Abstract
The BACTEC 9240 blood culture system with a standard aerobic medium, Plus Aerobic/F, was compared to the BacT/Alert system with a platelet-specific medium, BPA, as a means for detecting bacterial contamination of platelet preparations. One hundred thirteen platelet units seeded with low levels of different bacteria were examined with both systems. In 93 instances, growth was detected first in the BACTEC system; in 12 cases, growth registered first in the BacT/Alert system. Among all comparisons, growth was detected, on average, 1.7 h sooner with the BACTEC system. The differences in length of time to detection were statistically significant.
Each year, approximately 4 million platelet units are transfused in the United States (1). Platelet concentrates and apheresis platelets are stored in oxygen-permeable containers with agitation at 20 to 24°C for up to 5 days prior to use. These conditions permit growth of aerobic bacteria with the potential for transmission to patients receiving platelet preparations (12, 18). Indeed, as many as one per 1,000 to 2,000 platelet units is likely to have some form of bacterial contamination (2). Sepsis as a complication of platelet transfusions has been described frequently in the literature (3, 8, 13-17, 19, 20), the scope and magnitude of this problem having been recently reviewed by Brecher and Hay (3), Wagner et al. (19), and Burns and Werch (7).
In response to an apparently growing problem, a new standard was implemented by the American Association of Blood Banks (AABB) in March of 2004, requiring that all platelet units be assessed regarding the possibility of bacterial contamination prior to their use in patients (6, 11). A comparable requirement has also been promulgated by The College of American Pathologists (CAP) in its Transfusion Medicine Checklist (9).
The U.S. Food and Drug Administration (FDA) has approved two culture systems for the purpose of screening platelet units for bacterial contamination: the BacT/Alert system (bioMérieux, Durham, N.C.) and the eBDS system (Pall Corporation, East Hills, N.Y.) (3). The eBDS system is a culture-based method designed specifically for analysis of blood and blood products. The BacT/Alert platelet screening system utilizes a specialized medium, BPA, but employs the continuous monitoring BacT/Alert blood culture instrument as a testing platform. The FDA approval of the BacT/Alert platelet culture system as a means for screening platelets for bacterial contamination was predicated on two studies in which platelet units seeded with different bacteria were cultured (4, 5).
The BacT/Alert platelet screening method provides a convenient means for laboratories already utilizing the BacT/Alert blood culture system to screen platelets for contamination. The problem, however, is that only approximately one-third of clinical laboratories in the United States currently use this system for performing blood cultures, while the remainder use one of two other instrument-based continuous monitoring blood culture systems, i.e., either the BACTEC 9240 System (BD Microbiology, Cockeysville, MD) (ca. 60%) or the VersaTrek System (Trek Diagnostics, Westlake, OH) (ca. 5%). Laboratories that utilize the BACTEC 9240 or VersaTrek blood culture methods are forced to consider adopting a completely different technology in order to comply with AABB-CAP mandates for screening platelets.
In the current study, we compared the performance of the BACTEC 9240 continuous monitoring blood culture system with the BacT/Alert platelet culturing system for the detection of bacterial contaminants in platelet preparations. For the purposes of this comparison, we employed platelet units that had been seeded with various bacteria known to be common contaminants of platelet concentrates.
A total of 113 recent isolates of various bacteria (Table 1) were used. These were selected as being representative of those bacteria most frequently isolated from platelet products in routine clinical practice (3, 19). Suspensions of test strains approximately equivalent to 102 CFU/ml were prepared in Trypticase soy broth, and nonpooled single donor platelet preparations that had been stored as described above for >5 days were aseptically seeded with a volume of suspension sufficient to achieve an estimated final bacterial concentration in the platelet unit of approximately 10 CFU/ml. This target concentration was verified by culture quantitation of an aliquot from each seeded platelet unit. The seeded platelet preparations were gently agitated for ca. 5 min, and then a single 20-ml volume was removed aseptically with a 20-ml syringe equipped with a 21-gauge needle. Ten-milliliter aliquots of these samples were then immediately transferred aseptically into BacT/Alert BPA and BACTEC Plus Aerobic/F bottles. The order of bottle inoculation was random so as to ensure that each bottle was inoculated first approximately the same number of times. The bottles were immediately placed on their respective continuous monitoring instruments and incubated for a period of 5 days.
