Abstract
Yersinia pestis is the etiologic agent of plague, a disease that is transmitted from rodent to rodent and from rodent to humans by fleabites. Multiple copies of three insertion sequences (IS100, IS285, and IS1541) are scattered over the Y. pestis genome. The genomic instability generated by these insertion sequences (IS) creates a polymorphism of the hybridizing restriction fragments (restriction fragment length polymorphism [RFLP]) which can be used to subtype this relatively clonal species. The aim of this work was to evaluate and compare the potential of the three IS-RFLP techniques, individually or in combination, to define clusters of strains according to their focus of origin. The analysis of 61 Y. pestis isolates of worldwide origin indicated that no satisfactory strain clustering was observed with each IS-RFLP used individually. In contrast, the combination of the three IS-RFLP data (3IS-RFLP) resulted in both an efficient strain discrimination (D = 0.999) and a robust clustering of the isolates according to their biovar and geographical origin. This geographical clustering was observed even within the Orientalis group, although these strains had only a short period of time (one century) to diverge from the original clone that spread globally. Therefore, 3IS-RFLP is a technique that may be useful for addressing epidemiological problems and forensic issues. When plague reemerges after several decades of silence in a quiescent focus, it may help in determining whether the disease was reimported or reactivated. It may also be of value to identify the origin of a strain when plague cases appear in a previously plague-free region. Finally, this technique could be useful for the tracing of a Y. pestis isolate that has been used as a biological terrorism threat.
Yersinia pestis is a gram-negative bacillus which belongs to the family Enterobacteriaceae. This bacterium is the etiologic agent of plague, a disease that is transmitted from rodent to rodent by fleabites. Most often, humans become infected after an infectious fleabite and develop a bubonic form of plague. If the bacillus secondarily reaches the lungs of the patient, it causes a pneumonia which allows a direct human-to-human transmission through the spread of infected aerosols. Plague foci still persist that are endemic to Africa (mainly the Southern part), Central and East Asia, and North and South America (31).
Plague is considered a reemerging disease (27). This is due to the rise in human plague cases since the beginning of the 1990s and the reappearance of the disease in countries where no cases where reported for several decades, such as in the Northern seaport of Majunga in Madagascar in 1991 (26), in India (25) and Mozambique (4) in 1994, in Zambia in 1996 (30), and recently, in Algeria in 2003 (32). Whether these outbreaks were due to the reactivation of quiescent autochthon plague foci or were imported from distant countries by modern means of transportation is a question of key importance for the implementation of appropriate and efficient control measures. Having a molecular tool which could cluster Y. pestis strains based on their geographical focus, even when isolated at intervals of several years, would be of great help to answer this question. Furthermore, Y. pestis has been categorized as one of the major bacterial agents of bioterrorism (category A) (17). Having the possibility to trace an isolate that has been used as a biological weapon would also be critical.
Y. pestis has been shown to be a clone of Yersinia pseudotuberculosis that emerged less than 20,000 years ago (2). This recent clonal expansion accounts for the limited phenotypic and genetic diversity observed in this species. Phenotypically, the ability to ferment glycerol and to reduce nitrates to nitrites led to the subdivision of Y. pestis into three biovars: Antiqua, Medievalis, and Orientalis (10). Today, most of the strains isolated worldwide belong to biovar Orientalis, the biovar that spread globally from Hong Kong in 1894 during the third plague pandemic. The two other biovars have a geographically restricted distribution: Medievalis in Asia and Antiqua in some parts of Africa and in Central Asia. Y. pestis also displays a very low degree of genetic polymorphism. An analysis of six housekeeping genes by multilocus sequence typing demonstrated a complete lack of nucleotide polymorphism among 36 strains belonging to the three biovars and isolated from various countries at different times (2). A recent comparison of the three available Y. pestis genome sequences confirmed the very low genetic diversity of this species: as few as 80 synonymous single-nucleotide polymorphisms were detected among 3,250 orthologous coding sequences (1).
Despite the very high degree of conservation at the gene level, the genotypic subdivision of the species Y. pestis has been possible. Techniques using variations in the number of repeated sequences, usually located in noncoding chromosomal regions, such as variable-number tandem repeats (3), clustered regularly interspaced short palindromic repeats (24), or intergenic spacer sequences (11), have successfully allowed the discrimination of strains within the same biovar. Another group of techniques, including ribotyping (13) and pulsed-field gel electrophoresis (PFGE) (19), uses the restriction fragment length polymorphism (RFLP) generated by the high genome instability of Y. pestis (13, 22) to subdivide the species.
This genome instability is in large part attributable to the presence of multiple copies of insertion sequences (IS) scattered over the Y. pestis genome (9, 22, 29). Insertion sequences are simple genetic elements which can insert at multiple sites in a bacterial genome. Recombination between these IS may lead to chromosomal macro-rearrangements (20). The most numerous ISs are IS100 (30 to 44 chromosomal copies on the three Y. pestis sequenced genomes), IS1541 (43 to 62 copies), and IS285 (18 to 20 copies) (9, 22, 29). Variations in the chromosomal location of IS100 have been used to study the microevolution of Y. pestis (1, 21). Furthermore, RFLP analyses using one of these IS as a probe (IS-RFLP) have been applied to subtype Y. pestis (6, 15, 28). IS100-RFLP discriminated strains isolated from the same country (United States) more efficiently than IS285-RFLP, but both techniques were found to be inferior to PFGE for this purpose (15).
