Abstract
We have identified nine oligopeptide transporter (OPT) orthologs (AtOPT1 to AtOPT9) in Arabidopsis. These proteins show significant sequence similarity to OPTs of Candida albicans (CaOpt1p), Schizosaccharomyces pombe (Isp4p), and Saccharomyces cerevisiae (Opt1p and Opt2p). Hydrophilicity plots of the OPTs suggest that they are integral membrane proteins with 12 to 14 transmembrane domains. Sequence comparisons showed that the AtOPTs form a distinct subfamily when compared with the fungal OPTs. Two highly conserved motifs (NPG and KIPPR) were found among all OPT members. The identification of multiple OPTs in Arabidopsis suggests that they may play different functional roles. This idea is supported by the fact that AtOPTs have a distinct, tissue-specific expression pattern. The cDNAs encoding seven of the AtOPTs were cloned into a yeast vector under the control of a constitutive promoter. AtOPT4 expressed in S. cerevisiae mediated the uptake of KLG-[3H]L. Similarly, expression of five of the seven AtOPT proteins expressed in yeast conferred the ability to uptake tetra- and pentapeptides as measured by growth. This study provides new evidence for multiple peptide transporter systems in Arabidopsis, suggesting an important physiological role for small peptides in plants.
Peptide transport involves the translocation of peptides (2–6 residues in length) across the cellular membrane in an energy-dependent manner and has been well documented in bacteria, fungi, and mammals (Payne and Smith, 1994; Becker and Naider, 1995; Meredith and Boyd, 2000). After uptake, the internalized peptides are rapidly hydrolyzed by peptidases and used as a source of amino acids, nitrogen, or carbon. Compared with prokaryotes and animals, peptide transport in higher plants has received little attention.
There are few published reports dealing with small peptides in plants. Higgins and Payne (1982) reported that significant levels of peptides were found in phloem and xylem exudates. These included the non-protein-derived peptides (e.g. alanylaminobutyric acid and glycylketoglutaric acid). The xylem contains lower levels of nitrogen than the phloem, yet peptides were reported in the xylem exudates of several species, such as the vegetative organs and berries of grape (Khachidze, 1975) and the sap of corn (Fejer and Konay, 1958). Higgins and Payne (1982) suggested that the transport of peptides is a more efficient means of nitrogen distribution than the transport of individual amino acids. This would be especially true for long-distance transport during the bulk movement of protein-degradation products (e.g. leaf senescence, seed germination). As nitrogen carriers, peptides may also protect amino acids from catabolism by enzymes known to be present in the phloem during transport within the plant (Higgins and Payne, 1980). Glutathione (GSH), a modified tripeptide (γ-Glu-Cys-Gly), has also been suggested as a carrier of reduced sulfur in the phloem and xylem (Higgins and Payne, 1982).
γ-Glutamyl peptides frequently occur in considerable quantities, especially in seeds and storage organs (Higgins and Payne, 1982). For example, 34% of the non-protein amino nitrogen of a kidney bean seed is present as γ-glutamyl-S-methyl-l-Cys (Goore and Thompson, 1967). Upon seed germination, this peptide is degraded, suggesting that it plays an important role in the storage of nitrogen and/or sulfur. Plant growth factors, such as auxin and gibberellin, are frequently bound to small peptides and the peptide-hormone conjugates are present in many tissues, such as the vascular system and the endosperm of plant seeds (Salisbury and Ross, 1991). These conjugates may be involved in regulating hormone activity in response to plant growth and development, as well as facilitating hormone transport (Salisbury et al., 1991). In addition to endogenous plant peptides, several phytotoxins produced by plant pathogens are modified peptides (Walton, 1990). Gross (1991) suggested that plant peptide transport systems could be responsible for the recognition and transport of peptide phytotoxins into plant cells.
Peptide transporters can be grouped into three distinct families based on sequence similarity and mechanism: 1) the ATP-binding cassette (ABC) superfamily (Higgins, 1992); 2) the peptide transporter (PTR) or the proton-dependent oligopeptide transporter (POT) family (Paulsen and Skurray, 1994; Steiner et al., 1995); and 3) the recently identified oligopeptide transporter (OPT) family (Lubkowitz et al., 1997; Hauser et al., 2001).
Plant members of the ABC family of peptide transporters include the recently identified Arabidopsis GSH S-conjugate transporters, AtMRP1 to AtMRP4 (Lu et al., 1997, 1998; Sánchez-Fernández et al., 1998).
The first plant PTR-like peptide transporter, Arabidopsis AtPTR2, was identified by complementation of a yeast mutant defective in di/tripeptide transport (Steiner et al., 1994). Transgenic plants expressing antisense AtPTR2 exhibited a delayed flowering phenotype and an arrest in seed development, especially seed maturation (Song et al., 1997). In barley (Hordeumvulgare), peptide transfer occurs across the scutellum layer, a specialized absorptive tissue abutting the endosperm. The peptide transporter in barley seeds shares a number of similarities to members of the PTR family, including transport of di- and tripeptides (Sopanen et al., 1977; Higgins and Payne, 1978). A cDNA clone encoding a barley peptide transporter (HvPTR1) was recently isolated (West et al., 1998) and this protein was localized on the plasma membrane of the scutellar epithelium (Waterworth et al., 2000). Recent database comparisons revealed eight additional PTR orthologs in Arabidopsis, suggesting an important role for this family in plant growth and development (data not shown).