TABLE 1.
Detection of bacteria in 113 seeded platelet units with the BACTEC and BacT/Alert culture systemsa
| Organism (n) | Mean LTD [h (95% confidence interval)] in:
|
P value | |
|---|---|---|---|
| BACTEC | BacT/Alert | ||
| E. coli (33) | 11.2 (10.77-11.53) | 12.3 (11.98-12.68) | <0.0001 |
| Staphylococcus aureus (17) | 14.4 (12.97-15.92) | 14.8 (13.38-16.31) | NS |
| Cons (29) | 17.8 (16.42-19.27) | 19.0 (17.71-20.38) | 0.004 |
| Viridans group streptococci (13) | 16.7 (10.53-22.89) | 22.9 (17.6-28.2) | 0.019 |
| Serratia spp. (8) | 11.9 (11.31-12.54) | 13.4 (12.7-14.07) | <0.0001 |
| Micrococcus sp. (1) | 13.1 | 16.3 | ND |
| Diphtheroids (4) | 19.1 (3.864-34.37) | 19.4 (7.67-31.08) | NS |
| Enterobacter spp. (2) | 11.1 | 11.9 | ND |
| Total (107) | 14.5 | 16.2 | <0.0001 |
Abbreviations: LTD, length of time in hours to detection; NS, not significant; ND, not done; Cons, coagulase-negative Staphylococcus spp. In two instances, one each with the Micrococcus sp. and a Cons species, neither method was positive; in two instances, one each with a Cons species and a diphtheroid, the BACTEC system was uniquely positive; and in two instances, one each with the Micrococcus sp. isolate and a diphtheroid, the BacT/Alert was uniquely positive. These six comparisons have been excluded from this tabulation.
The time that all bottles first registered as being positive was recorded. Subcultures of positive bottles were performed to ensure that the organisms that grew were the same as the organisms used initially to seed the respective platelet samples. The lengths of time in hours to detection (LTD) for each system for all test strains were compared. Statistical analysis was performed using the paired-observation t test.
Among the 113 comparisons, growth was detected in both bottles of each culture pair in 107 instances. The mean LTD in the two culture systems for these 107 comparisons are listed in Table 1. On the average, growth was detected 1.7 h sooner in the BACTEC system than in the BacT/ALERT system. This difference was highly statistically significant (P value of <0.0001).
In 6 of the 113 comparisons, growth was not detected in both systems. In two instances, one with the Micrococcus sp. and one with a coagulase-negative Staphylococcus sp., neither system registered as positive. In two instances the BACTEC system was uniquely positive; these consisted of one coagulase-negative Staphylococcus sp. (LTD = 19.6 h) and one aerobic diphtheroid (LTD = 59.7 h). In the remaining two comparisons, the BacT/Alert system was the only system that registered as positive: one with the Micrococcus sp. (LTD = 67.2 h) and one with an aerobic diphtheroid (LTD = 16 h). Subcultures of all negative bottles in these six comparisons yielded no growth. Further, the initial inoculum concentrations in these six comparisons were determined to be <5 CFU/ml. We postulate that in our efforts to seed platelet concentrates with low levels of bacteria, in these six cases, the negative bottles simply did not receive viable inoculum initially. These six samples were excluded from further analysis.