The aim of this work was to evaluate and compare the potential of each of the three IS-RFLP techniques (IS100-, IS285-, and IS1541-RFLP), individually or in combination, to define clusters of strains according to their focus of origin.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Sixty-one strains of Y. pestis isolated between 1908 and 1994 from 17 countries on four continents (Table 1) were taken from the strain collection of the Yersinia Research Unit (Institut Pasteur) and analyzed. These strains belonged to the three classical biovars of Y. pestis (10): Antiqua (ferment glycerol [G+] and reduce nitrates to nitrites [N+]), Medievalis (G+, N−), and Orientalis (G−, N+). Bacterial suspensions were prepared from stock cultures kept at −80°C and streaked onto Luria Bertani agar plates containing 0.002% hemin or grown in peptone broth with shaking for 24 to 48 h at 28°C.
TABLE 1.
Characteristics of the 61 strains of Y. pestis analyzed by IS-RFLP
| Institut Pasteur no. (n) | Original designation | Geographical origina | Yr of isolation | Biotype | IS100 type | IS285 type | IS1541 type |
|---|---|---|---|---|---|---|---|
| Asia (19) | |||||||
| IP772 | Nhatrang 63-127 | Vietnam | 1963 | Orientalis | 17 | 11 | 22 |
| IP513 | Nhatrang 64-65 | Vietnam | 1964 | Orientalis | 13 | 11 | 24 |
| IP820 | Nhatrang 64-260 | Vietnam | 1964 | Orientalis | 14 | 12 | 23 |
| IP940 | Nhatrang 65-30 | Vietnam | 1965 | Orientalis | 19 | 13 | 22 |
| IP507 | Saigon 55-720 | Vietnam | 1955 | Orientalis | 21 | 11 | 25 |
| IP532 | Saigon 55-801 | Vietnam | 1955 | Orientalis | 21 | 16 | 20 |
| IP989 | Dalat 131 | Vietnam | 1964 | Orientalis | 15 | 11 | 22 |
| IP613 | 548 | Burma | 1970 | Orientalis | 30 | 11 | 22 |
| IP612 | 637 | Burma | 1970 | Orientalis | 30 | 19 | 26 |
| IP519 | PKH4 | Kurdistan | 1951 | Medievalis | 33 | 22 | 14 |
| IP562 | PKR6 | Kurdistan | 1947 | Medievalis | 32 | 22 | 14 |
| IP516 | PKR18 | Kurdistan | 1948 | Medievalis | 33 | 25 | 14 |
| IP564 | PKR25 | Kurdistan | 1948 | Medievalis | 32 | 23 | 13 |
| IP557 | PKR292 | Kurdistan | 1963 | Medievalis | 34 | 24 | 14 |
| IP565 | 10/5 | Turkey | 1952 | Medievalis | 35 | 26 | 14 |
| IP521 | 10/1 | Turkey | UNb | Orientalis | 1 | 1 | 19 |
| IP283 | Surat 9/95 | India | 1994 | Orientalis | 4 | 15 | 10 |
| IP1595 | Surat 4/95 | India | 1994 | Orientalis | 4 | 15 | 10 |
| IP579 | Bombay 195 | India | 1908 | Orientalis | 28 | 15 | 12 |
| America (8) | |||||||
| IP567 | Exu 21 | Brazil | 1967 | Orientalis | 24 | 5 | 12 |
| IP568 | Exu 53 | Brazil | 1967 | Orientalis | 24 | 5 | 18 |
| IP569 | Exu 56 | Brazil | 1967 | Orientalis | 24 | 5 | 15 |
| IP571 | Exu 184 | Brazil | 1967 | Orientalis | 24 | 5 | 11 |
| IP1747 | CO92 | United States | 1992 | Orientalis | 22 | 15 | 13 |
| IP573 | 7793 | United States | 1948 | Orientalis | 18 | 15 | 13 |
| IP574 | 193 | United States | 1950 | Orientalis | 8 | 15 | 13 |
| IP610 | Cuis 14 | Argentina | 1946 | Orientalis | 16 | 6 | 13 |
| Europe (3) | |||||||
| IP695 | Hamburg 9 | Germany | 1952 | Orientalis | 9 | 10 | 2 |
| IP685 | Hamburg 10 | Germany | 1952 | Orientalis | 9 | 5 | 13 |
| IP696 | Hamburg 26 | Germany | 1952 | Orientalis | 9 | 5 | 13 |
| Africa (31) | |||||||
| IP577 | 243 | Morocco | 1940 | Orientalis | 13 | 15 | 1 |
| IP578 | 48 | Morocco | 1940 | Orientalis | 10 | 15 | 21 |
| IP1867 | Arn | Algeria | 1945 | Orientalis | 23 | 20 | 27 |
| IP523 | Th | Senegal | 1944 | Orientalis | 12 | 17 | 3 |
| IP524 | Fa | Senegal | 1944 | Orientalis | 9 | 3 | 31 |
| IP550 | Ed | Kenya | UN | Antiqua | 41 | 27 | 30 |
| IP544 | Mi | Kenya | UN | Antiqua | 43 | 30 | 29 |
| IP677 | Béa | Kenya | UN | Antiqua | 42 | 33 | 28 |
| IP554 | Ros | Kenya | UN | Antiqua | 47 | 29 | 28 |
| IP545 | Da | Kenya | UN | Antiqua | 44 | 31 | 29 |
| IP552 | Ma | Kenya | UN | Antiqua | 46 | 28 | 28 |
| IP553 | Ky | Kenya | UN | Antiqua | 45 | 32 | 29 |
| IP542 | 144 | Kenya | 1952 | Antiqua | 40 | 36 | 7 |
| IP537 | 147 | Kenya | 1952 | Antiqua | 36 | 38 | 7 |
| IP539 | 164 | Kenya | 1952 | Antiqua | 38 | 34 | 8 |
| IP549 | logo 1/53 | Belgian Congo | 1953 | Antiqua | 39 | 37 | 5 |
| IP678 | Eli | Belgian Congo | 1950 | Orientalis | 2 | 4 | 6 |
