Abstract
The white rot basidiomycete Phanerochaete chrysosporium produces an array of nonspecific extracellular enzymes thought to be involved in lignin degradation, including lignin peroxidases, manganese peroxidases, and the H2O2-generating copper radical oxidase, glyoxal oxidase (GLX). Preliminary analysis of the P. chrysosporium draft genome had identified six sequences with significant similarity to GLX and designated cro1 through cro6. The predicted mature protein sequences diverge substantially from one another, but the residues coordinating copper and constituting the radical redox site are conserved. Transcript profiles, microscopic examination, and lignin analysis of inoculated thin wood sections are consistent with differential regulation as decay advances. The cro2-encoded protein was detected by liquid chromatography-tandem mass spectrometry in defined medium. The cro2 cDNA was successfully expressed in Aspergillus nidulans under the control of the A. niger glucoamylase promoter and secretion signal. The recombinant CRO2 protein had a substantially different substrate preference than GLX. The role of structurally and functionally diverse cro genes in lignocellulose degradation remains to be established.
The most abundant source of global carbon is plant biomass composed primarily of cellulose, hemicellulose, and lignin. White rot basidiomycetes, such as Phanerochaete chrysosporium, can degrade all of these cell wall components. White rot fungi are the only microbes convincingly shown to efficiently depolymerize and fully degrade lignin (reviewed in references 27 and 28).
Lignin encases cellulose fibers, providing strength and stability to plant cell walls (17). The complex water-insoluble polymer is recalcitrant to decay, and white rot fungi are thought to depolymerize lignin to gain access to cellulose. Indeed, no organism is known to use lignin as a sole carbon and energy source.
Oxidative enzymes involved in ligninolysis by P. chrysosporium include lignin peroxidase (LiP), manganese peroxidase (MnP), and glyoxal oxidase (GLX) (reviewed in reference 7). These extracellular, nonspecific enzymes are produced in submerged cultures during secondary metabolism (27). Size constraints prevent peroxidases from directly penetrating sound wood, and these enzymes are presumed to generate radical species that participate in further reactions to degrade the lignin polymer (27).
The proposed role of GLX in lignin decay is to generate H2O2 for LiP- and MnP-mediated reactions. P. chrysosporium possesses several oxidases that could potentially supply peroxide for these reactions, but only GLX appears to be secreted in ligninolytic cultures in liquid medium (27). In addition to the physiological connection to peroxidases, extracellular peroxide production may be involved in the generation of highly reactive hydroxyl radicals via a Fenton reaction. Fenton chemistry has been implicated in lignocellulose degradation in P. chrysosporium (11, 31), but it is generally thought to be more important in cellulose depolymerization by brown rot fungi (15, 22).
Glyoxal oxidase is a copper-radical oxidase, with broad substrate specificity for the oxidation of simple aldehydes, such as glyoxal and methylglyoxal, to the corresponding carboxylic acids (45). These substrates are found in ligninolytic cultures, suggesting a role as physiological substrates for GLX. GLX also has been implicated in the regulation of peroxidase activity, and it is activated in vitro by lignin peroxidase (24, 25). Based on similarities to the galactose oxidase from Dactylium dendroides (20, 45), the active site of GLX has been identified and includes Tyr377, His378, Tyr135, Tyr70, and His471 (23, 45). For many years, it was thought that P. chrysosporium possessed only a single glx gene with two alleles (26).
Automated gene predictions generated from P. chrysosporium genome assemblies v1.0 (32) and v2.0 (http://www.jgi.doe.gov/whiterot) include six incomplete sequences with partial but significant similarities to glx. Three of these putative copper radical oxidase genes (cro3, cro4, and cro5) are clustered within a larger cluster of lignin peroxidase genes (7, 32).
Our overall goal is to understand the biological role of these glx-like sequences. Specific objectives in this study were to determine (i) the structural relationships among cro genes, (ii) the transcript patterns of these genes in colonized wood, and (iii) the activity and substrate preference of a heterologously expressed cro gene. The expression of cro genes, particularly in decaying wood, together with the oxidation of a substrate produced in ligninolytic cultures is consistent with a role for these genes and the enzymes they encode in lignin degradation.
MATERIALS AND METHODS
Organisms.
Phanerochaete chrysosporium strain BKM-F-1767 and a homokaryotic derivative, RP78, were used throughout (37). Both strains are available from the Forest Mycology Center, Forest Products Laboratory, Madison, WI. Aspergillus nidulans IJFM A729 (argB2 biA1 methG1), obtained from A. T. Martinez (Centro de Investigaciones Biologicas, Madrid, Spain), was used for heterologous expression.
Culture conditions.
For RNA, 200 ml of defined medium (10) amended with 0.4% Avicel PH-101 (Fluka Chemie, Buchs, Switzerland) was inoculated with 1 × 107 RP78 spores in a 2-liter flask. Incubation was at 37°C and 250 rpm in a shaking incubator. The culture was harvested after 6 days by filtration through Miracloth (Calbiochem, La Jolla, CA). The mycelium was snap frozen in liquid N2 and stored at −90°C. RNA was also derived from 2-day-old carbon-limited cultures as previously described (28, 36). mRNA isolation and first-strand cDNA synthesis were as previously described (42).
For the analysis of extracellular protein from strain RP78, a medium containing Whatman CC41 microgranular cellulose was used (46). Each of three 2-liter flasks containing 1 liter of CC41 medium was inoculated with >107 RP78 spores. Inoculated flasks were maintained at 30°C and 150 rpm on an orbital shaker. On day 3, 1 g of autoclaved MnO2 was added directly to each flask. Cultures were harvested on day 12 by filtration through 100% nylon fabric (any sheer stocking is adequate; Hanes Corp., Winston-Salem, NC), followed by vacuum filtration through a Whatman GF/F 15-cm glass fiber filter (Whatman International Ltd, Maidstone, United Kingdom). Culture filtrates were stored at −20°C.
Wood wafer inoculation.