Members of the OPT family of peptide transporters have 12 to 14 predicted transmembrane domains and show no sequence similarity to ABC or PTR transporters. Until now, members of the OPT family were only characterized from yeast (i.e. Candida albicans, CaOpt1p [Lubkowitz et al., 1997]; Schizosaccharomyces pombe, Isp4p [Lubkowitz et al., 1998]; Saccharomyces cerevisiae, Opt1p and Opt2p; [Hauser et al., 2000]). These proteins were shown to mediate the uptake of tetra- and pentapeptides (e.g. KLGL, KLLG, or KLLLG). S. cerevisiae Opt1p was shown to transport Met-enkephalin (YGGFM) and Leu-enkephalin (YGGFL) with a Km of 310 μm for YGGFL (Hauser et al., 2000). In contrast, Opt1p did not transport various amino acids or di-/tripeptides tested (e.g. Tyr, L-L, and G-G-F; Hauser et al., 2000). Interestingly, Bourbouloux et al. (2000) reported that Opt1p was a high affinity GSH transporter, whereas Opt2p was not.
The discovery of the OPT family of transporters in yeast (Lubkowitz et al., 1997) led to a search of the sequence database for possible orthologs in Arabidopsis. We report here the identification of nine putative OPT family members that exhibit 49% to 53% sequence similarity to the yeast OPTs. Expression of seven of these proteins in yeast confirmed the ability of some to mediate the uptake of tetra- and pentapeptides.
RESULTS
Arabidopsis OPT Orthologs Can Be Identified by Sequence
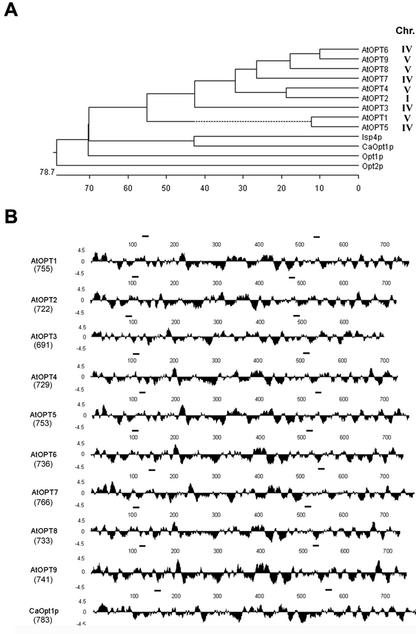
The GenBank database was searched using the TBLASTN 2.1.1 algorithm (Altschul et al., 1990, 1997) with the complete sequence of the C. albicans OPT, CaOpt1p (Lubkowitz et al., 1997). This analysis led to the identification of nine possible Arabidopsis OPT orthologs (Fig. 1) that exhibited 49% to 53% sequence similarity to CaOpt1p. The Arabidopsis OPTs formed a distinct subgroup when compared with the yeast OPT members and showed 61% to 85% sequence similarity when compared with each other (Fig. 1A). AtOPTs were positioned on the Arabidopsis genetic map based on the position of their corresponding BAC or P1 clones used for genome sequencing. AtOPT2 is located on chromosome 1, AtOPT3, 5, 6, and 7 are located on chromosome 4, whereas AtOPT1, 4, 8, and 9 are located on chromosome 5 (Fig. 1A).
Figure 1.
Comparison of OPTs. A, Dendogram showing a sequence comparison of the known members of the OPT family. Analysis was performed using the CLUSTAL method in MegAlign (DNASTAR, Madison, WI) using default parameters. Accession numbers are as follows: AtOPT1, AB026659 GI:9758213; AtOPT2, AAB60748 GI:2160185; AtOPT3, Z97341 GI:2244994; AtOPT4, AB008268 GI:9759417; AtOPT5, AL078465 GI:4938497; AtOPT6, AL035602.1 GI:4469024; AtOPT7, AF080119 GI:3600039; AtOPT8, BAB09728.1 GI:9759191; AtOPT9, AB015476 GI:9759190; Opt1p, Z49487; CaOpt1p, U60973; Isp4p, P40900; Opt2p, U25841. The mapped positions of each AtOPT are indicated. B, Hydrophilicity plots of AtOPT1–9 and CaOpt1p as predicted by Kyte and Doolittle (1982). The size (amino acids) of the each protein is shown below the name of the gene. Analysis was performed using Protean (DNASTAR) under default parameters. The bars over each sequence show the location of the two conserved motifs (NPG and KIPPR motifs) (i.e. NPG[P/A]F[N/T/S]XKEH[V/T/A][L/I/V][I/V]I[T/S/V][I/V/M] [F/M][A/S][N/S/A] and K[L/F][G/A][H/M/T]YMK[I/V/L][P/D/S]PR).
