Abstract
Sources of Mycobacterium bovis contamination remain unclear for many cases of animal and human disease. A major limitation is the lack of sufficiently informative or epidemiologically well evaluated molecular methods for typing. Here, we report an evaluation of a high-throughput method based on 29 mycobacterial interspersed repetitive unit-variable-number tandem-repeat (MIRU-VNTR) loci to genotype 127 M. bovis isolates from cattle from 77 different Belgian farms, representative of a nationwide collection obtained from 1995 to 2003. MIRU-VNTR stability was demonstrated by analyzing a series of 74 isolates in total, obtained from different animals from a single farm or from different farms with an identified epidemiological link. The genotyping results and the genotypic diversity (h) were compared with those obtained by IS6110 restriction fragment length polymorphism (RFLP) analysis and spoligotyping. Among 68 isolates with no known epidemiological link, MIRU-VNTR typing discriminated better than either RFLP analysis or spoligotyping, with isolates taken individually (32 versus 16 and 17 genotypes; h = 0.91 versus 0.73 and 0.85, respectively) or in combination (32 versus 28 genotypes; h = 0.91 versus 0.92). Maximal resolution was already achieved with a subset of 9 loci. The observed congruence of the genetic relationships based on IS6110 RFLP analysis, spoligotyping, and MIRU-VNTR markers is consistent with a clonal population structure of M. bovis. These results support MIRU-VNTR typing as a convenient and discriminatory technique for analysis of the population structure of M. bovis in much greater detail and for addressing some still unresolved issues in the epidemiology of the pathogen.
Mycobacterium bovis is the causative agent of tuberculosis in bovines. In addition to cattle, this pathogen has an exceptionally wide host range, extending to goats, cats, dogs, pigs, lions, deer, nonhuman primates, and humans. Many susceptible species, including humans, are putative spillover hosts, in which infection is not confined. Some countries, such as Great Britain and Ireland, are currently experiencing an exponential increase in the incidence of bovine tuberculosis. Moreover, M. bovis infection is highly prevalent in farmed animals in developing countries (5). Many of these countries have endemically infected wildlife, which is suspected of providing a reservoir for farmed animals. Under such conditions, eradication is not feasible, and control measures must be applied indefinitely (20). Therefore, M. bovis is a source of important economic damage and a potential threat to human health in many countries.
A few studies have estimated the proportion of M. bovis infection in human tuberculosis cases to be in the range of 0.3 to 1.5% in developed countries (5-7, 10, 24). This proportion may be higher in some developing countries, although little information is available (5). Cases of human tuberculosis with M. bovis are mainly associated with the consumption of unpasteurized dairy products or close contact with infected animals (3, 10). Therefore, these are most often suspected of being of zoonotic origin. Cases of interhuman transmission have only rarely been documented (10, 12, 19). However, the exact source of infection remains undetermined in most cases. Likewise, direct transmission between wildlife and farmed animals has not been demonstrated yet under natural conditions. Nevertheless, it has been demonstrated that experimentally infected deer can transmit M. bovis to cattle through the sharing of feed (22).
One main limitation results from the lack of sufficiently informative molecular methods for typing M. bovis. In contrast to Mycobacterium tuberculosis, restriction fragment length polymorphism typing using IS6110 (IS6110 RFLP) provides only limited discrimination among M. bovis isolates, since M. bovis strains usually only have a single or a few IS6110 copies. PCR-based spoligotyping, targeting the direct repeat region, also suffers from the same limitation. Analysis of multiple genomic regions that contain variable-number tandem repeats (VNTRs) of different families of genetic elements called mycobacterial interspersed repetitive units (MIRUs) (9, 18, 25, 27, 29, 31) has been recently proposed as an alternative tool for molecular epidemiological studies of M. bovis. In a sample of 47 isolates from Northern Ireland, these markers were shown to be more discriminatory than spoligotyping (26). However, the epidemiological interpretation of MIRU-VNTR genotyping data is difficult to assess accurately from studies in such a setting. Because of its high incidence, cattle movements, and the existence of potential environmental reservoirs (i.e., badgers), independent sources of infection may occur within given herds, and unsuspected transmission chains might often exist between farms. Therefore, it is difficult to evaluate the actual capacity to distinguish epidemiologically unrelated isolates and the stability of the markers among epidemiologically related isolates. Moreover, the performances of MIRU-VNTR typing data were not compared with those of IS6110 RFLP fingerprinting in that study.
Here, we have evaluated the clonal stability, epidemiological significance, and discrimination of 29 MIRU-VNTR loci (9, 18, 25, 27, 29, 31) using a sample of 127 M. bovis isolates from Belgium. This country has an extremely low incidence of bovine tuberculosis (0.02% of infected herds in 2003 and 2004) and no known environmental reservoir of M. bovis. The stability of the markers necessary to trace transmission chains over time was tested using several sets of isolates with identical spoligotypes and IS6110 fingerprints from a single farm and by investigating available cattle tracing data for clustered isolates obtained from different farms. The discriminatory power was evaluated using isolates representative of all of the different spoligotypes, IS6110 fingerprints available from a nationwide collection, and isolates with identical spoligotypes and IS6110 fingerprints from different farms with no obvious epidemiological link. Finally, the congruence of the genetic relationships as defined by IS6110 RFLP, spoligotyping, and MIRU-VNTR typing was analyzed.
MATERIALS AND METHODS
Control of bovine tuberculosis in Belgium and epidemiological investigation.
The control of bovine tuberculosis is currently based on skin testing in the context of animal contact tracing and trading. In addition, systematic postmortem examinations at slaughterhouses and individual postmortem examinations at the regional veterinary laboratories are performed. When a suspected lesion is identified, a sample is sent to the reference laboratory for analysis. Subsequently, all animals from the herd of origin are skin tested, and a complete epidemiological investigation is carried out, following standardized epidemiological procedures. The aim of this investigation is to determine the origin of the outbreak and to identify all farms in contact with infected animals or materials. This identification is possible by consulting SANITEL (Belgian databank for the registration and movement of cattle).
Study collection.
One hundred twenty-seven M. bovis isolates originating from cases of bovine tuberculosis diagnosed in 77 herds in Belgium during the period 1995 to 2003 were included in this study. The isolates were chosen among 451 samples from 204 herds characterized by IS6110 RFLP typing and spoligotyping methods from a total of 503 animal tuberculosis cases with positive cultures identified on 220 Belgian farms over this 9-year period. This collection is maintained at the Belgian National Reference Laboratory (CERVA-CODA, Brussels, Belgium). The isolates typed by the MIRU-VNTR method were chosen and grouped into three panels, named panels 1, 2, and 3, according to three analytical criteria (see Results and Table S in the supplemental material). Among these 127 samples, 26 were the result of pooled tissue samples from different animals from a single herd. In these cases, biopsy specimens of 2 to 33 different animals (usually from 2 to 5) from the same farm collected at the same time were pooled.
Bacteriology.
