Abstract
The susceptibilities to antimicrobial agents of and distributions of antiseptic resistance genes in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated between 1999 and 2004 in Japan were examined. The data of MRSA strains that are causative agents of impetigo and staphylococcal scalded skin syndrome (SSSS) were compared with those of MRSA strains isolated from patients with other diseases. The susceptibilities to antiseptic agents in MRSA isolates from patients with impetigo and SSSS were higher than those in MRSA isolates from patients with other diseases. The distribution of the qacA/B genes in MRSA strains isolated from patients with impetigo and SSSS (1.3%, 1/76) was remarkably lower than that in MRSA strains isolated from patients with other diseases (45.9%, 95/207). Epidemiologic typings of staphylococcal cassette chromosome mec (SCCmec) and pulsed-field gel electrophoresis (PFGE) showed that MRSA strains isolated from patients with impetigo and SSSS had type IV SCCmec (75/76), except for one strain, and 64.5% (49/76) of the strains had different PFGE types. In addition, the patterns of restriction digestion of all tested qacA/B plasmid in MRSA isolates having different PFGE types were identical. The results showed that a specific MRSA clone carrying qacA/B was not prevalent, but qacA/B was spread among health care-associated MRSA strains. Therefore, it was concluded that the lower distribution rate of qacA/B resulted in higher susceptibilities to cationic antiseptic agents in MRSA isolated from patients with impetigo and SSSS.
Methicillin-resistant Staphylococcus aureus (MRSA), which produces a penicillin-binding protein 2′ (PBP2′) with a low affinity to β-lactam antibiotics (11, 41, 48), is a major nosocomial pathogen throughout the world. The PBP2′ is encoded by the mecA gene that is located on a genetic element called the staphylococcal cassette chromosome (SCC) in Staphylococcus aureus (14, 17). SCCmec has been classified into five major types according to gene structure (15). Types I, II, and III of SCCmec are found in health care-associated MRSA (H-MRSA) strains, whereas types IV and V are found in community-associated MRSA (C-MRSA) strains (10, 15, 27). An increase in the number of C-MRSA strains carrying type IV SCCmec has became a matter of public concern.
Many antiseptic agents are used to prevent infections (25). Overuse of antiseptic agents has led to the emergence of MRSA with decreased antiseptic susceptibility, i.e., antiseptic-resistant MRSA (1, 2, 24, 32, 35). At least 12 antiseptic resistance genes (qacA to qacJ, smr, and norA) have been identified in Staphylococcus species (4, 8, 12, 13, 38, 43). Four antiseptic resistance genes, qacA, qacB, smr, and norA, are found mainly in clinical isolates of S. aureus (1, 2, 24, 35) and are associated with resistance to monovalent cationic agents such as quaternary ammonium compounds and ethidium bromide (1, 2, 23, 35, 38, 40). The qacA, qacB, and smr are mainly found on plasmids (22, 45), and norA is located on the S. aureus chromosome (30, 50). Antiseptic resistance in S. aureus is caused by proton motive force-dependent multidrug efflux (9, 29, 43). The qacA and qacB genes encode a 14-transmembrane-segment protein that belongs to the major facilitator superfamily (40, 43). Although qacA also confers more resistance to divalent cationic agents than qacB, the sequence of qacB is identical to that of qacA except for only seven or nine bases (2, 38). Therefore, it is difficult to distinguish qacA and qacB by simple PCR, and qacA and qacB are considered to be the same. The smr gene, which is identical to qacC, qacD, and ebr, encodes a small protein that belongs to a small multidrug resistance family (20, 22, 40). Therefore, the plasmid-borne antiseptic resistance genes are classified structurally into two families, qacA/B and smr (40). The chromosomal antiseptic resistance gene norA confers low-level resistance to hydrophilic fluoroquinolones such as norfloxacin and levofloxacin as well as to antiseptic agents (33, 50). The resistance of norA seemed to be due to the mutation(s) in the 5′-untranslated region that led to increases in norA transcription (7, 16, 33). At least seven mutations conferring antiseptic resistance have been identified (33). However, compared to qacA/B, the norA mutation(s) is considered to have a very low contribution to the resistance of cationic antiseptic agents. The qacA/B gene has been reported to be associated with high-level resistance to antiseptic agents and to be widely prevalent among MRSA isolates found in Europe and Asia (2, 24, 32, 35). Therefore, the qacA/B gene(s) has become a major antiseptic resistance gene in MRSA.
Impetigo and staphylococcal scalded skin syndrome (SSSS), which are diseases primarily of young children and neonates, are blistering skin diseases that are caused by exfoliative toxins (ETs) produced by S. aureus (19, 39). Serologically, ETs involved in human diseases consist of two types: ETA and ETB proteins (19, 39). The eta gene, encoding ETA, is located on a chromosome, whereas the etb gene, encoding ETB, is found on a plasmid (39). The etb gene might be transferred horizontally by transduction (42). Recently, an increasing number of refractory patients with impetigo and SSSS caused by ET-producing MRSA has became a serious problem (49). Almost all MRSA strains (i.e., C-MRSA) isolated from outpatients with impetigo and SSSS were reported to carry type IV SCCmec and were susceptible to various antibiotics except β-lactam antibiotics (3, 31, 36). However, the decreased susceptibility of C-MRSA to macrolides and aminoglycosides has also been reported (49). The spread of antiseptic resistance genes into MRSA leads to decreased susceptibility to antiseptic agents (35). The prevalence of antiseptic resistance genes, such as qacA/B, in C-MRSA is a cause for public health concern.