Wood wafers (1 cm by 1 cm by 2 mm) were cut from freshly harvested sapwood of Populus tremuloides, sterilized, and inoculated by contact with mycelium growing on malt extract agar (15 g malt extract [Difco, Detroit, MI] and 15 g agar liter−1) in petri dishes. Wood wafers were harvested after 1 h and then on days 1, 3, 10, 20, 90, and 120. Noninoculated wood wafers placed on malt extract agar in petri dishes served as controls. Three separate replicates of at least 10 wafers each were harvested for each time point. Wafers were snap frozen in liquid N2 and stored at −90°C until used. Lignin and carbohydrate analysis was performed as previously described (9).
Identification of genomic cro sequences and amplification of cDNAs.
The P. chrysosporium genome v1.0 database (http://www.jgi.doe.gov/whiterot) contains six sequences with limited similarities to GLX. Following manual curation of the incomplete v1.0 gene models, PCR primers (Table 1) were designed for the amplification of six full-length cro cDNAs. Proofreading polymerase PFU (Stratagene, La Jolla, CA) or GeneAmp XL (Applied Biosystems, Foster City, CA) was used according to the manufacturers' instructions. Pfu cycling parameters were 2 min for 1 cycle at 95°C, 30 s at 94°C, 30 s at 51°C, and 3 min at 72°C for 30 cycles, followed by a 15-min extension at 72°C. GeneAmp XL cycling parameters were 24 s at 94°C, 3 min for 7 cycles at 65°C, 25 s at 94°C, and 3 min at 67°C for 32 cycles, followed by a 7-min extension at 67°C. The fully sequenced cro2 cDNA was amplified from colonized wood, while cro1, cro3, cro4, and cro5 were amplified from C-starved cultures and cro6 was amplified from Avicel medium. Amplicons were cloned into pCR-Blunt (Invitrogen Corp., Carlsbad, CA) or pGEM-T Easy (Promega Corp., Madison, WI) and sequenced. Multiple alignments were performed using ClustalW with DNAStar MegAlign software (Madison, WI).
TABLE 1.
Primers for amplification of full-length cro cDNAa
| Gene | Upstream | Downstream | Coding region (bp) |
|---|---|---|---|
| cro1 | TTCGTGACCCTCCTCTTCTG | CTTGTCAAGACCAGCAGGCT | 1,947 |
| cro2 | GTAATCCTGTCCCTCGCCTA | CCATGCACACTTTCAGTACC | 2,019 |
| cro3 | ACTTGCTTCTCGGCGTTCAT | TACCCAAAACACCCGACAC | 2,295 |
| cro4 | TTTGCCATGGCACCCTACTC | AGGTTCATGGGTTGCAGATC | 3,072 |
| cro5 | TTCTCTCGGGTCATGGTCCG | ACACCGAGGCGTTTACACGC | 3,051 |
| cro6 | ATGGTCATCCGCTCTTTCTT | CTATTCGGAATCATCTTCGA | 2,355 |
Primers are listed 5′ to 3′.
Transcript patterns in colonized wood.
Wood wafers were placed in a mortar and pestle that was prechilled with liquid N2, ground to a fibrous mat, and transferred to 50-ml conical polypropylene tubes. Five chips were ground per replicate for samples taken at 1 h, 1 day, and 3 days. One chip was ground per replicate for the remaining samples. All time point isolations were repeated in triplicate. A total of 5 ml of extraction buffer (4 M guanidine thiocyanate, 100 mM Tris [pH 8.0], 1% dithiothreitol, 0.5% Sarkosyl) was added, and the tubes were placed on ice for 5 min. Extracts were filtered through Miracloth, and two volumes binding buffer (100 mM Tris, 0.4 M LiCl, 20 mM EDTA) were added. One hundred microliters of oligo(dT) Dynabeads (Dynal, Inc., Oslo, Norway) were washed once with binding buffer, resuspended in 200 μl binding buffer, added to the extract, and placed on ice for 30 min with occasional mixing. The tubes were placed on a Dynal MPC-1 magnetic stand for 5 min, and the supernatant was removed to a new tube and placed on the magnet for an additional 5 min. Beads from both magnetizing steps were washed with 500 μl washing buffer (10 mM Tris, 150 mM LiCl, 1 mM EDTA), combined, and washed three additional times in washing buffer. The beads were resuspended in 200 μl elution buffer (2 mM EDTA, pH 8.0) and placed in a 65°C water bath for 2 min. Tubes were immediately placed on the magnet, and the liquid was quickly transferred to a new 1.5-ml microcentrifuge tube. One hundred microliters of the suspension was used in the subsequent reverse transcription reaction, and the remainder was ethanol precipitated and stored at −20°C.
A 500-μl reverse transcription master mix, containing 1× PCR buffer (Promega, Inc., Madison, WI), 5 mM MgCl2, 4 mM dNTPs, 500 units RNasin (Promega), 105 pmol oligo(dT)15, 1,250 units Moloney murine leukemia virus-reverse transcriptase (Invitrogen), and 100 μl mRNA, was made for each mRNA replicate. Fifty-microliter aliquots were divided among 0.5-ml Eppendorf tubes and placed in a thermocycler. Reaction cycling was 23°C for 10 min, 42°C for 45 min, and 95°C for 5 min. All tubes for each replicate were combined and split into several aliquots for storage at −20°C.
Gene-specific cDNA levels were determined by competitive PCR as previously described (13, 40), with competitive template amounts ranging from 10−11 to 10−18 g plasmid/reaction. Genomic templates for competitive PCR were prepared by PCR amplification using the primers listed in Table 2 and cloned into pCR-Blunt or pGEM-T Easy. Using the primers listed in Table 2, competitive PCRs were performed on three separate first-strand syntheses for each time point. Gel band intensities were quantified and analyzed as described previously (5) by using NIH Image version 1.61.
TABLE 2.