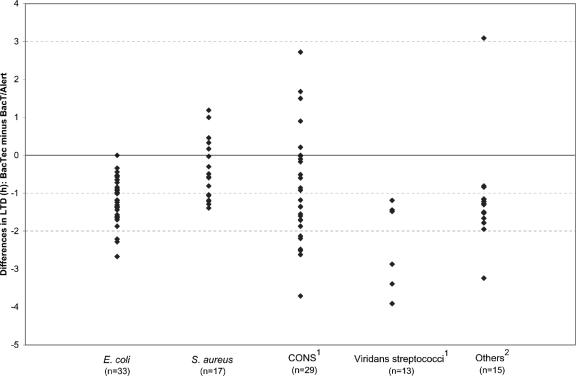
The difference in LTD in the two systems for individual organisms is presented in Fig. 1. In 93 instances (86.9%), growth was detected faster in the BACTEC system; in 12 cases (11.2%), growth registered first in the BacT/Alert system; and in the remaining two comparisons (1.9%), growth was recognized simultaneously in the two systems. Two observations are apparent from the distributions in Fig. 1. The shorter detection times observed in the BACTEC system were most conspicuous with Escherichia coli, various miscellaneous bacteria, and in particular, viridans group streptococci. Further, detection time differences for bacteria in a given organism group were extremely variable. For example, among 13 viridans group streptococci, in one instance, BACTEC was positive 1.2 h sooner than BacT/Alert. In another case, growth was detected in BACTEC 19 h faster.
FIG. 1.
Differences in lengths of time to detection of contaminants in platelet units for the BACTEC culture system versus the BacT/Alert culture system. The data points for eight isolates with detection time differences of greater than 5 h were not included in this figure: one coagulase-negative Staphylococcus sp. (CONS) (−8.9 h) and seven viridans group streptococci (+12.4, −6.4, −11.8, −12.6, −13.0, −15.7, and −19.0 h). The “Others” category includes eight Serratia spp., one Micrococcus sp., four aerobic diphtheroids, and two Enterobacter spp.
The results of our study clearly indicate that the BACTEC blood culture system, with a standard aerobic medium (Plus Aerobic/F), was at least comparable to the BacT/Alert system, employing a medium specifically designed for culturing platelet preparations, in the detection of bacteria in platelet units. Indeed, if LTD was used as a basis for comparison, the BACTEC system was statistically significantly superior to BacT/Alert, insofar as the BACTEC system detected positives significantly faster. These observations are consistent with the findings of one previous investigation by Dunne and colleagues (10), albeit not a comparative study with BacT/Alert, that demonstrated the functionality of the BACTEC system for rapid detection of bacteria in platelet preparations.
We conclude from the results of this study that the BACTEC system is a suitable method for the screening of platelet units for bacterial contamination in clinical microbiology laboratories. Furthermore, given the fact that the scope and design of our study were at least equivalent to if not more rigorous than the studies used previously to justify FDA approval of the BacT/Alert system for this application, it follows that the BACTEC system should also be considered by the FDA for approval.
REFERENCES
- 1.American Association of Blood Banks. 2003. Comprehensive report on blood collection and transfusion in the United States in 2001. National Blood Data Resource Center, Bethesda, Md.
- 2.Brecher, M. E. 2002. Bacterial contamination of blood products, p. 789. In T. L. Simon (ed.), Rossi's principles of transfusion medicine, 3rd ed. Lippincott Williams and Wilkins, Baltimore, Md.
- 3.Brecher, M. E., and S. N. Hay. 2005. Bacterial contamination of blood components. Clin. Microbiol. Rev. 18:195-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brecher, M. E., D. G. Heath, S. N. Hay, S. J. Rothenberg, and L. C. Stutzman. 2002. Evaluation of a new generation of culture bottle using an automated bacterial culture system for detecting nine common contaminating organisms found in platelet components. Transfusion 42:774-779. [DOI] [PubMed] [Google Scholar]
- 5.Brecher, M. E., N. Means, C. S. Jere, D. G. Heath, S. J. Rothenberg, and L. C. Stutzman. 2001. Evaluation of an automated culture system for detecting bacterial contamination of platelets: an analysis with 15 contaminating organisms. Transfusion 41:477-482. [DOI] [PubMed] [Google Scholar]
- 6.Brumit, M. C., S. N. Hay, and M. E. Brecher. 2003. Bacterial detection, p. 57-81. In M. E. Brecher (ed.), Bacterial and parasitic contamination of blood components. AABB Press, Bethesda, Md.