| IP543 | Lita | Belgian Congo | 1953 | Antiqua | 37 | 35 | 7 |
| IP1537 | 254 | Namibia | 1984 | Orientalis | 27 | 18 | 32 |
| IP1538 | 272 | Namibia | 1984 | Orientalis | 26 | 14 | 34 |
| IP1540 | 292 | Namibia | 1985 | Orientalis | 26 | 18 | 35 |
| IP1541 | 340 | Namibia | 1986 | Orientalis | 25 | 14 | 33 |
| IP1542 | 346 | Namibia | 1986 | Orientalis | 29 | 2 | 32 |
| IP1535 | 164 | South Africa | 1982 | Orientalis | 5 | 16 | 9 |
| IP304 | 6/69 | Madagascar | 1969 | Orientalis | 9 | 15 | 10 |
| IP528 | 62 | Madagascar | 1946 | Orientalis | 20 | 8 | 10 |
| IP529 | 112 | Madagascar | 1951 | Orientalis | 11 | 9 | 4 |
| IP530 | Ga | Madagascar | >1939 | Orientalis | 3 | 21 | 10 |
| IP241 | 3/89 | Madagascar | 1989 | Orientalis | 6 | 7 | 15 |
| IP644 | 6/89 | Madagascar | 1989 | Orientalis | 31 | 7 | 17 |
| IP666 | 100/92 | Madagascar | 1992 | Orientalis | 7 | 7 | 16 |
Name of the country at time of strain isolation.
UN, unknown.
DNA extraction, restriction, and transfer to nylon membrane.
Total DNA extraction from each Y. pestis strain was performed as described previously (7). Plasmid extracts were obtained by the method of Birnboim and Doly (5). Five micrograms of each sample was digested overnight at 37°C with various restriction enzymes before being loaded onto 0.8% agarose gels and subjected to electrophoresis (50 V in 1× Tris-Borate-EDTA buffer) for 24 h (IS285- and IS1541-RFLP) or 26 h (IS100-RFLP). The DNA of strain IP304 was systematically loaded on the flanking and middle lanes of each gel to serve for intra- and intergel normalization. DNA bands were stained with ethidium bromide. Alkaline denaturation, neutralization, and transfer of total DNA onto nylon filters (Hybond N+; Amersham, England) with a VacuGene apparatus (Pharmacia LKB Biotechnology, Uppsala, Sweden) were performed as previously described (13).
Preparation of IS probes and hybridization.
For IS fingerprinting, three sets of primers were designed and used to amplify a portion of the insertion sequences: IS100-F, 5′-AAAACGTTCGAAGAGTATGA-3′; IS100-R, 5′-GATGAGCAGGCGGGGGGCCA-3′ (255 bp); IS1541-F, 5′-AAAGCTTTCAGCTTTGGGTC-3′; IS1541-R, 5′-TCTTTCCCTTCAGGTACCCC-3′ (319 bp); IS285-F, 5′-AGCTTACCGAACACCTCGGG-3′; IS285-R, 5′-GTTGATGCCCAGCGCTAGGA-3′ (406 bp). The PCR amplification reactions were performed on the genomic DNA of Y. pestis strain IP304 as a template in a final volume of 50 μl with 1.25 U of Taq DNA polymerase (Roche/Cetus) used with the supplier's buffer, 2 mM MgCl2, and 200 μM concentrations of each of the four deoxynucleoside triphosphates. All primers were used at a final concentration of 1 μM. Each reaction involved a denaturing step at 94°C for 3 min, 25 cycles of amplification consisting of three steps each of 1 min at 94°C, 55°C, and 72°C, and a final extension step of 10 min at 72°C. PCR products were subjected to electrophoresis in 0.7% agarose gels and stained with ethidium bromide. The three IS probes were peroxidase labeled using the ECL direct nucleic acid labeling and detection system (Amersham) and used for Southern hybridization overnight at 42°C.
Bioinformatic analysis of IS-RFLP patterns.
The hybridization patterns obtained with each IS were scanned, and the computerized data were analyzed using the BioNumerics software, version 4.0 (Applied Maths, Kortrijk, Belgium). Bands automatically assigned by the computer were checked visually and corrected manually when necessary. A position tolerance of 1.8 was selected for each of the three IS. Cluster analysis of the individual or combined IS-RFLP patterns was done by the unweighted pair group method with average linkages (UPGMA), using the Dice coefficient to analyze the similarities of the banding patterns. The discriminatory power of each IS-RFLP was determined by calculating the discrimination index (D) based on Simpson's index of diversity (16). D depends on the number of types defined by the test method and the relative frequencies of these types: D = 1 − {[(1/(N × (N − 1)))] × [Σ nj (nj − 1)]}, where N is the total number of unrelated isolates, and nj is the number of strains that belong to the jth type. Two IS profiles were considered identical when their percent similarity was >98%.