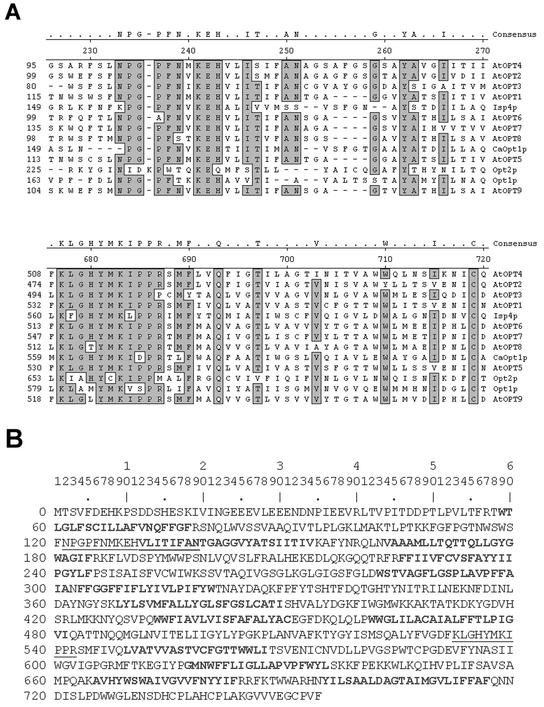
The hydrophilicity plots of the various AtOPTs were quite similar to that of CaOpt1p (Fig. 1B). The size of proteins predicted for each of the AtOPT varied from 696 amino acids (i.e. AtOPT3) to 766 amino acids (i.e. AtOPT7; Fig. 1B). With the exception of AtOPT3, which has shorter N and C terminus regions, all AtOPTs were similar in size to CaOpt1p (783 amino acids) and Opt1p (799 amino acids). Interestingly, sequence comparisons revealed two domains that were strongly conserved among all of the nine OPT family members. Both of these motifs (i.e. NPG motif, NPG[P/A]F[N/T/S]XKEH[V/T/A][L/I/V][I/V]I[T/S/V] [I/V/M][F/M][A/S][N/S/A] and KIPPR motif, K[L/F][G/A][H/M/T]YMK[I/V/L][P/D/S] PR; Fig. 2A) were found in regions of the protein predicted to be hydrophilic (Fig. 1B). As reported previously by Lubkowitz et al. (1998), none of the OPT family members showed any significant sequence similarity to the known ABC or PTR peptide transporters (data not shown). The putative transmembrane regions of the various AtOPTs were predicted either by hydrophilicity plots or the PRED-TMR algorithm (version 1.0, http://o2.db.uoa.gr/PRED-TMR), and the predicted transmembrane domains of AtOPT1 (as example) are shown in Figure 2B.
Figure 2.
Analysis of the OPT sequences. A, Two conserved motifs (NPG and KIPPR motifs) among the OPT members, including fungal OPTs, were determined based on the consensus of their sequences after analysis using the CLUSTAL method in the MegAlign (DNASTAR). Shaded area represents the consensus. B, The putative transmembrane domains of the AtOPTs were determined by the PRED-TMR algorithm. Predicted transmembrane regions of AtOPT1 are shown in bold and the underlined sequences represents the two conserved motifs found in all OPTs.
Tissue-Specific Expression of AtOPTs
Full-length cDNAs for the Arabidopsis OPT genes AtOPT1 to AtOPT7 were amplified by reverse transcriptase (RT)-PCR using gene-specific primers deduced from the DNA sequence in the database (see “Materials and Methods”). These cDNAs were completely sequenced to confirm that full-length cDNAs were obtained. The sequences of AtOPT8 and ATOPT9 only recently appeared in the database and, therefore, were not included in this study.
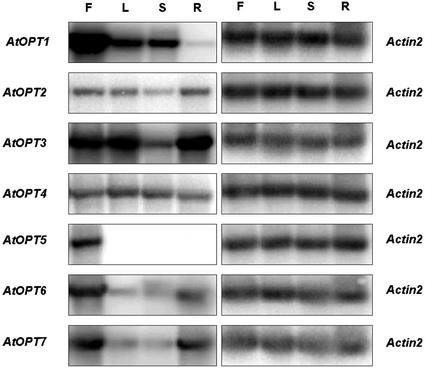
The tissue-specific expression pattern of each of the AtOPTs was determined by quantitative RT-PCR using gene-specific primers. This was deemed necessary because of the likelihood of cross-hybridization among the different family members. As an internal control, Actin2 mRNA was also amplified. The data shown in Figure 3 indicate that those OPTs that showed the greatest sequence similarity (Fig. 1A) also exhibited similar patterns of expression. For example, the levels of AtOPT2 and AtOPT4 mRNA were evenly expressed in all tissues. In contrast, the levels of AtOPT6 and AtOPT7 mRNA were highest in flower and root tissues but showed relatively low expression in leaf and stem. AtOPT1 was highly expressed in flower, and moderately expressed in leaf and stem. AtOPT5 showed the highest sequence similarity to AtOPT1 but its expression pattern was much more specific, being expressed predominantly in flowers. However, counts of 32P activity of AtOPT5 in the blot shown in Figure 3 confirmed very low expression of AtOPT5 in leaf and root (data not shown). AtOPT3 showed a unique pattern of expression being strongest in flower, leaf, and root. These data suggest that proteins with similar sequence, as indicated in the CLUSTAL analysis (Fig. 1A), may have similar physiological function, at least with regard to tissue specificity. The various expression patterns clearly suggest that the AtOPTs are likely playing a variety of physiological roles.