Competitive PCR primersa and product characteristics
| Gene | Primer pairs for generating genomic templatesb |
Primer pairs for competitive PCRc |
Productsd |
||||
|---|---|---|---|---|---|---|---|
| Upstream | Downstream | Upstream | Downstream | C | G | R | |
| cro1 | TTCGTGACCCTCCTCTTCTG | CTTGTCAAGACCAGCAGGCT | CTTGCTGGACAGATGACTACC | GTCGTCGTCCTCTTCATAGTC | 518 | 619 | 1.19 |
| cro2 | GACGAGAACACACGATCCCT | CCATGCACACTTTCAGTACC | GCCGTCACACATTTGCAGGCC | TCGGGTCGTCCTCGACGTAA | 578 | 622 | 1.08 |
| cro3 | GCACCAGCCCGCCAACACT | TACCCAAAACACCCCGACAC | CTCCAGGCTTGGCTCTACGAT | GAAGTCGGAGAAGTTAGGCCA | 539 | 641 | 1.19 |
| cro4 | GTCTTCTGCTCGGCGAGCAT | AGGTTCATGGGTTGCAGATC | TACCAGGCGGTACTCTACGAC | GACCCAATGCGAGTGAGATG | 497 | 593 | 1.19 |
| cro5 | ATTCGACAACTCGACTGGCG | ACACCGAGGCGTTTACACGC | TACAACGCGATCCTGTACGAT | AAAGTCAGGGAAGTTCGGCCA | 570 | 664 | 1.16 |
| cro6 | ATGGACGCTATCGTTGTTACT | AATAAGACGACTCTCCGGGA | GTCTTCACGCCCGTGCTGTA | GCCTTTGCTCGGAATCCCGTT | 491 | 550 | 1.12 |
| glx | TCACACCTTCGCTCTACACG | TATTTACTCCAGGGTCGGCG | 563 | 676 | 1.20 | ||
| gpd | ATGCCGGTCAGTACACCACAC | TTAGAGGGCACCGTCGACCT | TTCACGGAGACATTGACGTC | TGATCTCGTCGTAAGAAGCG | 693 | 901 | 1.30 |
Primers are listed 5′ to 3′.
Primers for generating competitive templates. The glx genomic clone was obtained earlier (26) and corresponds to coordinates 1198897 to 1205335 on scaffold 7.
Primers designed to coding regions and amplify both cDNA and genomic targets.
cDNA expression in Aspergillus.
Sequence overlap extension PCR (19) was used to construct a precise translational fusion between the cro2 cDNA encoding the mature peptide (amino acid 17) and the A. niger glucoamylase (gla) promoter plus secretion signal (12). A 200-bp region corresponding to the A. niger gla terminator was fused to the 3′ region, and the entire expression cassette was ligated into pCR-Blunt (Invitrogen). Nucleotide sequences of all junctions were confirmed by dideoxy sequencing. Plasmids were isolated by using the Wizard Midiprep kit (Promega).
Protoplasts from A. nidulans IJFM A729 were prepared as previously described by Ballance et al. (3), with modifications. Specifically, 100 ml YEG (0.5% yeast extract and 2% glucose) was inoculated with 2 × 108 spores, incubated overnight at 30°C, washed twice with 0.6 M KCl, and resuspended in Novozyme solution (0.24% Novozyme 234, 20 mM MgSO4, 0.1% bovine serum albumin in 0.6 M KCl). Tubes were incubated horizontally at 30°C for 2 h in an orbital shaker (50 rpm). The solution was filtered through sterile Miracloth, and protoplasts were collected by centrifugation (1,600 × g at room temperature for 10 min). The protoplast pellet was washed with 0.6 M KCl and resuspended in 500 μl sorbitol solution (0.8 M sorbitol, 50 mM Tris [pH 7.5], 50 mM CaCl2). To this, 125 μl polyethylene glycol (PEG) solution (40% PEG-4000, 50 mM Tris [pH 7.5], 50 mM CaCl2, 5% dimethyl sulfoxide) was added and mixed gently.
Cotransformation of protoplasts was performed as described previously (33) with 5 to 7 μg expression cassette, together with 3 μg of the pArgAns-1 selectable marker, with modifications. A total of 110 μl of the protoplast suspension was added to microcentrifuge tubes containing DNA. Tubes were incubated on ice for 30 min. Six hundred microliters PEG solution was added and mixed gently, and the suspension was incubated at room temperature for 10 min. Eight hundred microliters 0.6 M KCl was added and mixed, and the tubes were centrifuged (750 × g at room temperature [22°C to 25°C] for 5 min). All but 300 μl of the tube's contents were discarded, and the remaining protoplast suspension was plated directly onto two plates containing minimal medium (26) supplemented with 1 μg ml−1 biotin, 1 mg ml−1 l-methionine, 5% maltose, and 0.6 M KCl. Plates were incubated at 37°C for 3 to 7 days, and transformants were transferred to fresh minimal medium plates when visible.
To confirm cotransformation, 10 ml YEG in 100 by 15-mm petri dishes was inoculated with a loopful of spores. Following overnight incubation at 37°C, the mycelium was skimmed off the surface and snap frozen and DNA was isolated with the QIAGEN DNA plant mini kit (QIAGEN Sciences, Germantown, MD). The presence of cro2 and arg was confirmed by PCR amplification with primers specific to each gene.
For protein analysis, transformants were grown in 250-ml Erlenmeyer flasks, to which 50 ml minimal medium, amended as described above, was added. These cultures were inoculated with a loopful of spores and incubated for 3 days at 30°C on an orbital shaker (200 rpm). Culture fluid was collected by filtration through Miracloth.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) protein identification.
Mass spectrometry was used to confirm the presence of CRO2 in extracellular culture fluids of P. chrysosporium strain RP78 and an A. nidulans transformant. A total of 790 ml of P. chrysosporium CC41 culture filtrate was concentrated 63-fold by ultrafiltration with a 5,000 molecular weight cutoff polyethersulfone membrane (Millipore Corp., Bedford, MA). The concentrate was separated into fractions on a HiPrep 16/60, Sephacryl S-100, high-resolution gel filtration column (Amersham Biosciences Corp., Piscataway, NJ). Fractions containing cellulose dehydrogenase activity were pooled for further analysis. The Miracloth filtrate from the above-mentioned A. nidulans transformant was concentrated with a Microsep 10,000 polyethersulfone centrifugal device (Pall Life Sciences, Ann Arbor, MI).
Twenty-five microliters of the concentrated extracellular protein solutions was mixed with 20 μl Laemmli buffer (Bio-Rad Laboratories, Inc, Hercules, CA) and loaded onto a 12.5% Criterion Tris-HCl ready gel (Bio-Rad) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Electrophoresis was performed in a Bio-Rad Criterion cell, 200 V for 50 min at 23°C. Gels were stained with Coomassie blue R-250 (Bio-Rad) to estimate the protein abundance and molecular mass distribution. Gel regions defined by predicted molecular weights were excised with a scalpel, cut into ∼1-mm pieces, and placed in individual siliconized 1.5-ml microcentrifuge tubes (Fisher Scientific, Chestertown, MD) for subsequent enzymatic digestion.