Samples from animals suspected of suffering from tuberculosis were pooled, homogenized with sterile saline solution, and decontaminated with 5% oxalic acid for 20 to 30 min at 37°C (21). After centrifugation at 1,000 × g for 20 min, isolates were cultured on Coletsos solid medium (Bio-Rad, Belgium) at 37°C. Bacteriological identification of M. bovis was based on acid-fast staining, preferential growth at 37°C and 42°C, the nitrate reductase test (Becton Dickinson, Belgium), and the tiophene-2-carboxilic acid hydrazide assay (Biomérieux, Belgium).
DNA extraction.
Mycobacterial strains were grown in Middlebrook 7H9 liquid medium supplemented with oleic acid-albumin-dextrose-catalase (Becton Dickinson, Belgium), Tween 80, penicillin, and Fungizone for 3 weeks. Chromosomal DNA was isolated as described by van Embden et al. (34).
Genotyping. (i) IS6110 RFLP analysis.
DNA fingerprinting was performed as described by van Embden et al. (34). Briefly, purified mycobacterial DNA was digested with PvuII, separated on an agarose gel, transferred by Southern blotting to a nylon membrane, and hybridized with a peroxidase-labeled 245-bp IS6110 probe. Hybridizing restriction fragments were visualized using the ECL detection system (Amersham Biosciences, Belgium).
(ii) Spoligotyping.
The spoligotyping method was used as described by Kamerbeek et al. (16). PCR amplification of the direct repeat locus was performed on heat-treated cell suspensions. Biotin-labeled PCR products were detected by hybridization onto a spoligotyping membrane (Isogen Bioscience BV, Maarssen, The Netherlands). Purified sterile water and clinical isolates of M. tuberculosis and M. bovis were included as controls in every batch of tests. The results were recorded in SB (spoligotype bovis) code, followed by a field of 4 digits as defined on the Mycobacterium bovis Spoligotype Database website (www.Mbovis.org).
(iii) MIRU-VNTR.
MIRU-VNTR analysis was done by amplification of 29 genomic loci in 9 different multiplex PCRs and 2 different simplex PCRs with the previously described primers (9, 18, 25, 27, 29, 30) (Table 1). Multiplex PCR mixtures were prepared as follows, using 96-well plates and the Hotstart Taq DNA polymerase kit (QIAGEN, Benelux B.V.). Three nanograms of DNA was added to a final volume of 30 μl containing 0.1 μl of DNA polymerase (1 U); 6 μl of Q solution; 0.2 mM (each) dATP, dCTP, dGTP, dTTP (Fermentas GmbH, Germany); 3 μl of PCR buffer; 0.4 μM concentrations of each primer (Applied Biosystems, Belgium); and different concentrations of MgCl2, as shown in Table 1. For each multiplex mixture, one primer from each oligonucleotide pair was tagged with a different fluorescent dye. The thermocycler programs were identical for all of the reactions. The PCRs were carried out using a GeneAmp PCR system 2700 (Applied Biosystems, Belgium) starting with a denaturing step of 15 min at 95°C, followed by 40 (VNTR 3232 and 3336) or 30 (all other loci) cycles of 1 min at 94°C, 1 min at 59°C, and 1 min, 30 s at 72°C. Two microliters of PCR products was mixed with formamide and a MapMarker1000 ladder (23 fragments ranging from 50 to 1,000 bp; Eurogentec, Belgium). DNA fragments were separated by capillary electrophoresis using an ABI Prism 3100-Avant genetic analyzer. VNTR numbers were scored based on amplicon sizes using Genescan and customized Genotyper software packages (Applied Biosystems, Belgium).
TABLE 1.
Primer sequences and MgCl2 concentrations of the different PCR mixes
| Multiplex | Locus | Alias | MIRU-VNTR length (bp) | MgCl2 concn (mM)b | PCR primer pair (5′-3′, with labeling indicated)a |
|---|---|---|---|---|---|
| Mix A | MIRU 4 | VNTR 580 | 77 | GCGCGAGAGCCCGAACTGC (FAM) | |
| ETR-D | GCGCAGCAGAAACGTCAGC | ||||
| MIRU 26 | VNTR 2996 | 51 | 3 | TAGGTCTACCGTCGAAATCTGTGAC | |
| CATAGGCGACCAGGCGAATAG (VIC) | |||||
| MIRU 40 | VNTR 802 | 54 | GGGTTGCTGGATGACAACGTGT (NED) | ||
| GGGTGATCTCGGCGAAATCAGATA | |||||
| Mix B | MIRU 10 | VNTR 960 | 53 | GTTCTTGACCAACTGCAGTCGTCC | |
| GCCACCTTGGTGATCAGCTACCT (FAM) | |||||
| MIRU 16 | VNTR 1644 | 53 | 2 | TCGGTGATCGGGTCCAGTCCAAGTA | |
| CCCGTCGTGCAGCCCTGGTAC (VIC) | |||||
| MIRU 31 | VNTR 3192 | 53 | ACTGATTGGCTTCATACGGCTTTA | ||
| ETR-E | GTGCCGACGTGGTCTTGAT (NED) | ||||
| Mix C | MIRU 2 | VNTR 154 | 53 | TGGACTTGCAGCAATGGACCAACT | |
| TACTCGGACGCCGGCTCAAAAT (FAM) | |||||
| MIRU 23 | VNTR 2531 | 53 | 2.5 | CTGTCGATGGCCGCAACAAAACG (VIC) | |
| AGCTCAACGGGTTCGCCCTTTTGTC | |||||
| MIRU 39 | VNTR 4348 | 53 | CGCATCGACAAACTGGAGCCAAAC | ||
| CGGAAACGTCTACGCCCCACACAT (NED) | |||||
| Mix D | MIRU 20 | VNTR 2059 | 77 | TCGGAGAGATGCCCTTCGAGTTAG (FAM) | |
| GGAGACCGCGACCAGGTACTTGTA | |||||