Figure 3.
Tissue-specific mRNA expression of AtOPT1–7. RT-PCR analysis was performed as described in “Materials and Methods” using gene-specific primers. The level of Actin2 mRNA (right panel) was measured as an internal control. F, Flower; L, leaf; S, stem; and R, root.
The AtOPTs Are Functional Peptide Transporters
To test the biochemical function of the various AtOPT proteins, their cDNAs were cloned behind the constitutive ADH (alcohol dehydrogenase) promoter in the pDB20 vector (Becker et al., 1991) and transformed into S. cerevisiae strain BY4730. Transport activity was measured by the ability of various Leu-containing peptides to support prototrophic growth of this strain, which is a Leu auxotroph. In this assay system, S. cerevisiae strain BY4730 will only grow if the cells can transport and utilize the peptides provided as the sole source of Leu. Oligopeptides are not hydrolyzed extracellularly and remain intact until transported into the cell cytoplasm (Perry et al., 1994; Steiner et al., 1994; Song et al., 1996; Lubkowitz et al., 1997, 1998; Hauser et al., 2000).
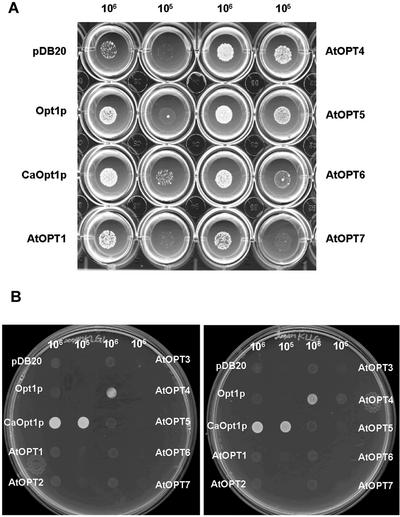
After transformation with each of the AtOPT plasmids, the S. cerevisiae BY4730 transformants SK101 to SK107 were tested for their ability to grow in the presence of the tetra-, and pentapeptides KLLG, KLGL, KLLLG, or YGGFL. As shown in Figure 4A and Table I, AtOPT1, 4, 5, 6, and 7 were able to support prototrophic growth in the presence of 200 μm KLLLG. In addition, AtOPT4 also mediated uptake of KLGL (at 200 μm) and KLLG (at 100 μm; Fig. 4B). The growth obtained was comparable with that shown by the positive control (i.e. BY4730 expressing CaOpt1p). These data show that AtOPTs (excluding AtOPT2 and AtOPT3) are functional tetra- and pentapeptide transporters.
Figure 4.
Peptide growth assays. S. cerevisiae BY4730 strain transformed with pDB20 (vector alone), or expressing CaOpt1p, Opt1p, or AtOPT1–7 were tested for their ability to use Leu-containing peptides (i.e. KLLG, KLGL, KLLLG) to fulfill the auxotrophic requirement for Leu. A, 200 μm KLLLG. The cell number added to each well is shown. B, 200 μm KLGL (left) and 100 μm KLLG (right).
Table I.
Peptide growth assay
| Yeast Transformantsa | Peptides
|
|||||
|---|---|---|---|---|---|---|
| −Leu | +Leu | KLLG | KLGL | KLLLG | YGGFL | |
| pDB20 | −b | +c | − | − | − | − |
| Opt1p | − | + | − | − | + | + |
| SK101 (AtOPT1) | − | + | − | − | + | − |
| SK102 (AtOPT2) | − | + | − | − | − | − |
| SK103 (AtOPT3) | − | + | − | − | − | − |
| SK104 (AtOPT4) | − | + | + | + | + | − |
| SK105 (AtOPT5) | − | + | − | − | + | − |
| SK106 (AtOPT6) | − | + | − | − | + | − |
| SK107 (AtOPT7) | − | + | − | − | + | − |
S. cerevisiae BY4730 (Met−, Leu−, Ura−).
−, No growth.
+, Growth.
Previously, Opt1p from S. cerevisiae was reported to transport Leu-enkephalin (YGGFL), Met-enkephalin (YGGFM; Hauser et al., 2000), and GSH (Bourbouloux et al., 2000). As shown in Table I, our experiments showed that BY4730 expressing Opt1p grew well in the presence of YGGFL. However, no growth was observed with YGGFL when cells were expressing any of the various AtOPTs (Table I). Uptake studies using [3H]GSH failed to reveal any uptake when yeast cells were expressing any of the AtOPTs, whereas, under similar conditions, Opt1p supported uptake of GSH (data not shown). Other studies using various di-and tripeptides (e.g. L-L, G-G-F) as sources of Leu showed no growth of yeast expressing the various AtOPTs (data not shown).