- 7.Burns, K. H., and J. B. Werch. 2004. Bacterial contamination of platelet units: a case report and literature survey with review of upcoming AABB requirements. Arch. Pathol. Lab. Med. 128:279-281. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, E. K., K. Y. Yuen, and A. K. Lie. 1994. A prospective study of symptomatic bacteremia following platelet transfusion and of its management. Transfusion 34:950-954. [DOI] [PubMed] [Google Scholar]
- 9.College of American Pathologists Commission on Laboratory Accreditation. 6 April 2006, revision date. Laboratory Accreditation Program transfusion medicine checklist. [Online.] http://www.cap.org/apps/docs/laboratory_accreditation/checklists/transfusion_medicine_april2006.doc.
- 10.Dunne, W. M., Jr., L. K. Case, L. Isgriggs, and D. M. Lublin. 2005. In-house validation of the BACTEC 9240 blood culture system for detection of bacterial contamination in platelet concentrates. Transfusion 45:1138-1142. [DOI] [PubMed] [Google Scholar]
- 11.Gilligan, P. H. 2004. Impact of clinical practice guidelines on the clinical microbiology laboratory. J. Clin. Microbiol. 42:1391-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heal, J. M., S. Singal, E. Sardisco, and T. Mayer. 1986. Bacterial proliferation in platelet concentrates. Transfusion 26:388-390. [DOI] [PubMed] [Google Scholar]
- 13.Hillyer, C. D., C. D. Josephson, M. A. Blajchman, J. G. Vostal, J. S. Epstein, and J. L. Goodman. 2003. Bacterial contamination of blood components: risks, strategies and regulation: joint ASH and AABB educational session in transfusion medicine. Hematology 2003:575-589. [DOI] [PubMed] [Google Scholar]
- 14.Kuehnert, M. J., V. R. Roth, and N. R. Haley. 2001. Transfusion transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 41:1493-1499. [DOI] [PubMed] [Google Scholar]
- 15.Morrow, J. F., H. G. Braine, T. S. Kickler, P. S. Ness, J. D. Dick, and A. K. Fuller. 1991. Septic reactions to platelet transfusions: a persistent problem. JAMA 266:555-558. [PubMed] [Google Scholar]
- 16.Ness, P., H. Braine, and K. King. 2001. Single donor platelets reduce the risk of septic platelet transfusion reactions. Transfusion 41:857-861. [DOI] [PubMed] [Google Scholar]
- 17.Perez, P., L. R. Salmi, and G. Follea. 2001. Determinants of transfusion-associated bacterial contamination: results of the French BACTHEM case-control study. Transfusion 41:862-872. [DOI] [PubMed] [Google Scholar]
- 18.Punsalang, A., J. M. Heal, and P. J. Murphy. 1989. Growth of gram-positive and gram-negative bacteria in platelet concentrates. Transfusion 29:596-599. [DOI] [PubMed] [Google Scholar]
- 19.Wagner, S. J., L. I. Friedman, and R. Y. Dodd. 1994. Transfusion-associated bacterial sepsis. Clin. Microbiol. Rev. 7:290-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yomtovian, R., H. M. Lazarus, L. T. Goodnough, N. V. Hirschler, A. M. Morrissey, and M. R. Jacobs. 1993. A prospective microbiological surveillance program to detect and prevent transfusion of bacterially contaminated platelets. Transfusion 33:902-909. [DOI] [PubMed] [Google Scholar]