. Chronica Botanica Press, Waltham, Mass.