RESULTS
Optimization of IS-RFLP conditions.
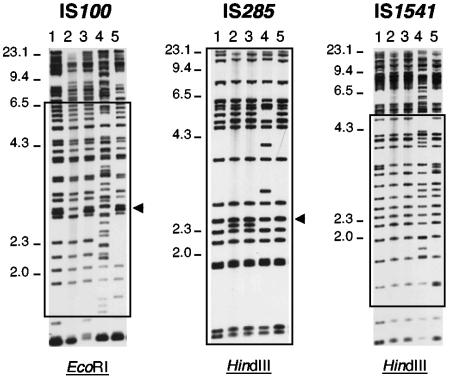
The three insertion sequences IS100, IS285, and IS1541 were chosen because they are present in the highest copy numbers on the genome of Y. pestis (9, 22, 29). Primers internal to each IS were designed and used to generate probes. To obtain the most distinct banding patterns, various restriction enzymes (EcoRI, HindIII, EcoRV, BamHI, PvuI, XbaI, XhoI, and XmaI) were tested on the DNA of a set of Y. pestis strains which were hybridized with each of the three IS probes. The best resolution was obtained when the genomic DNAs were digested with EcoRI for IS100-RFLP and with HindIII for IS285- and IS1541-RFLP. The duration of DNA electrophoresis was also varied to optimize band separation for each IS. Optimal separation of the hybridizing bands was obtained after 26 h of migration for IS100-RFLP and 24 h for IS285- and IS1541-RFLP. Because some hybridizing fragments with the largest or smallest sizes were not sufficiently resolved, a size window containing bands that were clearly and systematically separated on all Southern blots was defined for each IS (Fig. 1).
FIG. 1.
Examples of IS-RFLP profiles obtained after digestion and hybridization of the genomic DNA of various Y. pestis strains with each IS probe. Lanes: 1, IP304 (used as an intra- and intergel standard); 2, IP696; 3, IP685; 4, IP677; 5, IP666. Tick marks on the left indicate the sizes of the molecular mass markers (lambda DNA HindIII digest) in kilobases. Rectangles correspond to the size windows chosen for the analysis of the hybridization profiles for each IS. Arrows on the right indicate a hybridizing plasmid band (from pYV for IS100 and pFra for IS285).
Since the presence of IS copies on the three Y. pestis plasmids, pFra (101 kb), pYV (70.5 kb), and pPla (9.6 kb), may artificially introduce a variation in the hybridization profile due to the loss in vitro of one or several plasmids upon subcultures, the expected sizes of the plasmid-borne hybridizing fragments were determined based on the Y. pestis plasmid sequences available in the databases (9, 14, 18, 22). The plasmid bands were further identified by extracting the DNA of 11 different Y. pestis strains containing either one, two, or three of the Y. pestis plasmids and hybridizing them with each IS. For IS100-RFLP, a 2.75-kb pYV hybridizing fragment was present within the size window and was thus manually removed for cluster analysis (Fig. 1). Similarly, a 2.36-kb pFra fragment present in the IS285-RFLP size window was withdrawn before performing the UPGMA analysis. All other plasmid-borne IS sequences were located on fragments outside the size windows selected. The in vitro instability of the 102-kb pgm locus was another potential source of artificial profile variation (8, 12). The in silico analysis of the IS positions on the Y. pestis sequenced genomes indicated that deletion of the pgm locus does not modify the IS285- and IS1541-RFLP profiles. In contrast, deletion of this locus was predicted to result in the loss of a 1.5-kb EcoRI fragment on the IS100-RFLP pattern. This was confirmed by comparing the IS100 hybridization profiles of the wild-type strain 6/69 and its Δpgm derivative (data not shown). However, since this fragment is not in the size window selected, the instability of the pgm locus had no effect on the IS100-RFLP pattern.
The reproducibility of the clustering was evaluated by comparing the position on the UPGMA dendrograms of 12 Y. pestis strains studied independently (DNA extraction, digestion, migration, preparation of probes, and hybridizations) by two different investigators at intervals of several months. The same strains studied independently always fell into the same clusters with each IS-RFLP method.
IS100-RFLP profiles.
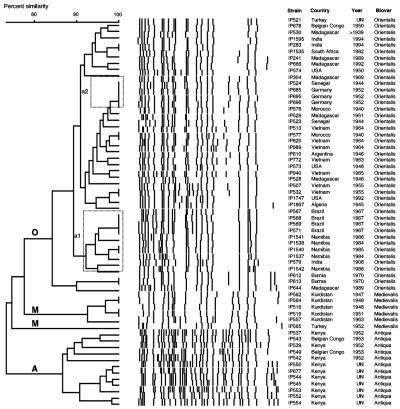
When IS100-RFLP was applied to the 61 Y. pestis strains analyzed, 47 profiles were identified (Table 1), leading to a discrimination index of 0.987. No dominant IS100 profile was noted. As previously observed (2), the three biovars formed distinct clusters in the UPGMA dendrogram (Fig. 2). The weight of the biovar was superior to that of the geographic origin in the clustering analysis. For instance, the three strains from Belgian Congo were separated into the Antiqua and Orientalis branches, although they were isolated from the same country. The same held true for the two strains from Turkey (biovar Medievalis and Orientalis) (Fig. 2). For the Orientalis strains, which are found worldwide because of their recent global spread, no strong geographical clustering was achieved by IS100-RFLP (Fig. 2). Nonetheless, a tendency of the strains from the same country to be grouped into some subbranches was sometimes observed. This was the case for all the strains from Brazil and Namibia, which were grouped in cluster a1, while cluster a2 contained all the isolates from Germany (Fig. 2). In several instances, strains isolated the same year from the same region, such as those from Burma in 1970, Hamburg (Germany) in 1952, Saigon (Vietnam) in 1955, Exu (Brazil) in 1967, and Surat (India) in 1994 displayed highly similar or identical IS100 types (Table 1 and Fig. 2).