“In-gel” digestion and mass spectrometric analysis were performed as previously described (41, 42) (www.biotech.wisc.edu/ServicesResearch/MassSpec/ingel.htm) with minor modifications. Specifically, 40 μl of each extracted peptide was automatically loaded onto a reverse-phase high-pressure liquid chromatography trap column and solvents were delivered at 20 μl min−1 to load the sample. Elution into the nanoelectrospray source was for 80 min.
A Spectrum Mill MS proteomics workbench (Agilent) and an in-house-licensed Mascot search engine (Matrix Science, London, United Kingdom) were used to identify peptides from a data set of 10,048 v2.1 gene sequences currently available (http://genome.jgi-psf.org/Phchr1/Phchr1.download.html [41]). Peptide sequences with Spectrum Mill and Mascot scores of >13 and >40, respectively, were considered significant matches. Throughout, protein similarity scores are based on the Smith-Waterman algorithm (35) with the BLOSUM62 matrix.
Oxidase activity assay.
Miracloth filtrates from six 50-ml, 3-day old cultures of Aspergillus transformant were combined. Phenylmethylsulfonyl fluoride was added to a final concentration of 0.1 mM, and filtrates were concentrated approximately 70-fold in an Amicon ultrafiltration unit (Millipore) using a 10,000-Dalton cutoff polyethersulfone membrane (Millipore). A total of 1.5 ml of concentrate was then buffer exchanged with 5 mM Na+ 2,2-dimethylsuccinate (pH 6.0) and 0.1 mM phenylmethylsulfonyl fluoride by using two HiTrap 5-ml desalting columns (Amersham) in tandem. One-milliliter fractions were collected, and 100 μl of each fraction was tested for oxidase activity in 200-μl reactions as previously described (16). Methylglyoxal and glycolaldehyde dimers (Sigma-Aldrich, St. Louis, MO) were tested as potential substrates at 10 mM, and control reactions were performed with no substrate. Concentrated filtrates from control transformant (pArgANS-1) were likewise screened but showed no activity. SDS-PAGE of cro2 and control transformants confirmed that similar protein levels were assayed.
P. chrysosporium RP78 grown on medium containing Whatman CC41 was also tested for oxidase activity. In this case, the culture filtrate of 12-day cultures was concentrated approximately 75-fold by ultrafiltration prior to buffer exchange.
RESULTS
Identification of P. chrysosporium cro sequences and cloning of cDNAs.
BLAST analysis of the P. chrysosporium genome identified six sequences with significant similarities to glx (32). As is commonly the case for automated gene predictions in eukaryotic genomes, inaccurate exon/intron boundaries were suspected. Accordingly, cDNAs corresponding to all six genes were PCR amplified, cloned, sequenced, and deposited (Table 3). These sequences differed from the automated gene predictions. All sequences featured predicted secretion signals (SignalP, version 3.0; www.cbs.dtu.dk/services/SignalP/) and multiple potential N-glycosylation sites (4).
TABLE 3.
Characteristics of P. chrysosporium copper radical oxidase genes
| Gene | Previous gene modelsa |
cDNA sequence analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| v1.0 | v2.1 | Molecular massb | pI | SSc | Best hitd | Accession no.e | Remarks | |
| cro1 | pc.120.7.1 + pc.120.6.1 | 124009 | 65 | 4.7 | 23/24 AHT-QT | C. neoformans gi57227801 (607) (ct) | DQ398767 | |
| cro2 | pc.46.103.1 | 134241 | 69 | 4.6 | 16/17 VLA-QG | U. maydis glo1 gi71013128 (522) | DQ398768 | |
| cro3 | pc.85.36.1 + gx.85.36.1 | 121818 | 80 | 4.6 | 20/21 ASA-FI | M. grisea gi39952025 (746) (ct) | DQ398769 | Two WSC domains |
| cro4 | gx.85.15.1 + pc.85.33.1 + pc.85.32.1 | 8882 | 106 | 4.6 | 21/22 SHA-SL | M. grisea gi39952025 (842) (ct) | DQ398770 | Four WSC domains |
| cro5 | gx.85.13.1 + pc.85.28.1 | 121730 | 106 | 4.4 | 21/22 VQA-SS | M. grisea gi39952025 (817) (ct) | DQ398771 | Four WSC domains |
| cro6 | pc.72.34.1 | 37905 | 82 | 5.3 | 21/22 VLA-SE | P. chrysosporium glx gi1050304 (412) | DQ398772 | |
| glx | pc.52.9.1 | 11068 | 57 | 5.1 | 22/23 ASD-AP | gi1050304 | ||
Automated gene models available at http://genome.jgi-psf.org/Phchr1/Phchr1.home.html, with v1.0 models archived. To directly access v2.1 model information, end the following URL with the model number: http://genome.jgi-psf.org/cgi-bin/dispGeneModel?db=Phchr1&id=___.
Predicted molecular mass of mature peptide in kilodaltons.
Secretion signals (SS) predicted by SignalP (www.cbs.dtu.dk/services/SignalP/) using both neural networks and hidden Markov models. Position and surrounding residues are shown. The glx signal was determined experimentally (23, 45).
BLASTP searches of nonredundant NCBI. ct, conceptual translation.
Deposited cDNA sequences were based on PCR-amplified products using RNA derived from standard carbon-starved cultures (cro1, cro3, cro4, and cro5), colonized wood wafers harvested on day 10 (cro2), and Avicel cultures harvested on day 6 (cro6). Subsequent RT-PCR and cDNA sequence analysis have shown that all seven copper radical oxidase transcripts are present in carbon-starved cultures and in colonized wood wafer after day 10.