| MIRU 24 | VNTR 2687 | 54 | 1.5 | CGACCAAGATGTGCAGGAATACAT | |
| GGGCGAGTTGAGCTCACAGAA (VIC) | |||||
| MIRU 27 | VNTR 3007 | 53 | TCGAAAGCCTCTGCGTGCCAGTAA | ||
| QUB5 | GCGATGTGAGCGTGCCACTCAA (NED) | ||||
| Mix E | VNTR 2347 | 57 | GCCAGCCGCCGTGCATAAACCT (FAM) | ||
| AGCCACCCGGTGTGCCTTGTATGAC | |||||
| ETR-B | VNTR 2461 | 57 | 1.5 | ATGGCCACCCGATACCGCTTCAGT (VIC) | |
| CGACGGGCCATCTTGGATCAGCTAC | |||||
| VNTR 3171 | 54 | GGTGCGCACCTGCTCCAGATAA (NED) | |||
| GGCTCTCATTGCTGGAGGGTTGTAC | |||||
| Mix F | VNTR 0424 | 51 | CTTGGCCGGCATCAAGCGCATTATT | ||
| GGCAGCAGAGCCCGGGATTCTTC (FAM) | |||||
| ETR-C | VNTR 0577 | 58 | 1.5 | CGAGAGTGGCAGTGGCGGTTATCT (VIC) | |
| AATGACTTGAACGCGCAAATTGTGA | |||||
| VNTR 1895 | QUB 1895 | 57 | GTGAGCAGGCCCAGCAGACT (NED) | ||
| CCACGAAATGTTCAAACACCTCAAT | |||||
| Mix G | VNTR 2401 | 58 | CTTGAAGCCCCGGTCTCATCTGT (FAM) | ||
| ACTTGAACCCCCACGCCCATTAGTA | |||||
| VNTR 3690 | 58 | 3.0 | CGGTGGAGGCGATGAACGTCTTC (VIC) | ||
| TAGAGCGGCACGGGGGAAAGCTTAG | |||||
| VNTR 4156 | QUB 4156 | 59 | TGACCACGGATTGCTCTAGT | ||
| GCCGGCGTCCATGTT (NED) | |||||
| Mix I | QUB 11a | VNTR 2163a | 69 | CCCATCCCGCTTAGCACATTCGTA | |
| TTCAGGGGGGATCCGGGA (FAM) | |||||
| QUB 26 | VNTR 4052 | 111 | 1.5 | AACGCTCAGCTGTCGGAT (VIC) | |
| GGCCAGGTCCTTCCCGAT | |||||
| ETR-A | VNTR 2165 | 75 | AAATCGGTCCCATCACCTTCTTAT (NED) | ||
| CGAAGCCTGGGGTGCCCGCGATTT | |||||
| Mix J | QUB 11b | VNTR 2163b | 69 | CGTAAGGGGGATGCGGGAAATAGG | |
| CGAAGTGAATGGTGGCAT (FAM) | |||||
| VNTR 1955 | 57 | 1.5 | AGATCCCAGTTGTCGTCGTC (VIC) | ||
| CAACATCGCCTGGTTCTGTA | |||||
| VNTR 1982 | QUB 18 | 78 | CCGGAATCTGCAATGGCGGCAAATTAAAAG | ||
| TGATCTGACTCTGCCGCCGCTGCAAATA (NED) | |||||
| Individual | VNTR 3232 | QUB 3232 | 56/57 | 1.5 | TGCCGCCATGTTTCATCAGGATTAA |
| GCAGACGTCGTGCTCATCGATACA (FAM) | |||||
| Individual | VNTR 3336 | QUB 3336 | 59 | 1.5 | ATCCCCGCGGTACCCATC (VIC) |
| TTCTACGACTTCGCAACCAAGTATC |
Forward and reverse primers, respectively. FAM, blue dye label (6-carboxy-fluorescein); NED, yellow dye label; VIC, green dye label.
Final concentration (Hotstart buffer already contains 1.5 mM MgCl2).
(iv) RD4 analysis.
PCR analysis of the RD4 deletion region, defining classical M. bovis strains, was performed using primers and conditions, as described by Brosch et al. (4).
Allelic and genotypic diversity.
The allelic diversity of each VNTR locus was calculated using the following equation: h = 1 − Σxi2 [(n/n − 1)], where n is the number of tested isolates and xi is the frequency of the ith allele. Genotypic diversity was used as a measure of the discriminatory power of the three genotyping methods. It was defined as 1 − Σxi2 [(n/n − 1)], where n is the total number of genotypes obtained and xi is the frequency of the ith genotype.
Genetic relationship analysis.
IS6110 RFLP patterns were recorded and analyzed using the Bionumerics package (Applied Maths, St-Martin-Latem, Belgium) as fingerprint data. The Dice coefficient was used to plot dendrograms using the unweighted pair group method with arithmetic averages (UPGMA). Spoligotyping and MIRU-VNTR profiles were recorded as character data and were analyzed using the categorical character option and UPGMA to align branch tips for better visualization in Fig. 1 and 2 or the neighbor-joining algorithm for congruence analysis in Fig. 4. Composite data sets were created to analyze clustering resulting from a combination of the different genotyping methods.
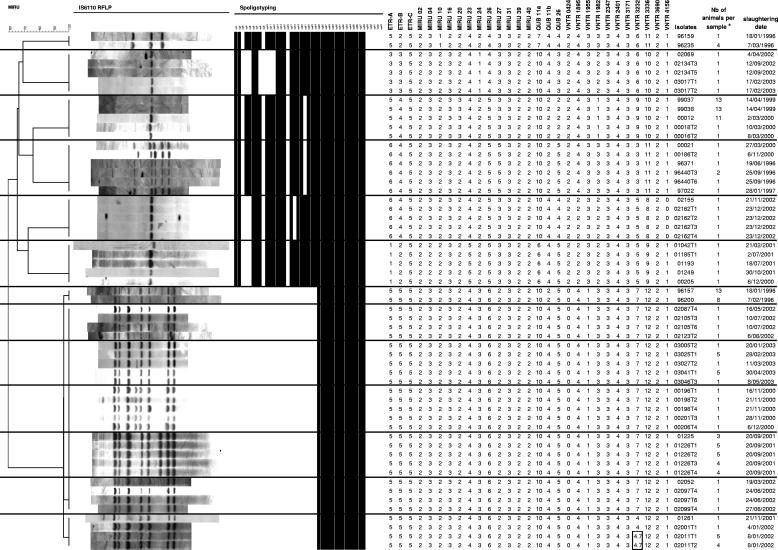
FIG. 1.
IS6110 RFLP, spoligotyping, and MIRU-VNTR patterns of isolates obtained from different animals from a single farm. The dendrogram at the left is based on MIRU-VNTR profiles and was built by using the UPGMA algorithm, as described in Materials and Methods. Double alleles (4 and 7) detected in locus 3232 of two pooled samples (02011T1 and 02011T2) are boxed. a, a number (Nb) of >1 indicates a sample obtained by pooling tissues of n different animals from the same herd before mycobacteriological culture.
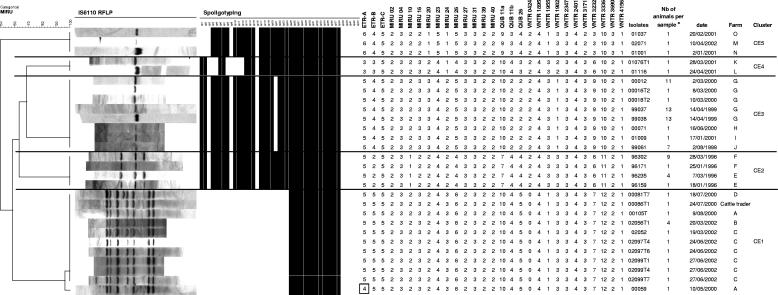
FIG. 2.
IS6110 RFLP, spoligotyping, and MIRU-VNTR patterns of isolates obtained from animals from different farms included in five distinct clusters with defined epidemiological links. The dendrogram at the left is based on MIRU-VNTR profiles and was built by using the UPGMA algorithm, as described in Materials and Methods. The single repeat change in locus ETR-A of sample 00059 compared to the same locus of the other isolates of cluster CE1 is boxed. a, a number (Nb) of >1 indicates a sample obtained by pooling tissues of n different animals from the same herd before mycobacteriological culture. See text and Fig. 3 for descriptions of farms and clusters.