FIG. 2.
Dendrogram generated from the IS100-RFLP patterns of the 61 Y. pestis strains studied using the UPGMA clustering analysis with the BioNumerics software. A position tolerance of 1.8% was chosen. Dotted rectangles outline clusters of strains of interest. A, Antiqua branch; M, Medievalis branch; O, Orientalis branch.
IS285-RFLP profiles.
Thirty-eight IS285-RFLP types were identified among the 61 Y. pestis strains analyzed (D = 0.963). Three IS285 types represented 33% of the strains: IS285 type 15 (9 strains), IS285 type 5 (6 strains), and IS285 type 11 (5 strains) (Table 1). The UPGMA dendrogram delineated three main branches (Fig. 3). One branch was entirely composed of Antiqua strains. The second branch was split into two subbranches, one corresponding to biovar Antiqua and the other to biovar Medievalis strains (Fig. 3). The Orientalis strains were all found in a third branch (Fig. 3). Within the Orientalis group, no clear clustering of the strains according to their focus of origin was noted. Some strains isolated from the same country were found in different subbranches. Grouping of strains from the same geographic origin could nonetheless be observed, as for the strains from Brazil which were all found in cluster b1 (Fig. 3). Clusters b2, b3, and b4 each contained only strains from Madagascar, Southeast Asia, and Namibia, respectively (Fig. 3). Of note was the fact that identical IS285 types were found for the two strains from Morocco. This was also the case for the three strains from India and for the three strains from the United States (Table 1 and Fig. 3).
FIG. 3.
Dendrogram generated from the IS285-RFLP patterns of the 61 Y. pestis strains studied using the UPGMA clustering analysis with the BioNumerics software. A position tolerance of 1.8% was chosen. Dotted rectangles outline clusters of strains of interest. A, Antiqua branch; M, Medievalis branch; O, Orientalis branch.
IS1541-RFLP profiles.
IS1541-RFLP analysis allowed the delineation of 35 profiles (D = 0.968). Three of them, IS1541 type 13 (7 strains), IS1541 type 14 (5 strains), and IS1541 type 10 (5 strains), represented 28% of the Y. pestis isolates analyzed (Table 1). The UPGMA dendrogram did not exhibit biovar-specific branches (Fig. 4). Most of the Antiqua strains were grouped in two clusters and the Medievalis strains were grouped in one cluster, but these clusters were interspersed among Orientalis clusters. Furthermore, one Antiqua strain (IP549) and one Medievalis strain (IP564) were found within Orientalis branches (Fig. 4). In contrast to the two other IS-RFLPs, the two strains from Belgian Congo, although of different biovars, were found in the same cluster. Among the Orientalis strains, three clusters exhibiting some geographic specificity were identified. Cluster c1 was composed of strains from Africa (except one strain from Germany), cluster c2 was exclusively constituted of strains from Southeast Asia (Vietnam and Burma), and cluster c3 contained all strains from Namibia (Fig. 4).
FIG. 4.
Dendrogram generated from the IS1541-RFLP patterns of the 61 Y. pestis strains studied using the UPGMA clustering analysis with the BioNumerics software. A position tolerance of 1.8% was chosen. Dotted rectangles outline clusters of strains of interest. A, Antiqua branch; M, Medievalis branch; O, Orientalis branch.
Combination of two IS-RFLP profiles.
Although each IS-RFLP technique allowed, to some extent, a geographical grouping of strains, this clustering remained limited and varied with each IS. To determine whether a combination of two IS would improve the geographically based Y. pestis clustering, the IS-RFLP data were combined two by two and analyzed. The discrimination indexes increased for the three combinations: IS100/IS285-RFLP, D = 0.995; IS1541/IS285-RFLP, D = 0.993; and IS100/IS1541-RFLP, D = 0.998. While IS100/IS285-RFLP and IS100/IS1541-RFLP allowed a clear delineation of the three biovars into distinct main branches, this distinction was less pronounced with IS1541/IS285-RFLP (see Fig. S1, S2, and S3 in the supplemental material). Within the Orientalis group, combining two IS-RFLPs improved strain grouping. With each of the three IS-RFLP combinations, all isolates from Namibia and all those from the United States fell into specific clusters. This was also true for the isolates from Burma, but only IS1541/IS285-RFLP grouped these strains with those from the neighboring Vietnam. IS100/IS285-RFLP and IS100/IS1541-RFLP, but not IS1541/IS285-RFLP, allowed the clustering of Brazilian isolates. Interestingly, the strains from Germany fell into the same cluster as the isolate from Argentina with each of the three combined IS-RFLPs. IS100/IS285-RFLP was the only combination which grouped the three strains from India into the same cluster (which also contained isolates from other geographical origins). In contrast, strains from Madagascar fell into different clusters with each of the three IS-RFLP combinations (see Fig. S1, S2, and S3 in the supplemental material).