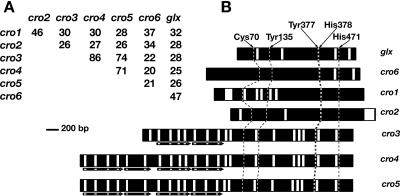
Deduced mature proteins had identities ranging from 20 (cro4 versus cro6) to 86% (cro3 versus cro4) (Fig. 1A). Overall sequence similarity to GLX was relatively low, but multiple alignments identified conserved residues constituting the Cu-coordinating active site of GLX (Tyr135, Tyr377, His378, and His471) (44, 45) (Fig. 1B). In addition, a cysteine cross-linked with Tyr135 forms the radical redox site and also is conserved in all six sequences (Cys70). Thus, based on structure, these cDNAs probably encode copper radical oxidases.
FIG. 1.
Percent identity among mature peptides predicted from cDNA (A) and schematic alignment (B) of glyoxal oxidase (glx) and six new copper radical oxidases (cro1 through cro6). Sequences were aligned by ClustalW (38). Introns and exons are shown in white and black boxes, respectively. Residues involved in glyoxal oxidase catalysis are indicated, as are conserved positions within cro1 though cro6. Repeats of the highly conserved WSC domain, potentially involved in carbohydrate binding (IPR288; www.ebi.ac.uk/interpro/), are indicated by arrows underlying cro3, cro4, and cro5. The genes are arbitrarily aligned at the His471 position.
cro1.
Initially predicted to be two separate genes (pc.120.7.1 and pc.120.6.1 [32]), the v2.0 gene model, 124009, is identical to the full-length cDNA except for one six-nucleotide exon. In pairwise comparisons of all P. chrysosporium CRO proteins, CRO1 is most similar to CRO2 and least similar to CRO5 (Fig. 1A). BLASTP of NCBI shows CRO1 most closely related to conceptual translations from the basidiomycetes Cryptococcus neoformans genomes (e.g., gi57227801, score 607) and Ustilago maydis glo2 (gi33386644, score 519). The latter gene belongs to a family of glx-like genes recently studied by Leuthner et al. (29). cro1 is located on scaffold 6 (coordinates 592442 to 594775), distantly linked to glx (coordinates 1201617 to 1203348).
Cro2.
The v2.0 sequence corresponding to cro2, gene model 132241, has a complete N terminus in contrast to the corresponding v1.0 model pc.46.103.1 (32). However, gene model 132241 incorrectly identifies a second intron, resulting in a seven-amino-acid deletion. Ustilago maydis glo1 (gi71013128, score 522) is the most closely related sequence in the current NCBI database. The membrane-bound Glo1 protein is involved in filamentous growth and pathogenicity of U. maydis (29). A variant cDNA clone extending the coding region by 102 amino acids also was amplified by reverse transcription-PCR (RT-PCR) from P. chrysosporium CC41 cultures and from colonized wood (GenBank DQ400693). BLASTP and BLASTX analyses of this sequence identified no significant similarity to any known sequences.
cro3, cro4, and cro5.
Initial analysis of the v1.0 draft genome identified a cluster of three glx-like sequences surrounded by well-characterized lignin peroxidase genes (32, 36). Careful manual inspection and cDNA sequencing revealed complex N-terminal regions which were not predicted in v1.0. Specifically, cro3, cro4, and cro5 contain two to four tandem copies of a WSC (cell wall integrity and stress component) domain (Fig. 1B). The function of the WSC domain is unclear; two copies are found in a β-1,3 exoglucanase of the mycoparasite Tricoderma harzianum, and this protein has been implicated in the degradation of the host cell wall (6). In Saccharomyces cerevisiae, a family of WSC-containing proteins is required for heat shock response and maintenance of cell wall integrity (30). Classified as putative carbohydrate binding domains (www.ebi.ac.uk/interpro/), the WSC regions contain up to eight conserved cysteine residues that may be involved in the formation of disulfide bridges. The P. chrysosporium CRO WSC domains contain five to six of these conserved cysteines. The function of the WSC in P. chrysosporium remains unclear.
Based on structure and organization, cro3 through cro5 form a distinct subfamily among the copper radical oxidases. In addition to the common feature of repeated WSC domains, their intron positions are highly conserved (Fig. 1B). Moreover, if the WSC domains are excluded from alignments, cro3, cro4, and cro5 are 79 to 90% identical in pairwise comparisons. Beyond these structural considerations, the current assembly places cro3-5 within a 45-kb region on scaffold 19. This gene clustering raises the question of whether these genes are coordinately regulated. If so, it is not apparent from our analysis of transcript patterns in colonized wood (see below) or in defined ligninolytic cultures (data not shown).
cro6.
Of the six new cro genes, cro6 is most closely related to glx (Fig. 1A). BLASTP of NCBI shows C. neoformans conceptual translations (e.g., gi57227801, score 330) as the next most closely related sequences. The N-terminal region (∼200 amino acids) of the mature peptide is absent from all other copper radical oxidases and distantly related to functionally disparate sequences such as Propionibacterium acnes β-galactosidase fused to β-N-acetylhexosaminidase (gi50843277, score 63.5) and Solibacter usitatus family 2 glucosyl transferase (gi67932023, score 59.7). TBLASTN of the CRO6 protein against the unpublished Coprinus cinereus genome (www.broad.mit.edu/annotation/fungi/coprinus_cinereus/) clearly showed a homologous translation on scaffold 5, contig 1.103 (coordinates 529357 to 531543). Thus, this two-domain copper radical oxidase structure is conserved, at least in these two filamentous basidiomycetes.
Analysis of colonized wood.
Earlier investigations had quantified cellulase and peroxidase transcripts from P. chrysosporium grown on wood chips under standard biopulping conditions (21, 39). Those studies had shown that transcript patterns in defined media bore little resemblance to more “natural” substrates. However, biopulping wood chips are nonuniform with respect to size and anatomical source. To reduce this sample variation, chips in “bioreactors” were replaced with thin wood wafers placed directly on actively growing mycelia. Ultimately, colonized wafers permitted an integrated view of transcript patterns, microscopic assessments of decay, and chemical composition over time.
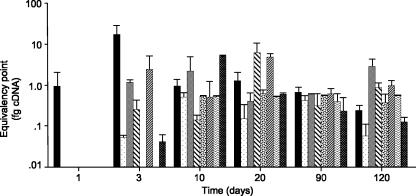
Competitive PCR results showed differential regulation among the cro genes over a 120-day time course (Fig. 2). No transcript was detected at the 1-h sampling (data not shown). However, by 24 h (day 1), gpd transcripts could be detected, and by day 3, transcript levels were measurable for all but cro4 and cro6. Microscopic analysis of wafers during the time course showed that the fungus had colonized wood surfaces and had begun cell penetration by day 20, and by day 120, cell walls were completely degraded leaving large voids (data not shown). Chemical analyses of wood wafers removed 20 and 120 days after inoculation showed 16 and 60% lignin loss, respectively (data not shown).