AtOPT4 Mediates the Uptake of KLG-[3H]L
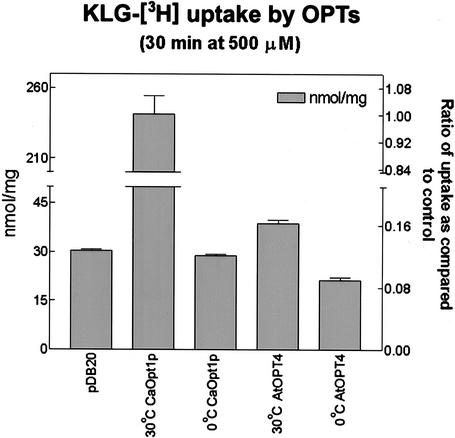
Few of the possible 160,000 (204) tetra- and 3,200,000 (205) pentapeptides containing the 20 naturally occurring amino acids are commercially available. Therefore, based on the results in Table I, we synthesized KLG-[3H]L and tested the ability of AtOPT4 to uptake this tetrapeptide when expressed in yeast. As shown in Figure 5, yeast cells expressing AtOPT4 accumulated KLG-[3H]L to a level significantly above the controls. However, this level of uptake was much less than that mediated by CaOpt1p from C. albicans. Transport of substrate at 0°C by both CaOpt1p and AtOPT4 was attenuated and similar to that of the negative control (empty vector, pDB20). These data indicate that AtOPT4 does mediate the uptake of KLG-[3H]L to a level significantly above background and confirm the results from the growth assays (Fig. 4; Table I). However, this peptide appears to be a poor substrate. Given the large number of possible tetrapeptides and the few available to test, it is would have been surprising to find that KLGL is an ideal substrate.
Figure 5.
Uptake of KLG-[3H]L by S. cerevisiae BY4730 transformants. Uptake of KLG-[3H]L after 30 min at 30°C or 0°C for cells transformed with pSK104 (AtOPT4), pCaOPT1 (CaOpt1p from C. albicans), and 30°C for cells containing the empty vector pDB20. Uptake is expressed in nanomoles per milligram dry weight on the left y axis, and fraction of uptake as compared with the positive control of CaOpt1p at 30°C on the right y axis. Data shown represent the average of three replicates ± sd.
DISCUSSION
Searching of the available Arabidopsis sequences using the CaOpt1p sequence identified nine possible OPT orthologs (AtOPT1–AtOPT9). Both the ability of AtOPT4 to take up KLG-[3H]L and the ability of AtOPT1, 4, 5, 6, and 7 to promote growth of a yeast strain defective in peptide transport demonstrates that these are bonafide OPT transporters. Sequence comparisons suggest that the Arabidopsis proteins comprise a distinct subfamily of OPT peptide transporters. All OPT family members (plant and fungus) appear to be integral membrane proteins with 12 to 14 predicted transmembrane domains. The availability of these AtOPT sequences allowed us to identify two highly conserved sequence motifs found in all OPT members. These motifs are found in regions predicted to be hydrophilic suggesting that they are probably critical to function. As is the case for the yeast OPTs, no significant sequence similarity was found between the AtOPTs and members of the ABC or PTR families of peptide transporters. Analysis of the expression of the various AtOPTs suggests that these proteins likely play distinct roles in the plant. Those proteins showing the highest sequence similarity also appeared to have comparable tissue-specific expression patterns, suggesting that they have related function.
Unfortunately, very few peptide substrates applicable to the yeast strain utilized are commercially available. Therefore, we synthesized the majority of the substrates used in this study, none of which appeared to be excellent substrates for transport. The nature of the physiological substrates for the various AtOPTs remains an important, unanswered question. Of all of the substrates tested (including di-and tripeptides), the AtOPTs were only able to mediate the uptake of selected tetra-and pentapeptides. Therefore, the data would suggest that their physiological substrates are likely small peptides, larger than a tripeptide. The presence of several OPT transporters in Arabidopsis suggests that these proteins are important and, therefore, small peptides may play an important physiological role in plants. Although the function of these transporters may be strictly nutritional, it is interesting to speculate that they could also mediate the transport of important regulatory molecules (e.g. hormone-peptide conjugates).
MATERIALS AND METHODS
Sequence Comparisons
The Arabidopsis orthologs were identified by comparison of the CaOpt1p sequence (Lubkowitz et al., 1997) to the data in the GenBank database using the TBLASTN 2.1.1 algorithm (Altschul et al., 1990, 1997). These orthologs were named AtOPT1 to AtOPT9. The map positions of these Arabidopsis orthologs were determined based on the genomic sequence information of the BAC clones or P1 clones that harbor the Arabidopsis orthologs (Fig. 1A).
All of the OPT sequences were compared using MegAlign (DNASTAR). This led to the identification of two conserved motifs found in all OPT proteins. A dendogram comparing the various OPT sequences was generated using the CLUSTAL method in MegAlign under the default parameters. Hydrophilicity plots for AtOPT1 through -7 were generated based on the Kyte and Doolittle (1982) method using Protean sequence analysis software (DNASTAR) under default parameters. The putative transmembrane domains were predicted by the PRED-TMR algorithm (version 1.0; Pasquier et al., 1999).