Combination of three IS-RFLP profiles.
When the data obtained with the three IS-RFLP techniques were combined (3IS-RFLP), strain discrimination became extremely efficient (D = 0.999). Only two pairs of strains displayed identical 3IS-RFLP patterns (Table 1 and Fig. 5). The three biovars formed three distinct main branches on the UPGMA dendrogram. The Antiqua branch split into two subbranches, and interestingly, the two isolates from Belgian Congo clustered into the same Antiqua subbranch (Fig. 5). Similarly, in the Medievalis branch, the five isolates from Kurdistan were much more closely related to each other than to the Turkish strain. Most importantly, within the Orientalis branch, the majority of the subbranches typified the geographic origin of the strains: all Namibian isolates were in cluster d1, all but one (IP532) strain from Southeast Asia were in cluster d2 (with a subdivision of this cluster into two subbranches corresponding to the Vietnamese and Burmese isolates), all Brazilian isolates were in cluster d3, all strains from West Africa were in cluster d4 (with the two strains from Morocco and the two from Senegal being into distinct subbranches), and all strains from the United States were in cluster d5 (Fig. 5). Furthermore, cluster d6 contained the two strains from Surat (India), and cluster d7 contained the three strains from Germany. Single Orientalis strains from a given country (Turkey, Belgian Congo, Algeria, and South Africa) were scattered in the dendrogram. Madagascar was the only focus for which no marked strain clustering was observed. The three most recent isolates from this country were grouped in cluster d8, while the four others were in different subbranches (Fig. 5).
FIG. 5.
3IS-RFLP dendrogram. This dendrogram was generated after combination of the three individual RFLP patterns (IS100, IS285, and IS1541). Dotted rectangles indicate clusters of strains of interest. A, Antiqua branch; M, Medievalis branch; O, Orientalis branch.
DISCUSSION
IS-RFLP and molecular typing.
Two extremes may be found among the molecular typing methods used to analyze Y. pestis isolates. One corresponds to ribotyping, which provides a global view of the circulating strains but has a low discriminatory power, especially within the Orientalis group. The other extreme is PFGE which, because of the high number of restriction fragments generated and the complexity of the patterns, has a high discriminatory power but is hardly applicable to Y. pestis strains of worldwide origin (unpublished results). IS-RFLP may represent an intermediate between these two techniques because the number of fragments to be analyzed is higher than with ribotyping, but the bands are better resolved than with PFGE, allowing a more reliable strain-to-strain comparison. Of the three IS-RFLP techniques used in this study, we found that IS100-RFLP had the highest discriminatory power and IS285-RFLP had the lowest discriminatory power. Similar results were obtained when Y. pestis isolates from the United States were analyzed by IS100- and IS285-RFLP (15). This may not be entirely attributable to the difference in IS copy numbers on the Y. pestis chromosome because a size window was defined for each IS pattern, and the number of bands analyzed within these windows was comparable for the three IS. This suggests that variations in the position of the IS on the chromosome are more frequent for IS100 and that IS285 is the most stable. Combination of two IS-RFLPs increased the discriminatory power of the technique. The highest discrimination index (D = 0.998) was obtained with the IS100/ IS1541-RFLP combination, which allowed the delineation of 58 IS types among the 61 strains analyzed. Combination of the three IS-RFLP patterns further increased the discriminatory power of the technique (D = 0.999). Although accurate comparison of the discriminatory power of different techniques would need them to be applied to the same set of strains, it appears from this study that 3S-RFLP can discriminate Y. pestis strains of worldwide origin at least as efficiently as two other highly sensitive typing methods such as variable-number tandem repeats and PFGE. From a practical viewpoint, since the gain in sensitivity was relatively limited when 3IS-RFLP was used instead of IS100/IS1541-RFLP (59 IS types instead of 58), it may be simpler and faster to simply use IS100/IS1541-RFLP for the purpose of discriminating global isolates of Y. pestis. Remarkably, IS100/IS1541-RFLP could differentiate most strains having the same biovar and country of origin and sometimes even having been isolated the same year. This relatively easy to perform technique may thus be a valuable tool for molecular typing of global Y. pestis strains.
IS-RFLP and Y. pestis microevolution.
The Antiqua and Medievalis biovars have a specific geographic distribution. The Medievalis plague focus is situated in Central Asia. Two main Antiqua plague foci exist, one in Central Asia and the other one in Central Africa. It has been recently shown that these two Antiqua foci belong to two distinct evolutionary branches termed 1.ANT and 2.ANT, respectively (1). Only the African branch (1.ANT) was studied here because almost all of the Antiqua isolates in our strain collection belong to this branch. We found that, as previously observed (2), IS100-RFLP grouped Y. pestis strains based on their biovar. This study further shows that IS285-RFLP allowed the clustering of the three classical Y. pestis biovars in independent parts of the dendrogram. In contrast, this grouping was less effective with IS1541-RFLP. The biovar-based clustering became even more pronounced when the IS100/IS1541-RFLP or 3IS-RFLP combinations were used. Therefore, IS-RFLP techniques may be useful for the analysis of Y. pestis microevolution.
IS-RFLP and Y. pestis tracing.