FIG. 2.
Relative transcript levels for cro1 through cro6, glx, and gpd on inoculated wood wafers over time. Wood wafers (1 cm by 1 cm by 1 mm) were inoculated by direct contact with P. chrysosporium growing on agar plates, and Poly(A) RNA was extracted with oligo(dT) magnetic beads. cDNA synthesis and competitive PCR were performed in triplicate as previously described (21, 39, 40). On day one, only the “housekeeping” gpd transcripts were detected. Genes and corresponding fill patterns were as follows: ▪, gpd; ░⃞, cro1; ▩, cro2; ▧, cro3;  , cro4; ▨, cro5;
, cro4; ▨, cro5;  , cro6;
, glx. Error bars indicate standard deviations.
, cro6;
, glx. Error bars indicate standard deviations.
Identification of native copper radical oxidases.
Shotgun LC-MS/MS of strain RP78 cultures had previously identified GLX peptides in extracellular fluids of carbon and nitrogen-limited cultures (41, 42). No copper radical oxidases were detected in cellulolytic medium containing Avicel as the sole carbon source (42). In this investigation, we examined a medium initially designed for the expression of cellobiose dehydrogenase (2, 46). The CRO2-specific peptide KGEIGDTNNGAYDSSYGDLAGQTSVRV was detected in this CC41 medium (Spectrum Mill score, 15.6). The detection of the CRO2 peptide and the presence of the cro2 transcripts in colonized wood prompted further characterization of the protein.
Heterologous expression.
Based on previous success expressing GLX in Aspergillus (26), the cro2 cDNA was placed under the control of the Aspergillus niger glucoamylase promoter, secretion signal, and terminator and cotransformed with pArgANS-1 into the A. nidulans argB auxotrophic mutant strain IJFM A729. Controls were transformed with pArg ANS-1 (8). Cotransformants were verified by PCR amplification of arg and cro2 sequences from isolated DNA.
Transformants were grown in 50-ml cultures and concentrated. SDS-PAGE revealed a band in the cro2 transformant, and mass spectroscopy analysis confirmed the identity. Specifically, five CRO2-specific sequences with Spectrum Mill scores of >13, including the CC41-derived peptide, were detected. These sequences were distributed over the protein's full length and accounted for 12% coverage. The observed protein was >100 kDa, significantly larger than the predicted 69 kDa for the mature protein. This discrepancy is probably due to glycosylation, a common process during protein secretion in Aspergillus (1).
The Aspergillus-produced CRO2 product had a substrate preference substantially different from that of GLX, and a screening protocol based on oxidase activity with methylglyoxal, the prototypical substrate for GLX, did not detect CRO2 transformants that were positive by SDS-PAGE. A preliminary screening for possible substrates identified the glycolaldehyde dimer as a substrate based on peroxide formation in a coupled reaction with horseradish peroxidase. Under conditions where oxidase activity was easily detected with glycolaldehyde dimer, e.g., 1.6 absorbance units h−1 under optimized conditions, no activity was observed with methylglyoxal. On the basis of this information, activity was tested in crude, buffer-exchanged enzyme from P. chrysosporium RP78 grown on medium containing Whatman CC41, a condition where CRO2 peptide was identified in 12-day cultures. The same preference for glycolaldehyde dimer was observed, while methylglyoxal oxidase activity was absent, indicating that CRO2 activity exceeds GLX activity in these cultures. Approximately fourfold-higher CRO2 activity per unit volume culture filtrate was found with the Aspergillus CRO2 transformant than with the RP78 filtrate, indicating that the Aspergillus expression system should be useful in further characterization of the oxidase.
DISCUSSION
A number of oxidases have been proposed to play a role in hydrogen peroxide production in ligninolytic cultures of P. chrysosporium, but until now, GLX was the only one that appeared to be secreted (27). The temporal correlation of GLX, peroxidase, and oxidase substrate in liquid cultures is consistent with a close physiological connection between these components (24, 25). However, the nonspecificity of GLX, together with the possibility that the simple substrates may be derived from carbohydrate (24, 25), lignin (16), and lipid (43), suggests a complex role for GLX in metabolism. Alternatively, a family of related enzymes of similar but distinct specificities may function under various metabolic regimes.
In support of this possibility, here we demonstrate the existence of six sequences structurally related to GLX. All six contain conserved active site residues and predicted secretion signals, but they are diverse with regard to other structural features (Fig. 1). The clustered genes cro3, cro4, and cro5 show remarkable conservation of exon/intron positions and form a subfamily of sequences that contain N-terminal WSC domains of unknown function. cro6 is most closely related to the GLX-encoding gene glx, but it also features a 200-amino-acid N-terminal region of unknown function.
The expression of cro genes is consistent with a role(s) in lignocellulose degradation. cDNAs of all six genes were identified in extensively decayed wood wafers. Transcript patterns determined by competitive RT-PCR showed differential regulation among the cro genes over the 120-day time course (Fig. 2). The absence of cro4 and cro6 transcripts at 3 days of colonization suggests that these genes are not essential for hyphal penetration and early decay. Concentrated filtrates of P. chrysosporium grown in defined submerged medium were analyzed by LC-MS/MS and shown to contain a CRO2-specific peptide. The apparent absence of other CRO proteins in extracellular fluids may reflect low protein concentrations, compartmentalization, or, particularly in the case of WSC-containing genes, cell wall binding.
In addition to the cro genes, complex gene families are well known in P. chrysosporium, particularly among sequences encoding secreted proteins. Examples include LiPs (36), certain glycosyl hydrolases (32), and peptidases (34, 41). The role of such genetic multiplicity remains poorly understood. Structurally, the 10 LiP genes are highly conserved and all are believed to encode high-oxidation potential, nonspecific enzymes. However, slight differences in isozyme activities (14) and differential transcriptional regulation (18, 36) suggest that the LiP genes are not merely redundant. Perhaps subtle differences in specificities enhance the efficiency of cell wall degradation under a broad range of environmental conditions. Targeted disruption or suppression would help clarify the role of individual genes, but suitable transformation systems are not yet available for P. chrysosporium.