Analysis of Tissue-Specific Expression
Total RNA was isolated from 2- to 5-week-old whole Arabidopsis (Landsberg erecta) plants grown under 16-h light/8-h dark at 21°C. RNA was isolated using the Trizol reagent (Invitrogen, San Diego) as described by the manufacturer.
Gene-specific primers from the 5′-untranslated region of each AtOPT were designed by analyzing the DNA sequence of each gene using Primer3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The gene-specific primers used for full length cDNA cloning in this study are as follows: AtOPT1-RC3, 5′-GCCAGGACGGGAAAAAGGAGCTTGAAGAC-3′; AtOPT2-RC2, 5′-TGACGTCGTCTCCTTCACCAAATTCCA-3′; AtOPT3-RC2, 5′-GGCCAACAACGCAACTTTGTCTGGTACTTCA-3′; AtOPT4-RC1, 5′-TCACACACAAACTAAACCGGAATGG-3′; AtOPT5-RC2, 5′-AATGTCAGGTCATTAACACAGGTTGCT-3′; AtOPT6-RC1, 5′-GGCCCCAAAGAACGGAACACTCACTCT-3′; AtOPT7-RC1, 5′-GGCCGTCAGATTCACATCTCCCCAAAA-3′. First strand cDNA synthesis was performed using M-MLV reverse transcriptase (Promega, Madison, WI) using the AP adapter primer (Invitrogen), according to the manufacturer's protocol. The products of this reaction were then used in PCR reactions containing the gene-specific primers and the Abridged Universal Amplification Primer (Invitrogen) primer under the following conditions: 96°C for 5 min (1 cycle), 94°C for 15 s, 65°C for 30 s, 72°C for 2 min (36 cycles), and 72°C for 4 min (1 cycle). The PCR products obtained were cloned into the pCR2.1 TOPO cloning vector following the instructions provided in the TOPO TA cloning kit (Invitrogen). The complete DNA sequence (both strands) was determined for each of the cDNA clones and checked to confirm the presence of both the translational start and stop codons.
Because of the sequence similarity among the various AtOPTs, RT-PCR, using gene-specific primers was used to measure mRNA levels in various tissues. Total RNA from 3- to 5-week-old plant flower, leaf, bolting stem, and root was isolated using the Trizol reagent. The PCR conditions were identical to those described above. However, as an internal control, a 629-bp cDNA fragment of Actin2 was also amplified using specific primers (Actin2-for, 5′-GTTGGTGATGAAGCACAATCCAAG-3′, and Actin2-rev, 5′-CTGGAACAAGACTTCTGGGCATCT-3′). Amplification of the Actin2 and AtOPT cDNA was done in the same tube. After PCR, the products were electrophoresed in agarose gels and then blotted. AtOPT expression was visualized by Southern hybridization using a 32P-labeled probe made from the cDNA clone of the respective AtOPT gene. Likewise, the level of Actin2 expression was visualized by hybridizing to a labeled actin gene probe. The CPM/mm2 values for each hybridization were obtained with an instant imager (Packard Instrument Co., Meriden, CO) and calculated using the following equation: relative gene expression = cpm/mm2 of AtOPTs/the cpm/mm2 of Actin2.
Synthesis of Peptides
KLGL, KLLG, and KLLLG were prepared by conventional automated solid phase peptide synthesis on an synthesizer (model 433A, Applied Biosystems, Foster City, CA). Peptides were cleaved from the resin with trifluoroacetic acid and purified using a C18 reversed phase column (19 × 300 mm) to >99% homogeneity with a 5% to 20% linear gradient of acetonitrile in water over 60 min. The Mr and amino acid composition were verified using mass spectrometry.
Growth Assays
Full-length cDNAs of AtOPT1 to AtOPT7 were cloned into the pDB20 vector, under the control of the constitutive ADH promoter (Becker et al., 1991) using either the in vivo ligation method (Gietz et al., 1991) or in vitro cloning method.
AtOPT1 and AtOPT2 full-length cDNAs including the translation start and stop codons were amplified with the respective forward and reverse gene-specific primers including the intact NotI sites, and cotransformed with the BstXI digested pDB20 (URA3) vector into Saccharomyces cerevisiae BY4730 (MATα leu2Δ0 met15Δ0 ura3Δ0). Yeast transformants with in vivo ligated plasmids (pSK101 [AtOPT1] and pSK102 [AtOPT2]) were selected on 0.2% (w/v) casamino acid medium lacking uracil. Plasmids were isolated from cells that grew on this medium and re-transformed into Escherichia coli. NotI enzyme digestions of re-isolated plasmids from E. coli were used to confirm the cloning of AtOPT1 and AtOPT2 in pSK101 and pSK102, respectively. Plasmids pSK101 and pSK102 were transformed into S. cerevisiae BY4730 to create strains SK101 and SK102, respectively.