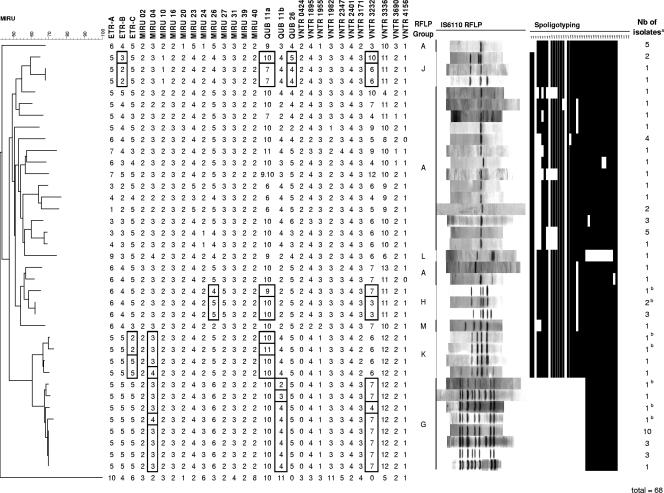
FIG. 4.
Congruence of genetic relationships between MIRU-VNTR typing, IS6110 RFLP, and spoligotyping. The dendrogram was constructed based on MIRU-VNTR genotypes using the neighbor-joining method, as described in Materials and Methods. The MIRU-VNTR tree was rooted using the Mycobacterium canetti clonal group C MIRU-VNTR genotype (11). The figure displays 37 different patterns among the isolates in panel 3, identified by the three genotyping methods. Boxed alleles correspond to MIRU-VNTR differences observed among samples within the G, H, J, and K lineages (see text). a, the number (Nb) of isolates with an identical genotype using the three genotyping methods. b, isolates with an identical IS6110 fingerprint within RFLP groups which were distinguished by MIRU-VNTR typing.
Congruence between the genetic distances based on MIRU-VNTR types and IS6110 fingerprints and spoligotypes was evaluated with Bionumerics, using the Pearson correlation. The g test (32) was used to evaluate the statistical significance of this congruence. This test takes random association as a null hypothesis and was done by evaluating the significance of the correlation between the genetic distances inferred from the above methods for any possible pair of isolates, with a nonparametric Mantel test based on a Monte Carlo simulation with 104 iterations.
RESULTS
Selection of isolates.
One hundred twenty-seven M. bovis isolates originating from cases of bovine tuberculosis diagnosed on 77 Belgian farms between 1995 and 2003 were included in this study. The isolates were selected from a nationwide collection of 451 isolates collected between 1995 and 2003 at the Belgian National Reference Laboratory, representing 89.6% of all animal tuberculosis cases with a positive culture identified in Belgium over this 9-year period. The isolates were chosen according to three criteria: panel 1 included 13 series of 2 to 6 isolates collected from different animals coming from a single farm, panel 2 included 5 series of 2 to 11 isolates obtained from different farms but with a defined epidemiological link, while panel 3 included isolates representative of all the different spoligotypes and/or IS6110 fingerprints available in the nationwide collection as well as isolates with identical spoligotypes and IS6110 fingerprints from different farms without obvious epidemiological links. With this system, some isolates were represented in more than one panel, as for instance, a sample representative of a defined spoligotype (panel 3) could be found with other isolates from the same farm (panel 1) and also with isolates from another epidemiologically linked farm (panel 2) (see Table S in the supplemental material).
Stability of MIRU-VNTR loci. (i) Multiple isolates from individual farms (panel 1).
The stability of the 29 MIRU-VNTR loci was assessed by analyzing 13 groups including 57 M. bovis isolates in total (panel 1) (Fig. 1). Each group included between 2 and 6 isolates with identical IS6110 RFLP patterns and spoligotypes, obtained from different animals from a single farm over periods of up to 4 years. We postulated that the cows within these groups were probably infected from a single source and thus involved a single transmission chain. Consistent with this hypothesis, the 29 loci remained unchanged within all groups except one. In this group, the simultaneous occurrence of two different alleles (4 and 7) was clearly detected in locus 3232 of two samples (02011T1 and 02011T2), while the two other isolates of the group displayed a single allele 4 in this locus. Samples 02011T1 and 02011T2 were each the result of a pool of tissue samples from 5 and 4 different animals, respectively. Therefore, this observation may indicate either the presence of a second strain or, alternatively, of a clonal variant in a limited number of animals in this herd. There was no such evidence of genotypic heterogeneity in any of the other pooled samples in this panel or in the other 2 panels (see below). This was consistent with complete genotypic homogeneity among all the isolates obtained from several different animals from individual farms, suggesting that such isolates share a common clonal origin in most cases.
Therefore, in accordance with our earlier hypothesis, these results indicate that the 29 loci were stable in transmission chains spanning periods of up to 4 years on at least 12 farms of the 13 examined.
(ii) Epidemiologically linked isolates from different farms (panel 2).
Panel 2 included samples obtained from different farms among which epidemiological links could be clearly established by analysis of complete cattle tracing data. This panel comprised five clusters (named CE1 to CE5), within which IS6110 RFLP and spoligotyping patterns were identical or almost identical (Fig. 2).
In cluster CE1, a link could be found between four farms and a cattle trader (Fig. 3). Farm A had a tuberculosis outbreak identified in 2000 but was suspected of having hosted diseased animals since at least 1996. The cattle trader bought animals from farmer A and sold them to farmer B. In turn, animals from farm B were transferred to farm C. One isolate (00105T) from farm A displayed a fully identical genotype by the three genotyping methods when compared to all the isolates obtained from the trader and from farms B and C. Interestingly, it was found that the cattle trader also sold animals to farm D. There was a difference in the RFLP pattern in the isolate from this farm due to an additional IS6110 band, but the MIRU-VNTR code did not differ from the above samples (Fig. 2). Conversely, one isolate (00059) from farm A displayed a single repeat change in one MIRU-VNTR locus, together with a single band change in its IS6110 fingerprint, compared to the other members of the cluster.
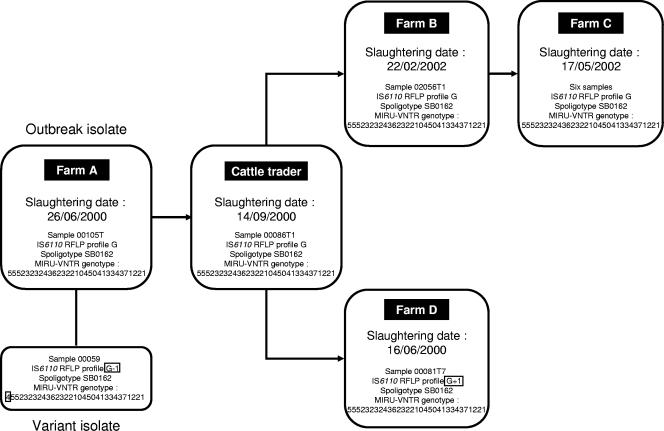
FIG. 3.