Beyond structural diversity and differential regulation of the cro genes, substrate preferences argue in favor of distinct biological roles, at least for glx and cro2. Our results show that heterologously produced CRO2 oxidizes glycolaldehyde dimer, but not methylglyoxal, the prototypical substrate for GLX. Therefore, the two oxidases are distinguished by catalytic differences. Clarification of their physiological roles, as well as those of the other CRO enzymes, will require comprehensive characterizations, including extensive substrate specificity screenings.
Acknowledgments
This work was supported by U.S. Department of Energy grant no. DE-FG02-87ER13712.
We thank Angel Martinez for the gift of Aspergillus nidulans IJFM A729, Mark Davis for chemical analysis, and Benjamin Held for laboratory assistance.
REFERENCES
- 1.Archer, D. B., and J. F. Peberdy. 1997. The molecular biology of secreted enzyme production by fungi. Crit. Rev. Biotechnol. 17:273-306. [DOI] [PubMed] [Google Scholar]
- 2.Ayers, A. R., S. B. Ayers, and K.-E. Eriksson. 1978. Cellobiose oxidase, purification and partial characterization of a hemoprotein from Sporotrichum pulverulentum. Eur. J. Biochem. 90:171-181. [DOI] [PubMed] [Google Scholar]
- 3.Ballance, D. J., F. P. Buxton, and G. Turner. 1983. Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem. Biophys. Res. Commun. 112:284-289. [DOI] [PubMed] [Google Scholar]
- 4.Blom, N., T. Sicheritz-Ponten, R. Gupta, S. Gammeltoft, and S. Brunak. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4:1633-1649. [DOI] [PubMed] [Google Scholar]
- 5.Bogan, B., B. Schoenike, R. Lamar, and D. Cullen. 1996. Manganese peroxidase mRNA and enzyme activity levels during bioremediation of polycyclic aromatic hydrocarbon-contaminated soil with Phanerochaete chrysosporium. Appl. Environ. Microbiol. 62:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Kupiec, R., K. E. Broglie, D. Friesem, R. M. Broglie, and I. Chet. 1999. Molecular characterization of a novel beta-1,3-exoglucanase related to mycoparasitism of Trichoderma harzianum. Gene 226:147-154. [DOI] [PubMed] [Google Scholar]
- 7.Cullen, D., and P. J. Kersten. 2004. Enzymology and molecular biology of lignin degradation, p. 249-273. In R. Brambl and G. A. Marzulf (ed.), The mycota III biochemistry and molecular biology. Springer-Verlag, Berlin, Germany.
- 8.Cullen, D., L. Wilson, D. Henner, G. Turner, and J. Ballance. 1987. High frequency transformation of Aspergillus nidulans conferred by novel sequence ANS-1. Nucleic Acids Res. 15:9163-9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, M. 1998. A rapid method for compositional carbohydrate analysis of lignocellulosics by high pH anion-exchange chromatography with pulsed amperometric detection (HPAEC/PAD). J. Wood Chem. Technol. 18:235-252. [Google Scholar]
- 10.Eriksson, K.-E., and S. G. Hamp. 1978. Regulation of endo-1,4-b-glucanase production in Sporotrichum pulverulentum. Eur. J. Biochem. 90:183-190. [DOI] [PubMed] [Google Scholar]
- 11.Forney, L. J., C. A. Reddy, M. Tien, and S. D. Aust. 1982. The involvement of hydroxyl radical derived from hydrogen peroxide in lignin degradation by the white rot fungus Phanerochaete chrysosporium. J. Biol. Chem. 257:11455-11462. [PubMed] [Google Scholar]
- 12.Fowler, T., R. Berka, and M. Ward. 1990. Regulation of the glaA gene of Aspergillus niger. Curr. Genet. 18:537-545. [DOI] [PubMed] [Google Scholar]
- 13.Gilliland, G., S. Perrin, and H. Bunn. 1990. Competitive PCR for quantitation of mRNA, p. 60-69. In M. Innis, D. Gelfand, J. Sninsky, and T. White (ed.), PCR protocols. Academic Press, New York, N.Y.
- 14.Glumoff, T., P. J. Harvey, S. Molinari, M. Goble, G. Frank, J. M. Palmer, J. D. G. Smit, and M. S. A. Leisola. 1990. Lignin peroxidase from Phanerochaete chrysosporium: molecular and kinetic characterization of isozymes. Eur. J. Biochem. 187:515-520. [DOI] [PubMed] [Google Scholar]
- 15.Goodell, B. 2003. Brown rot fungal degradation of wood: our evolving view, p. 97-118. In B. Goodell, D. Nicholas, and T. Schultz (ed.), Wood deterioration and preservation. American Chemical Society, Washington, D.C.