Full-length cDNAs of AtOPT3, 4, 5, 6, and AtOPT7 were cloned into the pDB20 vector using a normal in vitro cloning method. AtOPT3, AtOPT6, and AtOPT7 full-length cDNAs including the translation start and stop codons were amplified using the respective gene-specific primers with the NotI sites. The resulting PCR products were digested with NotI and ligated into the NotI site of the pDB20 vector. AtOPT4 and AtOPT5 full-length cDNAs were also PCR amplified using 5′-untranslated region gene-specific primers and the AUAP-NotI primer as 3′ primer and also cloned into the NotI site of the pDB20 vector. The orientation of each gene in the pDB20 vector was determined by digestion with various restriction enzymes. The resulting clones were transformed into S. cerevisiae BY4730 and transformants were selected on 0.2% (w/v) casamino acid medium lacking uracil. The resulting transformants were named SK103 to SK107, corresponding to AtOPT3 to AtOPT7, respectively.
Growth assays were performed as described previously (Hauser et al., 2000). SK101 to SK107 were grown overnight in a Pro liquid medium containing yeast nitrogen base without amino acids and ammonium sulfate, 2% (w/v) Glc, 0.1% (w/v) Pro, 228 μm Leu, and 191 μm Met (Hauser et al., 2000). The cells were harvested by centrifugation, washed twice with sterile distilled water, and resuspended in sterile distilled water to 2 × 107 cells/mL or 2 × 106 cells/mL. Five microliters of each suspension (1 × 106 cells or 1 × 105 cells, respectively) was applied as a small spot to a solid growth medium supplemented with a specific tetra- and pentapeptides (i.e. KLLG, KLGL, KLLLG, or YFGGL) instead of Leu and incubated at 30°C for 110 h. Growth was scored every 24 h as uniform colony formation compared with both negative (pDB20 vector only) and positive controls (CaOpt1p and Opt1p). The medium for the growth assay used Pro (0.1%) as a nitrogen source and was supplemented with either 100 or 200 μm of a specifically synthesized tetra-or pentapeptide (KLLG, KLGL, YFGGL, or KLLLG) as indicated. 191 μm Met was included in the medium to fulfill the auxotrophic requirement of the strain BY4730 for this amino acid. Leu-enkephalin (YGGFL) was purchased from Sigma (St Louis). Met-enkephalin (YGGFM) from Sigma was also purchased but found to contain a high level of Met amino acid contamination that prevented its use.
Radiolabeled Peptide Uptake Assays
Radioactive uptake assays with KLGL or GSH were initiated by combining equal volumes of prewarmed cells in 2% (w/v) Glc (30°C) and 2× uptake assay medium (2% [w/v] Glc, 40 mm sodium citrate/potassium buffer, pH 5.5, and 500 μm KLGL or 500 μm GSH [Sigma; New England Nuclear, Boston] at 0.5 μCi/mL of [3H]labeled substrate). Cells were incubated either with medium at 30°C or 0°C for 30 min with KLGL or 12 min with GSH. Medium containing GSH was constantly kept under nitrogen until it was mixed with cells, to prevent its oxidation. At the end of the incubation period, aliquots (90 μL) were removed, and placed on a membrane filter (HAWP, Millipore, Bedford, MA). The filter was immediately washed four times by vacuum filtration with 1 mL of ice water. Filters were counted by liquid scintillation spectrometry, and results were reported as nanomoles per milligram dry weight. KLGL uptake assays were done in triplicate, whereas GSH uptake was measured in quadruplicate.
ACKNOWLEDGMENT
We acknowledge the contribution of Dr. Chengdong Zhang for cloning of the AtOPT1 gene.
Footnotes
This work was supported by the U.S. Department of Agriculture (grant no. 99–35304–8194).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010332.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DM, Fikes JD, Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci USA. 1991;88:1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JM, Naider F. Controlling transport and metabolism. In: Tayler MD, Amidon GL, editors. Peptide-Based Drug Design. Washington, DC: American Chemical Society; 1995. pp. 369–384. [Google Scholar]
- Bourbouloux A, Shahi P, Chakladar A, Delrot S, Bachhawat AK. Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J Biol Chem. 2000;275:13259–13265. doi: 10.1074/jbc.275.18.13259. [DOI] [PubMed] [Google Scholar]
- Fejer D, Konay E. Occurrence of two additional peptides in the fine sap of corn. Naturwissenschaften. 1958;45:387–388. [Google Scholar]
- Gietz D, Andrew J, Woods R, Schiestl R. Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1991;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goore MY, Thompson JF. Gamma-glutamyl transpeptidase from kidney bean fruit: I. Purification and mechanism of action. Biochim Biophys Acta. 1967;132:15–26. doi: 10.1016/0005-2744(67)90187-8. [DOI] [PubMed] [Google Scholar]
- Gross DC. Molecular and genetic analysis of toxin production by pathovars of Pseudomonas syringae. Annu Rev Phytopathol. 1991;29:247–278. [Google Scholar]
- Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM. Enkephalins are transported by a novel eukaryotic peptide uptake system. J Biol Chem. 2000;275:3037–3041. doi: 10.1074/jbc.275.5.3037. [DOI] [PubMed] [Google Scholar]