Bovine tuberculosis transmission chain between different farms involving animals sold by one cattle trader. The cattle trader bought animals from farmer A and sold them to farmer B. In turn, animals from farm B were transferred to farm C. Samples from farms B and C and the cattle trader displayed completely identical genotypes (from the three genotyping methods) to that of isolate 00105T from farm A. Presumed clonal variants with a single band change in the IS6110 RFLP pattern (see boxed RFLP profiles for isolates 00059 and 00081T7) accompanied by a single repeat change in the ETR-A locus for isolate 00059 (see boxed allele) were found on farms A and D, respectively.
In cluster CE2, the four isolates from farms E and F all shared the same MIRU-VNTR genotype over the 29 loci, although one isolate (96171) from farm F had one less IS6110 band than the pattern shared with the other isolate from this farm and the two isolates from farm E (Fig. 2). A direct epidemiological link was clearly established between the two farms involved, as it was found that the two farmers worked together on both farms and their animals shared the same pastures.
The other clusters showed the same profiles by the three genotyping methods (Fig. 2). In cluster CE3, the farms were epidemiologically linked through the purchase of animals (farm G to farm J) or by being neighbors (farms H, I, and J). In cluster CE4, a direct epidemiological link was established between the two farms involved, as the discovery of the outbreak on farm L was actually traced back to animals originating from farm K and led to the identification of an unsuspected outbreak on the latter farm. Likewise, a direct epidemiological link was established between the three farms in cluster CE5 because the discovery of outbreaks on farms M and O was traced back to animals originating on farm N and led to the identification of an unsuspected outbreak on this farm.
Discriminatory power and genotypic diversity (panel 3).
The discriminatory power of MIRU-VNTR typing was compared to that of IS6110 RFLP and spoligotyping by analyzing 68 isolates in panel 3 (Table 2 and Fig. 4). This panel was designed to include isolates assumed to be without epidemiological links. These isolates were representative of all the different IS6110 fingerprints as well as of all the different spoligotypes identified in this nationwide collection. In addition, panel 3 included isolates with identical spoligotypes and IS6110 fingerprints from different farms without obvious epidemiological links based on cattle tracing.
TABLE 2.
Number of different patterns observed among the M. bovis isolates in panel 3 and calculated genotypic diversities generated by IS6110 RFLP, spoligotyping, and MIRU-VNTR typing, taken alone or in combination
| Technique(s) | No. of different patterns | Genotypic diversity |
|---|---|---|
| IS6110 RFLP | 16 | 0.73 |
| Spoligotyping | 17 | 0.85 |
| MIRU-VNTR 29 loci | 32 | 0.91 |
| IS6110 RFLP + spoligotyping | 28 | 0.92 |
| Spoligotyping + VNTR 29 loci | 32 | 0.91 |
| IS6110 RFLP + VNTR 29 loci | 37 | 0.95 |
| IS6110 RFLP + spoligotyping + VNTR 29 loci | 37 | 0.95 |
| Spoligotyping + VNTR 6 locia | 29 | 0.90 |
| IS6110 RFLP + VNTR 6 locia | 35 | 0.94 |
| IS6110 RFLP + spoligotyping + VNTR 6 locia | 35 | 0.94 |
The 6 loci are VNTR 3232, ETR-B, ETR-A, MIRU-26, QUB11b, and QUB11a.
In this collection, different RFLP groups were named according to the IS6110 fingerprints of the isolates, based on a code described by Rigouts et al. (23). Groups A and G included M. bovis isolates with a single IS6110 band and high (8 to 11 bands) IS6110 copy numbers, respectively. These groups were the most prevalent in Belgium with 43% and 48% of the isolates, respectively. The remaining 9% comprised five minor and distinct fingerprint groups, including isolates with intermediate numbers of IS6110 copies, designated H (4 to 5 bands), J (2 to 3 bands), K (4 to 6 bands), L (3 bands), and M (4 bands).
Overall, MIRU-VNTR typing discriminated 32 different patterns in this panel, corresponding to a genotypic diversity of 0.91. In comparison, only 16 different patterns were obtained with IS6110 RFLP and 17 patterns were obtained with spoligotyping, corresponding to genotypic diversities of 0.73 and 0.85, respectively. MIRU-VNTR typing performed even better than a combination of spoligotyping and IS6110 RFLP, which discriminated 28 patterns (Table 2).
Spoligotypes were often distinguished by MIRU-VNTR typing. For instance, the single SB0162 spoligotype characterizing the highly prevalent RFLP group G corresponded to 5 distinct MIRU-VNTR types. In contrast, spoligotyping did not discriminate any MIRU-VNTR genotypes. Likewise, none of the MIRU-VNTR genotypes were distinguished by IS6110 fingerprinting in RFLP group A. In contrast, isolates with no known epidemiological links within three MIRU-VNTR clusters corresponding to the RFLP groups H, J, and G with intermediate or high numbers of IS6110 bands were distinguished by single-band differences in their respective IS6110 fingerprints. Therefore, maximal discrimination was apparently achieved by combining MIRU-VNTR and IS6110 RFLP typing, resulting in 37 patterns and a genotype diversity of 0.95.
Allelic diversity of VNTR loci (panel 3).
Compared with each other, the 29 VNTR loci did not display equal discriminatory power in panel 3. In this test panel, MIRU 39 and 40 and VNTR 2401 displayed only a single allele, while at the other extremity VNTR 3232 displayed 8 different alleles (Table 3). Moreover, for a given locus, some alleles were clearly much more frequent than others (for instance, the allele with 10 repeats in QUB11a was encountered in 67% of the isolates, although 6 different alleles were identified in this locus). The resolution provided by each locus was thus quantified by calculating its allelic diversity, depending upon both the number and the distribution of the alleles. Allelic diversities ranged from 0.00 for MIRU 39 and 40 and VNTR 2401 to 0.76 for VNTR 3232.
TABLE 3.