- 16.Hammel, K. E., M. D. Mozuch, K. A. Jensen, and P. J. Kersten. 1994. H2O2 recycling during oxidation of the arylglycerol β-aryl ether lignin structure by lignin peroxidase and glyoxal oxidase. Biochemistry 33:13349-13354. [DOI] [PubMed] [Google Scholar]
- 17.Higuchi, T. 1990. Lignin biochemistry: biosynthesis and biodegradation. Wood Sci. Technol. 24:23-63. [Google Scholar]
- 18.Holzbaur, E., and M. Tien. 1988. Structure and regulation of a lignin peroxidase gene from Phanerochaete chrysosporium. Biochem. Biophys. Res. Commun. 155:626-633. [DOI] [PubMed] [Google Scholar]
- 19.Horton, R., H. Hunt, S. Ho, J. Pullen, and L. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 20.Ito, N., S. E. Phillips, C. Stevens, Z. B. Ogel, M. J. McPherson, J. N. Keen, K. D. Yadav, and P. F. Knowles. 1991. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature 350:87-90. [DOI] [PubMed] [Google Scholar]
- 21.Janse, B. J. H., J. Gaskell, M. Akhtar, and D. Cullen. 1998. Expression of Phanerochaete chrysosporium genes encoding lignin peroxidases, manganese peroxidases, and glyoxal oxidase in wood. Appl. Environ. Microbiol. 64:3536-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerem, Z., and K. E. Hammel. 1999. Biodegradative mechanism of the brown rot basidiomycete Gloeophyllum trabeum: evidence for an extracellular hydroquinone-driven Fenton reaction. FEBS Lett. 446:49-54. [DOI] [PubMed] [Google Scholar]
- 23.Kersten, P., and D. Cullen. 1993. Cloning and characterization of a cDNA encoding glyoxal oxidase, a peroxide-producing enzyme from the lignin-degrading basidiomycete Phanerochaete chrysosporium. Proc. Natl. Acad. Sci. USA 90:7411-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersten, P. J. 1990. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc. Natl. Acad. Sci. USA 87:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kersten, P. J., and T. K. Kirk. 1987. Involvement of a new enzyme, glyoxal oxidase, in extracellular H2O2 production by Phanerochaete chrysosporium. J. Bacteriol. 169:2195-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kersten, P. J., C. Witek, A. Vanden Wymelenberg, and D. Cullen. 1995. Phanerochaete chrysosporium glyoxal oxidase is encoded by two allelic variants: structure, genomic organization and heterologous expression of glx1 and glx2. J. Bacteriol. 177:6106-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirk, T. K., and R. L. Farrell. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465-505. [DOI] [PubMed] [Google Scholar]
- 28.Kirk, T. K., E. Schultz, W. J. Conners, L. F. Lorentz, and J. G. Zeikus. 1978. Influence of culture parameters on lignin metabolism by Phanerochaete chrysosporium. Arch. Microbiol. 117:277-285. [Google Scholar]
- 29.Leuthner, B., C. Aichinger, E. Oehmen, E. Koopmann, O. Muller, P. Muller, R. Kahmann, M. Bolker, and P. H. Schreier. 2005. A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol. Genet. Genomics 272:639-650. [DOI] [PubMed] [Google Scholar]
- 30.Lodder, A. L., T. K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansfield, S. D., E. deJong, and J. N. Saddler. 1997. Cellobiose dehydrogenase, an active agent in cellulose depolymerization. Appl. Environ. Microbiol. 63:3804-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez, D., L. F. Larrondo, N. Putnam, M. D. Sollewijn Gelpke, K. Huang, J. Chapman, K. G. Helfenbein, P. Ramaiya, J. C. Detter, F. Larimer, P. M. Coutinho, B. Henrissat, R. Berka, D. Cullen, and D. Rokhsar. 2004. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat. Biotechnol. 22:695-700. [DOI] [PubMed] [Google Scholar]
- 33.Oakley, B. R., J. E. Rinehart, B. L. Mitchell, C. E. Oakley, C. Carmona, G. L. Gray, and G. S. May. 1987. Cloning, mapping and molecular analysis of the pyrG (orotidine-5′-phosphate decarboxylase) gene of Aspergillus nidulans. Gene 61:385-399. [DOI] [PubMed] [Google Scholar]
- 34.Sims, A. H., N. S. Dunn-Coleman, G. D. Robson, and S. G. Oliver. 2004. Glutamic protease distribution is limited to filamentous fungi. FEMS Microbiol. Lett. 239:95-101. [DOI] [PubMed] [Google Scholar]
- 35.Smith, T. F., and M. S. Waterman. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147:195-197. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, P., and D. Cullen. 1999. Organization and differential regulation of a cluster of lignin peroxidase genes of Phanerochaete chrysosporium. J. Bacteriol. 181:3427-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, P., J. Gaskell, and D. Cullen. 2000. A homokaryotic derivative of a Phanerochaete chrysosporium strain and its use in genomic analysis of repetitive elements. Appl. Environ. Microbiol. 66:1629-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. ClustalW improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallim, M. A., B. J. Janse, J. Gaskell, A. A. Pizzirani-Kleiner, and D. Cullen. 1998. Phanerochaete chrysosporium cellobiohydrolase and cellobiose dehydrogenase transcripts in wood. Appl. Environ. Microbiol. 64:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanden Wymelenberg, A., S. Denman, D. Dietrich, J. Bassett, X. Yu, R. Atalla, P. Predki, U. Rudsander, T. T. Teeri, and D. Cullen. 2002. Transcript analysis of genes encoding a family 61 endoglucanase and a putative membrane-anchored family 9 glycosyl hydrolase from Phanerochaete chrysosporium. Appl. Environ. Microbiol. 68:5765-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanden Wymelenberg, A., P. Minges, G. Sabat, D. Martinez, A. Aerts, A. Salamov, I. Grigoriev, H. Shapiro, N. Putnam, P. Belinky, C. Dosoretz, J. Gaskell, P. Kersten, and D. Cullen. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveals complex mixtures of secreted proteins. Fungal Genet. Biol. 43:343-356. [DOI] [PubMed] [Google Scholar]
- 42.Vanden Wymelenberg, A., G. Sabat, D. Martinez, A. S. Rajangam, T. T. Teeri, J. Gaskell, P. J. Kersten, and D. Cullen. 2005. The Phanerochaete chrysosporium secretome: database predictions and initial mass spectrometry peptide identifications in cellulose-grown medium. J. Biotechnol. 118:17-34. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe, T., N. Shirai, H. Okada, Y. Honda, and M. Kuwahara. 2001. Production and chemiluminescent free radical reactions of glyoxal in lipid peroxidation of linoleic acid by the ligninolytic enzyme, manganese peroxidase. Eur. J. Biochem. 268:6114-6122. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker, M. M., P. J. Kersten, D. Cullen, and J. W. Whittaker. 1999. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J. Biol. Chem. 274:36226-36232. [DOI] [PubMed] [Google Scholar]
- 45.Whittaker, M. M., P. J. Kersten, N. Nakamura, J. Sanders-Loehr, E. S. Schweizer, and J. W. Whittaker. 1996. Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J. Biol. Chem. 271:681-687. [DOI] [PubMed] [Google Scholar]
- 46.Wood, J. D., and P. M. Wood. 1992. Evidence that cellobiose:quinone oxidoreductase from Phanerochaete chrysosporium is a breakdown product of cellobiose oxidase. Biochim. Biophys. Acta 1119:90-96. [DOI] [PubMed] [Google Scholar]