The primary and major aim of this study was to determine whether various IS-RFLP techniques, used individually or in combination, could efficiently group Y. pestis strains according to their geographical focus. For the Antiqua and Medievalis strains, since they have a specific and precise geographical distribution, the simple determination of their biovar may be sufficient to identify their main focus of origin. This is clearly different for biovar Orientalis strains which are now the most commonly isolated Y. pestis strains worldwide because of their global spread during the third pandemic. The major challenge was thus to identify a technique which could cluster strains within the Orientalis group according to their geographical foci. When the three IS-RFLP techniques were used individually, some degree of geographical clustering was observed with each technique, but none gave reliable and satisfactory results. IS100-RFLP displayed nonetheless a slightly better clustering capacity than the other two IS-RFLPs. A two-by-two combination of the IS-RFLP techniques significantly improved the geographical clustering of the Orientalis strains. The least effective combination was IS1541/IS285-RFLP, while the two others were comparable, with a slight advantage for IS100/IS1541-RFLP. With a combination of the three IS-RFLP methods, a clear clustering of most of the Orientalis strains according to their geographic origin was obtained. Indeed, strains from the Namibian, Southeast Asian (Burma and Vietnam), U.S., Brazilian, and North East African (Morocco and Senegal) foci each formed distinct clusters. One exception was the strains from Madagascar, which were found in different clusters. However, it is noteworthy that the three strains that were grouped in a single cluster (d8) were recent isolates (1989 to 1992) from the same region (Ambositra). The other Malagasy strains were more ancient (1946 to 1969) and were isolated from different regions of the island. This indicates that the clustering potential of 3IS-RFLP may not be limited to distantly related foci but may also be applicable at a regional or local scale. One possible reason for the greatest IS-RFLP diversity of the Malagasy isolates, compared to the other isolates worldwide, may be that Madagascar is one of the most active plague foci in the world, and therefore, the higher rate of bacterial generations has allowed a faster accumulation of genetic diversity in the strains from this country.
The 3IS-RFLP dendrogram displayed a clustering of the strains by country or geographical focus but not by continent. For instance, the Namibian and Southeast Asian clusters were closer to each other than to the other clusters from Africa or Asia. This can be explained by the fact that a single Y. pestis clone (biovar Orientalis and ribotype B) spread over the world in a short period of time during the third pandemic. Most of the current Orientalis plague foci worldwide result from the establishment and local spread of the original strain imported by steamships at the end of the 19th century (23). However, no large extension of these foci over a continent has been reported. Each focus evolved independently from the others regardless of the continent where they became established.
Tracing of Y. pestis may be important to implement appropriate public health measures when a plague outbreak occurs in a country after several decades of silence. Indeed, if the epidemic clone has been imported, a better control at the frontiers may be necessary. In contrast, if the outbreak results from the resurgence of a local and transiently dormant focus, the national surveillance and control system needs to be reactivated. For instance, when plague reemerged in India in 1994 after almost 30 years of silence, it was not clear whether the causative agent was of local or foreign origin. The ribotype (S) of the strains isolated during the 1994 pneumonic outbreak in Surat was different from the classical ribotype B found in an older strain (Bombay 195) isolated in India in 1908 (25). The recent and older Indian strains also had different pulsotypes (unpublished data). The results of the 3IS-RFLP analysis performed here demonstrate that the two strains recently isolated in Surat have identical 3IS types, confirming that they both derive from the epidemic clone which caused the pneumonic outbreak of 1994. These results also confirm that the Bombay and Surat strains are different clones. However, the fact that the three Indian strains are found in the same higher-order branch (which includes clusters d5 and d6 and additional strains) may suggest that, despite the 88 years which separated their isolation, the strains from Bombay and Surat are related.
Although the plague arrived with steamships in numerous harbors over the world during the third pandemic, it did not systematically form permanent foci. In the seaport of Hamburg, for instance, the disease was imported during the 20th century but did not become established. The 3IS-RFLP analysis of the isolates from Hamburg grouped them into the same cluster, supporting the hypothesis that all three originated from the same focus. Furthermore, the fact that these strains clustered with a strain from Argentina suggests either that this country was the source of the imported strains in Germany or that a same clone of unknown origin was imported simultaneously in both countries.
The application of IS-RFLP techniques to the analysis of global isolates of Y. pestis has shown that this tool has a high discriminatory power and can thus be used for molecular typing of this species. For this purpose, IS100/IS1541-RFLP and 3IS-RFLP are the most appropriate techniques. This study also indicates that the IS100 and IS285 fingerprints reflect the divergence of the species into the three classical biovars and, therefore, that not only IS100 but also IS285 could be useful for the study of the microevolution of Y. pestis. But the major goal of this work was to determine whether IS-RFLP, individually or in combination, would be applicable to the identification of the origin of an unknown isolate. Our results indicate that 3IS-RFLP may indeed be a powerful tool to trace Y. pestis strains, even within the Orientalis group, which had only a short period of time (one century) to diverge from the original clone. We now dispose of a database of global Y. pestis isolates in which any new strain may be incorporated and compared with those of known geographic origin. In the future, this database will need to be enlarged and enriched with isolates as varied as possible (including pestoides/microtus strains [branch 0] and Antiqua strains from Asia [branch 2.ANT]) to reflect the most accurately the global diversity of the species. Altogether, the results of this study show that 3IS-RFLP may be of great utility when plague reemerges in an ancient focus to help determine whether the disease has been imported or results from the reactivation of the ancient local focus. It may also be valuable when plague cases appear in a previously plague-free region, to identify the source of importation. Finally, this technique may also prove useful for the tracing of a Y. pestis isolate used as a biological terrorism threat.