MIRU-VNTR allelic distribution among 68 isolates in panel 3b
| Locus | No. of isolates with MIRU allele:
|
Allelic diversity (h) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0a | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 9 and 10 | ||
| MIRU 39 | 68 | 0.00 | ||||||||||||||
| MIRU 40 | 68 | 0.00 | ||||||||||||||
| VNTR 2401 | 68 | 0.00 | ||||||||||||||
| MIRU 02 | 67 | 1 | 0.01 | |||||||||||||
| MIRU 31 | 1 | 67 | 0.01 | |||||||||||||
| VNTR 2347 | 1 | 66 | 1 | 0.01 | ||||||||||||
| MIRU 10 | 4 | 64 | 0.10 | |||||||||||||
| VNTR 4156 | 5 | 63 | 0.12 | |||||||||||||
| MIRU 04 | 6 | 61 | 1 | 0.18 | ||||||||||||
| MIRU 20 | 4 | 61 | 3 | 0.18 | ||||||||||||
| ETR-C | 2 | 3 | 2 | 61 | 0.18 | |||||||||||
| VNTR 3690 | 3 | 59 | 6 | 0.23 | ||||||||||||
| MIRU 23 | 10 | 58 | 0.24 | |||||||||||||
| VNTR 1895 | 1 | 58 | 9 | 0.24 | ||||||||||||
| VNTR 3171 | 7 | 4 | 57 | 0.27 | ||||||||||||
| MIRU 16 | 12 | 56 | 0.28 | |||||||||||||
| VNTR 1982 | 4 | 3 | 8 | 50 | 2 | 1 | 0.43 | |||||||||
| QUB 11a | 1 | 21 | 46 | 0.44 | ||||||||||||
| VNTR 424 | 25 | 43 | 0.46 | |||||||||||||
| QUB 11b | 13 | 9 | 46 | 0.48 | ||||||||||||
| VNTR 1955 | 31 | 1 | 36 | 0.50 | ||||||||||||
| QUB 26 | 26 | 37 | 5 | 0.55 | ||||||||||||
| MIRU 27 | 1 | 6 | 16 | 42 | 3 | 0.55 | ||||||||||
| MIRU 24 | 12 | 35 | 21 | 0.60 | ||||||||||||
| MIRU 26 | 1 | 13 | 33 | 21 | 0.63 | |||||||||||
| ETR-A | 2 | 9 | 2 | 33 | 19 | 2 | 1 | 0.66 | ||||||||
| ETR-B | 6 | 13 | 22 | 27 | 0.69 | |||||||||||
| VNTR 3336 | 1 | 1 | 4 | 4 | 20 | 12 | 25 | 1 | 0.74 | |||||||
| VNTR 3232 | 10 | 4 | 6 | 17 | 25 | 2 | 3 | 1 | 0.76 | |||||||
Corresponds to amplified alleles devoid of any repeat unit.
The 29 loci are listed on the left, the alleles are at the top and the calculated allelic diversities on the right. Loci are presented by increasing calculated allelic diversity.
We tested various combinations of loci to identify a minimal subset that provided the same resolution as the total set of 29 loci. In our test panel, the maximal resolution of the 32 patterns obtained with the 29 loci was already achieved using a subset of 9 loci (Table 4). This subset thus corresponded to the minimal combination required to achieve nonredundant discrimination. These loci all displayed an allelic diversity greater than 0.10 in panel 3. However, they were not all ranked in the top 9 in the hierarchy of allelic diversities among the 29 loci. For instance, VNTR 4156, MIRU 4, and ETR-C displayed allelic diversities of 0.12, 0.18, and 0.18, respectively, which were much lower than those of VNTR 1955 (0.50) or MIRU 24 (0.60). These three former loci were included in the minimal subset because, in some instances, one of these three markers was involved in a single-locus variation, i.e., it was the only locus out of the 29 to distinguish an isolate from other closely related isolates. In contrast, neither VNTR 1955 nor MIRU 24 was included in the minimal subset in our collection because they were never involved in single-locus variations. Therefore, the discrimination provided was always redundant with that generated by the loci in the minimal subset. VNTR 3336, which had the second highest allelic diversity, was repeatedly difficult to amplify. Therefore, this locus was also not included in the subset of 9 nonredundant loci and we do not recommend its routine use.
TABLE 4.
Number of different patterns observed among the 68 M. bovis isolates in panel 3 and calculated genotypic diversities of different subsets of VNTRs
| Combination of loci | No. of different patterns | Genotypic diversity |
|---|---|---|
| VNTR 3232, ETR-A and B | 21 | 0.86 |
| VNTR 3232, ETR-A and B, MIRU 26 | 24 | 0.87 |
| VNTR 3232, ETR-A and B, MIRU 26, QUB11b | 27 | 0.89 |
| VNTR 3232, ETR-A and B, MIRU 26, QUB11b, QUB11a | 28 | 0.90 |
| VNTR 3232, ETR-A and B, MIRU 26, QUB11b, QUB11a, ETR-C, VNTR 4156, MIRU 4 | 32 | 0.91 |
| MIRU-VNTR 29 loci | 32 | 0.91 |
The above minimal subset was defined by analyzing all isolates and genotypes included in panel 3. We further analyzed the relative variability of the 29 MIRU-VNTR loci by identifying specifically those that varied within each of the four best identified lineages in this panel, as defined by the congruence of their MIRU-VNTR, spoligotyping, and IS6110 RFLP profiles (corresponding to groups G, H, J, and K, see below). We reasoned that loci that were variable within given lineages were more likely to have higher evolutionary rates because the isolates distinguished were mutually closely related. The 8 loci that varied in either of these lineages (Fig. 4) consistently comprised the 9 loci in the discriminatory subset defined in the whole of panel 3.
The simultaneous occurrence of two different alleles (9 and 10) was clearly detected for locus QUB 11a, included in the discriminatory subset, for one isolate obtained from an individual animal (96427). This observation indicates the presence of a clonal variant in the bacterial population from this isolate.
Genetic relationships (panel 3).
The congruence of the genetic relationships, as defined by IS6110 fingerprints, spoligotyping, and MIRU-VNTR markers, was analyzed for the isolates of panel 3 on the basis of the MIRU-VNTR genotypes (corresponding to the most discriminatory method) using the neighbor-joining distance algorithm (Fig. 4). The groupings defined by MIRU-VNTR typing were the most clearly congruent with those apparent from RFLP patterns and spoligotyping for the isolates of RFLP groups G, H, J, and K. For instance, group G was initially defined by similar RFLP patterns that varied from 8 to 11 IS6110 elements. Samples of this group were distinguished by MIRU-VNTR into 5 different patterns but were grouped together based on variations involving at most a one-repeat difference in a single locus. The isolates of this group all had a single specific spoligotype, SB0162. The consistency of RFLP group K, initially defined by a 4- to 6-band RFLP pattern, was likewise confirmed by mutual MIRU-VNTR variations involving at most 3 loci and by identification of a single, albeit nonspecific, spoligotype, SB0120. In contrast and not surprisingly, the isolates previously assigned to group A on the basis of their single-banded IS6110 RFLP pattern appeared heterogeneous both by spoligotyping and by MIRU-VNTR typing (13 and 17 different patterns, respectively).
We quantified the congruence of the genetic relationships based on MIRU-VNTR typing on the one hand and the combination of IS6110 RFLP and spoligotyping on the other to reduce biases due to the much lower resolution of the two latter methods when taken individually. When the whole of panel 3 was considered, these methods were found to correlate (r = 0.30) with a high degree of significance (P < 10−4). Again, it was no surprise to find that when the single-banded IS6110 fingerprints (group A) were excluded from the comparison, the correlation increased to r = 0.63 with an identical threshold of significance.
DISCUSSION
MIRU-VNTR typing is a useful tool for genotyping M. tuberculosis (2, 8, 15). Such a method has recently been shown to be more discriminatory than spoligotyping for M. bovis when using a limited number of isolates from Northern Ireland (25) or from Chad (14). However, the epidemiological relevance of this method has been difficult to appreciate so far, notably because of the lack of detailed epidemiological information about the isolates involved. Here, we have identified the most informative markers from a screen of 29 MIRU-VNTR loci by using a larger panel of isolates, representative of the genotypic diversity in a nationwide collection, together with cattle tracing information. Also in contrast to previous studies, a more stringent analysis of the resolution power and stability of the markers was possible by comparing them with both IS6110 RFLP and spoligotyping, especially as half of our collection included M. bovis isolates with high IS6110 copy numbers rarely reported elsewhere.