- Hauser M, Narita V, Donhardt AM, Naider F, Becker JM. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Mol Membrane Biol. 2001;18:105–112. [PubMed] [Google Scholar]
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW. Peptide transport by germinating barley embryo: uptake of physiological di- and oligopeptides. Planta. 1978;138:211–216. doi: 10.1007/BF00386813. [DOI] [PubMed] [Google Scholar]
- Higgins CF, Payne JW. Transport and utilization of amino acids and peptides by higher plants. In: Payne JW, editor. Microorganisms and Nitrogen Sources. New York: John Wiley & Sons Ltd.; 1980. pp. 609–637. [Google Scholar]
- Higgins CF, Payne JW. Plant peptides. In: Boulder D, Parthier B, editors. Encyclopedia of Plant Physiology. 14A. New York: Springer Verlag; 1982. pp. 438–458. [Google Scholar]
- Khachidze OT. Peptides in the vegetative organs and berries of grape plants and their formation path. In: Oparin AI, editor. Vopr Biokhim Vinograda Vina. Moscow, Russia: Tr Uses Konf 2nd; 1975. pp. 18–122. [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lu Y-P, Li Z-S, Drozdowicz YM, Hörtensteiner S, Martinoia E, Rea PA. AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with AtMRP1. Plant Cell. 1998;10:267–282. doi: 10.1105/tpc.10.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y-P, Li Z-S, Rea PA. AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci USA. 1997;94:8243–8248. doi: 10.1073/pnas.94.15.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowitz MA, Barnes D, Breslav M, Burchfield A, Naider F, Becker JM. Schizosaccharomyces pombe isp4encodes a transporter representing a novel family of oligopeptide transporters. Mol Microbiol. 1998;28:729–741. doi: 10.1046/j.1365-2958.1998.00827.x. [DOI] [PubMed] [Google Scholar]
- Lubkowitz MA, Hauser L, Breslav M, Naider F, Becker JM. An oligopeptide transport gene from Candida albicans. Microbiology. 1997;143:387–396. doi: 10.1099/00221287-143-2-387. [DOI] [PubMed] [Google Scholar]
- Meredith D, Boyd CA. Structure and function of eukaryotic peptide transporters. Cell Mol Life Sci. 2000;57:754–758. doi: 10.1007/s000180050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS, Hamodrakas SJ. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 1999;12:381–385. doi: 10.1093/protein/12.5.381. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Skurray RA. The POT family of transport proteins. Trends Biochem Sci. 1994;10:404. doi: 10.1016/0968-0004(94)90087-6. [DOI] [PubMed] [Google Scholar]
- Payne JW, Smith MW. Peptide transport by microorganisms. Adv Micro Physiol. 1994;36:52–69. doi: 10.1016/s0065-2911(08)60176-9. [DOI] [PubMed] [Google Scholar]
- Perry JR, Basrai MA, Steiner HY, Naider F, Becker JM. Isolation and characterization of a Saccharomyces cerevisiaepeptide transport gene. Mol Cell Biol. 1994;14:103–115. doi: 10.1128/mcb.14.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury FB, Ross CW. Hormones and growth regulators: auxins and gibberellins. In: Salisbury FB, Ross CW, editors. Plant Physiology. Belmont, CA: Wadsworth Publishing Company; 1991. pp. 357–381. [Google Scholar]
- Sánchez-Fernández R, Ardiles-Díaz W, Van Montagu M, Inzé D, May MJ. Cloning and expression analyses of AtMRP4, a novel MRP-like gene from Arabidopsis thaliana. Mol Gen Genet. 1998;258:655–662. doi: 10.1007/s004380050779. [DOI] [PubMed] [Google Scholar]
- Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G. Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsisplants. Plant Physiol. 1997;114:927–935. doi: 10.1104/pp.114.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Steiner HY, Zang L, Naider F, Becker JM, Stacey G. Cloning of a second Arabidopsispeptide transport gene. Plant Physiol. 1996;110:171–178. doi: 10.1104/pp.110.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopanen T, Burston D, Matthews DM. Uptake of small peptides by the scutellum of germinating barley. FEBS Lett. 1977;79:4–7. doi: 10.1016/0014-5793(77)80337-2. [DOI] [PubMed] [Google Scholar]
- Steiner HY, Naider F, Becker JM. The PTR family: a new group of peptide transporters. Mol Microbiol. 1995;16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- Steiner HY, Song W, Zhang L, Naider F, Becker JM, Stacey G. An Arabidopsispeptide transporter is a member of a new class of membrane transport proteins. Plant Cell. 1994;6:1289–1299. doi: 10.1105/tpc.6.9.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD. Peptide phytotoxins from plant pathogenic fungi. In: Kleinkauf H, Dohren HV, editors. Biochemistry of Peptide Antibiotics. New York: Walter de Gruyter; 1990. pp. 179–203. [Google Scholar]
- Waterworth WM, West CE, Bray CM. The barley scutellar peptide transporter: biochemical characterization and localization to the plasma membrane. J Exp Bot. 2000;51:1201–1209. [PubMed] [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM. Cloning and functional characterization of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. Plant J. 1998;15:221–230. doi: 10.1046/j.1365-313x.1998.00199.x. [DOI] [PubMed] [Google Scholar]