Supplementary Material
Acknowledgments
This work was supported in part by grants 97/25200/DCE/CEB (Centre d'Etude du Bouchet) and 93811-77/A000/DRET/DS (Direction de la Recherche et de la Technologie, Ministère de la Défense, France).
We thank Mark Achtman for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., K. Zurth, C. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreto, A., M. Aragon, and P. R. Epstein. 1995. Bubonic plague outbreak in Mozambique, 1994. Lancet 345:983-984. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobrov, A. G., and A. A. Filippov. 1997. Prevalence of IS285 and IS100 in Yersinia pestis and Yersinia pseudotuberculosis genomes. Mol. Genet. Mikrobiol. Virusol. 2:36-40. [PubMed] [Google Scholar]
- 7.Carniel, E., O. Mercereau-Puijalon, and S. Bonnefoy. 1989. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect. Immun. 57:1211-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Almeida, A. M. P., A. Guiyoule, I. Guilvout, I. Iteman, G. Baranton, and E. Carniel. 1993. Chromosomal irp2 gene in Yersinia: distribution, expression, deletion and impact on virulence. Microb. Pathog. 14:9-21. [DOI] [PubMed] [Google Scholar]
- 9.Deng, W., V. Burland, G. Plunkett, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. G. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devignat, R. 1951. Variétés de l'espèce Pasteurella pestis. Nouvelle hypothèse. Bull. W. H. O. 4:247-263. [PMC free article] [PubMed] [Google Scholar]
- 11.Drancourt, M., W. Roux, L. V. Dang, L. Tran-Hung, D. Castex, V. Chenal-Francisque, H. Ogata, P. E. Fournier, E. Crubezy, and D. Raoult. 2004. Genotyping, orientalis-like Yersinia pestis, and plague pandemics. Emerg. Infect. Dis. 10:1585-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fetherston, J. D., P. Schuetze, and R. D. Perry. 1992. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol. Microbiol. 6:2693-2704. [DOI] [PubMed] [Google Scholar]
- 13.Guiyoule, A., F. Grimont, I. Iteman, P. A. D. Grimont, M. Lefevre, and E. Carniel. 1994. Plague pandemics investigated by ribotyping of Yersinia pestis strains. J. Clin. Microbiol. 32:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, P., J. Elliott, P. McCready, E. Skowronski, J. Garnes, A. Kobayashi, R. R. Brubaker, and E. Garcia. 1998. Structural organization of virulence-associated plasmids of Yersinia pestis. J. Bacteriol. 180:5192-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, X. Z., M. C. Chu, D. M. Engelthaler, and L. E. Lindler. 2002. Genotyping of a homogeneous group of Yersinia pestis strains isolated in the United States. J. Clin. Microbiol. 40:1164-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortepeter, M. G., and G. W. Parker. 1999. Potential biological weapons threats. Emerg. Infect. Dis. 5:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindler, L. E., G. V. Plano, V. Burland, G. F. Mayhew, and F. R. Blattner. 1998. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect. Immun. 66:5731-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucier, T. S., and R. R. Brubaker. 1992. Determination of genome size, macrorestriction pattern polymorphism, and nonpigmentation-specific deletion in Yersinia pestis by pulsed-field gel electrophoresis. J. Bacteriol. 174:2078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahillon, J., C. Leonard, and M. Chandler. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675-687. [DOI] [PubMed] [Google Scholar]
- 21.Motin, V. L., A. M. Georgescu, J. M. Elliott, P. Hu, P. L. Worsham, L. L. Ott, T. R. Slezak, B. A. Sokhansanj, W. M. Regala, R. R. Brubaker, and E. Garcia. 2002. Genetic variability of Yersinia pestis isolates as predicted by PCR-based IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J. Bacteriol. 184:1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeño-Tárraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 23.Pollitzer, R. 1954. Plague. W.H.O. Monograph Series 22. World Health Organization, Geneva, Switzerland.
- 24.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653-663. [DOI] [PubMed] [Google Scholar]
- 25.Ramalingaswami, V. 1995. Plague in India. Nat. Med. 1:1237-1239. [DOI] [PubMed] [Google Scholar]
- 26.Rasolomaharo, M., B. Rasoamanana, Z. Andrianirina, P. Buchy, N. Rakotoarimanana, and S. Chanteau. 1995. Plague in Majunga, Madagascar. Lancet 346:1234. [DOI] [PubMed] [Google Scholar]
- 27.Schrag, S. J., and P. Wiener. 1995. Emerging infectious diseases: what are the relative roles of ecology and evolution? Trends Evol. Ecol. 10:319-324. [DOI] [PubMed] [Google Scholar]
- 28.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Yu, H. Yang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization. 2003. Human plague in 2000 and 2001. Wkly. Epidemiol. Rec. 78:130-135. [PubMed] [Google Scholar]
- 31.World Health Organization. 2004. Human plague in 2002 and 2003. Wkly. Epidemiol. Rec. 79:301-306. [PubMed] [Google Scholar]
- 32.World Health Organization. 2003. Plague, Algeria. Wkly. Epidemiol. Rec. 78:253. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.