As a prerequisite to using them to trace transmission chains, genetic markers must be adequately stable to identify isolates of the same strain. As Belgium has an extremely low incidence of bovine tuberculosis (declared officially free of cattle tuberculosis in June 2003 according to the European Commission decision 2003/467/EC) and has no known environmental reservoir for M. bovis, it is likely that an outbreak on a farm is most often caused by a single source and thus involves a single strain or transmission chain. In our study, all but one series of samples from outbreak episodes covering up to 4 years on the same farm were perfectly matched by fully identical MIRU-VNTR alleles over the 29 target loci. The only exception was observed for two pooled samples, both displaying 2 alleles simultaneously in locus 3232. This observation could reflect the presence of two independent strains in these samples or the stochastic emergence of a clonal variant for this locus.
Furthermore, we also found that the MIRU-VNTR loci were stable in transmission chains covering at least 2 years and up to 5 different herds. Again, a single exception was observed for one isolate in one cluster, which had both a one-repeat difference in ETR-A and one IS6110 band less in relation to the other isolates in this cluster. Interestingly, this isolate was obtained in 2000 from the farm presumably at the origin of this cluster (CE1) and where an outbreak was suspected to have started in 1996 or earlier. Therefore, this isolate could correspond to a glimpse into progressive clonal diversification over longer periods of time. In contrast, a couple of isolates with fully identical MIRU-VNTR types had slightly distinct multibanded IS6110 RFLP patterns, although they were clearly epidemiologically linked. Such a phenomenon, which has also been observed for epi-linked M. tuberculosis isolates (1), suggests that IS6110 RFLP may evolve very fast in certain M. bovis strains, and therefore, the use of IS6110 fingerprinting alone might in some cases underestimate ongoing transmission of tuberculosis in cattle.
Among the isolates without known epidemiological links, MIRU-VNTR typing provided a higher discriminatory power than the two other methods, taken individually or in combination. As expected, most spoligotyping- or low-IS6110-copy-number-fingerprint-based clusters were distinguished by MIRU-VNTR. MIRU-VNTR typing was even able to distinguish a total of 7 isolates in IS6110 RFLP fingerprint-based clusters with intermediate or high IS6110 copy numbers. Similar observations have been recently obtained for epidemiologically unlinked M. tuberculosis isolates clustered by identical high-IS6110-copy-number fingerprints (17, 33). Conversely, none of the MIRU-VNTR clusters was split by spoligotyping. The epidemiological significance of three isolates in MIRU-VNTR clusters, distinguished by a single-band change in their intermediate- or high-IS6110-copy-number fingerprint, as well as other isolates with identical genotypes from the three methods, remains unknown.
The observed congruence between MIRU-VNTR, IS6110 RFLP, and spoligotyping was found to be highly significant. This congruence is consistent with the clonal population structure of M. bovis (28) and indicates that the three independent markers are generally phylogenetically informative. Not surprisingly, however, IS6110 RFLP appeared to be less reliable for identifying consistent genetic groups in cases of fingerprints with a single band. One lineage accounting for about half of all isolates collected in Belgium was clearly identified by the three methods. The corresponding spoligotype is quite specific, as it has only been reported so far in one French region (named as type F002 in reference 13). We verified that these isolates had the RD4 genomic deletion (data not shown) typical of classical M. bovis (4). In contrast, not a single representative of the SB0140 spoligotype, which is, according to www.mbovis.org, the most common spoligotype in the United Kingdom, was found in our collection. Thus, the M. bovis population differs greatly between the cattle from Belgium and Northern Ireland.
Our study collection includes more than 20% of the samples from bovine tuberculosis cases with positive culture collected over a 9-year period in Belgium and includes isolates representative of all the different spoligotypes and/or IS6110 fingerprints available from a nationwide collection. The degree of epidemiological representation of this collection is difficult to ascertain, as information about potential links between the farms involved was not systematically available. This limitation also applies to the previous studies evaluating molecular typing methods of M. bovis. Despite these limitations, it is remarkable that some loci systematically emerge among the most discriminatory markers across different settings. Our set of MIRU-VNTR loci included 25 loci in common with the set of 30 tested in Ireland (26) and incorporated the 16 loci recently tested on 67 isolates in a Chadian setting (14). In our case, the maximal resolution of MIRU-VNTR typing was already achieved using a subset of 9 loci (VNTR 3232, ETR-B, ETR-A, MIRU-26, QUB11b, QUB11a, ETR-C, VNTR 4156, and MIRU 4). Consistently, the differences among closely related isolates within well-identified clonal lineages were virtually all confined to these loci, indicating a generally higher evolutionary rate compared to the other loci. Moreover, about ninety percent of this resolution was concentrated in a core set of only 6 loci with comparatively high allelic diversities (VNTR 3232, ETR-A and B, MIRU 26, and QUB 11a and 11b). Interestingly, these 6 loci were all ranked among the markers with the highest allelic diversities in Ireland (25-27). In the Chadian study, high allelic diversities were also found for MIRU 26, ETR-A, and ETR-B, although a comparatively lower allelic diversity was observed for VNTR 3232, and neither QUB 11a nor 11b was tested. The three remaining loci (ETR-C, VNTR 4156, and MIRU 4) were marginally more polymorphic in our study, as in the Irish studies (25-27), but their contribution resided in the nonredundant discrimination of a few specific genotypes by single locus variations. Alternative, albeit larger, combinations of loci common to both studies were, however, possible to reach nearly maximal resolution. The core set (VNTR 3232, ETR-A and B, MIRU 26, and QUB 11a and 11b) of highly discriminatory loci, both in Ireland and Belgium, with radically distinct bacterial populations (see above), appear to be the most interesting candidates for testing the discrimination of M. bovis isolates on a more general scale. A future multicenter study should determine the performance of this core set for the general first-line discrimination of M. bovis isolates in different countries. The above comparisons suggest that secondary typing with a few additional loci potentially specific to some settings may provide additional limited discrimination of a few particular genotypes.
In conclusion, this study indicates that MIRU-VNTR markers provide a good balance between stability and variability compared to spoligotype and IS6110 RFLP analysis and may be useful molecular tools for epidemiological studies of M. bovis. Their stability in virtually all epidemiologically related isolates analyzed so far support their use, at least as an exclusion method, i.e., even small differences in MIRU-VNTR genotypes can be interpreted as evidence of the absence of a link, with a high degree of confidence. In countries with a low incidence of disease, such as Belgium, the resolution may even be sufficient to identify different transmission chains. In addition, MIRU-VNTR typing is a convenient, rapid, and reproducible technique, especially compared to IS6110 RFLP. Eventually, the portability of MIRU-VNTR data and the development of a standardized format integrated into an international database could be very useful for tracing the worldwide dissemination of the pathogen.
Supplementary Material
Acknowledgments
We thank Vanessa Vandenpoorte, Sylvie Malbrecq, Philippe Vannoorenberghe, and Damien Desqueper for their excellent technical skills. We thank Leen Rigouts and Françoise Portaels for transmitting their RFLP and spoligotyping expertise of mycobacteria to VAR. We thank all members of the Federal Agency for the Safety of the Food Chain (FASFC) who cooperated in this study, especially regarding epidemiological inquiries.
P.S. is a chercheur du Centre National de la Recherche Scientifique.
This work was financially supported by Brussels-Capital Region; Institut Scientifique de Santé Publique, Institut Pasteur, Belgium; and Federal Agency for the Safety of the Food Chain (FASFC).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alito, A., N. Morcillo, S. Scipioni, A. Dolmann, M. I. Romano, A. Cataldi, and D. van Soolingen. 1999. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant Mycobacterium tuberculosis strains may evolve too fast for reliable use in outbreak investigation. J. Clin. Microbiol. 37:788-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allix, C., P. Supply, and M. Fauville-Dufaux. 2004. Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39:783-789. [DOI] [PubMed] [Google Scholar]
- 3.Ayele, W. Y., S. D. Neill, J. Zinsstag, M. G. Weiss, and I. Pavlik. 2004. Bovine tuberculosis: an old disease but a new threat to Africa. Int. J. Tuberc. Lung Dis. 8:924-937. [PubMed] [Google Scholar]
- 4.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosivi, O., J. M. Grange, C. J. Daborn, M. C. Raviglione, T. Fujikura, D. Cousins, R. A. Robinson, H. F. Huchzermeyer, I. de Kantor, and F. X. Meslin. 1998. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cousins, D. V., and D. J. Dawson. 1999. Tuberculosis due to Mycobacterium bovis in the Australian population: cases recorded during 1970-1994. Int. J. Tuberc. Lung Dis. 3:715-721. [PubMed] [Google Scholar]
- 7.Cowan, L. S., and J. T. Crawford. 2002. Genotype analysis of Mycobacterium tuberculosis isolates from a sentinel surveillance population. Emerg. Infect. Dis. 8:1294-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowan, L. S., L. Diem, T. Monson, P. Wand, D. Temporado, T. V. Oemig, and J. T. Crawford. 2005. Evaluation of a two-step approach for large-scale, prospective genotyping of Mycobacterium tuberculosis isolates in the United States. J. Clin. Microbiol. 43:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 10.Gibson, A. L., G. Hewinson, T. Goodchild, B. Watt, A. Story, J. Inwald, and F. A. Drobniewski. 2004. Molecular epidemiology of disease due to Mycobacterium bovis in humans in the United Kingdom. J. Clin. Microbiol. 42:431-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez, M. C., S. Brisse, R. Brosch, M. Fabre, B. Omais, M. Marmiesse, P. Supply, and V. Vincent. 2005. Ancient origin and gene mosaicism of the progenitor of mycobacteriumtuberculosis. PLoS Pathog. 1:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez, M. C., J. C. Galan, J. Blazquez, E. Bouvet, and V. Vincent. 1999. Molecular markers demonstrate that the first described multidrug-resistant Mycobacterium bovis outbreak was due to Mycobacterium tuberculosis. J. Clin. Microbiol. 37:971-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haddad, N., A. Ostyn, C. Karoui, M. Masselot, M. F. Thorel, S. L. Hughes, J. Inwald, R. G. Hewinson, and B. Durand. 2001. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 39:3623-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hilty, M., C. Diguimbaye, E. Schelling, F. Baggi, M. Tanner, and J. Zinsstag. 2005. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet. Microbiol. 109:217-222. [DOI] [PubMed] [Google Scholar]
- 15.Kam, K. M., C. W. Yip, L. W. Tse, K. L. Wong, T. K. Lam, K. Kremer, B. K. Au, and D. van Soolingen. 2005. Utility of mycobacterial interspersed repetitive unit typing for differentiating multidrug-resistant Mycobacterium tuberculosis isolates of the Beijing family. J. Clin. Microbiol. 43:306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 41:2683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Fleche, P., M. Fabre, F. Denoeud, J. L. Koeck, and G. Vergnaud. 2002. High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LoBue, P. A., W. Betancourt, L. Cowan, L. Seli, C. Peter, and K. S. Moser. 2004. Identification of a familial cluster of pulmonary Mycobacterium bovis disease. Int. J. Tuberc. Lung Dis. 8:1142-1146. [PubMed] [Google Scholar]
- 20.O'Reilly, L. M., and C. J. Daborn. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber. Lung Dis. 76(Suppl. 1):1-46. [DOI] [PubMed] [Google Scholar]
- 21.Palmer, D. N. M. A. 23 July 2004, revision date. Chapter 2.3.3. Bovine tuberculosis. In OIE manual of diagnostic tests and vaccines for terrestrial animals, 5th ed. World Organisation for Animal Health, Paris, France. [Online.] http://www.oie.int/eng/normes/en_mmanual.htm.
- 22.Palmer, M. V., W. R. Waters, and D. L. Whipple. 2004. Investigation of the transmission of Mycobacterium bovis from deer to cattle through indirect contact. Am. J. Vet. Res. 65:1483-1489. [DOI] [PubMed] [Google Scholar]
- 23.Rigouts, L. D. M., J. Dufey, C. Saegerman, K. Fissette, K. Kremer, H. Traore, D. van Soolingen, K. Walravens, J. Godfroid, and F. Portaels. 2000. Third International Conference on Mycobacterium bovis, p. 39.
- 24.Robert, J., F. Boulahbal, D. Trystram, C. Truffot-Pernot, A. C. de Benoist, V. Vincent, V. Jarlier, J. Grosset, et al. 1999. A national survey of human Mycobacterium bovis infection in France. Int. J. Tuberc. Lung Dis. 3:711-714. [PubMed] [Google Scholar]
- 25.Roring, S., A. Scott, D. Brittain, I. Walker, G. Hewinson, S. Neill, and R. Skuce. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roring, S., A. N. Scott, R. Glyn Hewinson, S. D. Neill, and R. A. Skuce. 2004. Evaluation of variable number tandem repeat (VNTR) loci in molecular typing of Mycobacterium bovis isolates from Ireland. Vet. Microbiol. 101:65-73. [DOI] [PubMed] [Google Scholar]
- 27.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 28.Smith, N. H., J. Dale, J. Inwald, S. Palmer, S. V. Gordon, R. G. Hewinson, and J. M. Smith. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. USA 100:15271-15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smittipat, N., and P. Palittapongarnpim. 2000. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80:69-74. [DOI] [PubMed] [Google Scholar]
- 30.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 32.Tibayrenc, M., F. Kjellberg, and F. J. Ayala. 1990. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc. Natl. Acad. Sci. USA 87:2414-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Deutekom, H., P. Supply, P. de Haas, E. Willery, S. Hoijng, C. Locht, R. Coutinho, and D. van Soolingen. 2005. Molecular typing of Mycobacterium tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat analysis, a more accurate method for identifying epidemiological links between patients with tuberculosis. J. Clin. Microbiol. 43:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, et al. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.