The aim of this study was to understand the susceptibility of MRSA strains isolated from patients with impetigo and SSSS to antimicrobial agents, including antiseptics. A secondary aim of our study was to find potential effective antiseptic agents to MRSA that cause impetigo. To this end, we determined the susceptibilities of antimicrobial agents including antiseptics and distribution of antiseptic resistance genes in MRSA isolated between 1999 and 2004 and compared the data of MRSA strains isolated from patients with impetigo and SSSS with those of MRSA strains isolated from patients other diseases. In addition, we performed molecular epidemiological typings of SCCmec and pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
Bacterial strains.
A total of 283 isolates of MRSA were collected at Kori Hospital (250 isolates) and Rakusai Newtown Hospital (14 isolates), both associated with Kansai Medical University, and Hyogo Prefecture Tsukaguchi Hospital (19 isolates) in Japan between July 1999 and December 2004. Seventy-six strains were isolated from outpatients with impetigo (66 strains) and SSSS (10 strains), and 207 strains were isolated from the following clinical departments: otolaryngology, 70 strains; dermatology, 49 strains; pediatrics, 36 strains; urology, 21 strains; surgery, 16 strains; internal medicine, 13 strains; and gynecology, 2 strains. All strains were isolated from different patients. S. aureus N315 (18) was used for a typical strain of MRSA, and JCM2874 (ATCC 29213) was used as a reference strain and for quality control during susceptibility testing. The following strains were used as SCCmec type strains: S. aureus NCTC10442 (type I), N315 (type II), 85/2082 (type III), and JCSC4744 (type IV) (15).
Bacterial identification.
All clinical isolates were identified as S. aureus by a positive Gram stain, the utilization of mannitol salt agar (Oxoid, Hampshire, England), and a production of coagulase (PS LATEX; Eiken Chemical, Tokyo, Japan). MRSA strains were identified by proliferation on Muller-Hinton agar (Oxoid) including 6 μg/ml of oxacillin and 4% of NaCl and the detection of the mecA gene by PCR (47). The MRSA strains isolated from patients with impetigo and SSSS were judged by the production of ET and detection of an ET gene(s), in addition to the identification of MRSA (39).
Antimicrobial susceptibility testing.
MICs were determined by the agar doubling dilution method according to the CLSI (formerly the National Committee for Clinical Laboratory Standards) guidelines (28). Cefmetazole, clarithromycin, levofloxacin, and arbekacin were kindly provided by their manufacturers. Vancomycin, gentamicin, minocycline, benzalkonium chloride, benzethonium chloride, chlorhexidine digluconate, cetyltrimethylammonium bromide, and ethidium bromide were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan), and oxacillin was from Sigma-Aldrich (Tokyo, Japan), and oxacillin was from Sigma-Aldrich (Tokyo, Japan). The breakpoints of these antimicrobial agents were determined by the interpretation criteria of the CLSI (28).
PCR amplification.
PCR assays for detection of various genes were performed using the modified colony direct method (35). Briefly, the point of a toothpick was gently placed into 100 μl of H2O. A small sample of cells was suspended, and 1 μl of the cell suspension was added directly into 20 μl of the PCR mixture, containing primers and 10 μl of PCR master mix (Promega). Searches for mecA, eta, etb, qacA/B, and smr were performed with the following sets of primers: for mecA, 5′-GTGGAAGTTAGATTGGGATCATAGC-3′ and 5′-GTCAACGATTGTGACACGATAGC-3′ (product size, 544 bp) (GenBank accession no. X52593); for eta, 5′-ATATCAACGTGAGGGCTCTAGTAC-3′ and 5′-ATGCAGTCAGCTTCTTACTGCTA (product size, 1,155 bp) (GenBank accession no. AP001553); for etb, 5′-CACACATTACGGATAATGCAAG-3′ and 5′-TCAACCGAATAGAGTGAACTTATCT-3′ (product size, 604 bp) (GenBank accession no. AP003088); for qacA/B, 5′-GCAGAAAGTGCAGAGTTCG-3′ and 5′-CCAGTCCAATCATGCCTG-3′ (product size, 361 bp); and for smr, 5′-GCCATAAGTACTGAAGTTATTGGA-3′ and 5′-GACTACGGTTGTTAAGACTAAACCT-3′ (product size, 195 bp). PCR was performed in the following cycles: for mecA, 25 cycles (30 s of denaturation at 95°C, 30 s of annealing at 58°C, and 30 s of extension at 72°C); and for eta, etb, qacA/B, and smr, 25 cycles (30 s of denaturation at 95°C, 30 s of annealing at 52°C, and 1.5 min of extension at 72°C). PCR products were analyzed by agarose gel electrophoresis. All results were confirmed by at least two independent experiments.
SCCmec and PFGE typings.
SCCmec typing was performed by the multiplex PCR method described by Ito et al. and by Oliveira and de Lencastre (15, 37). PFGE of SmaI-digested chromosomal DNA was performed as described previously (26, 34, 35, 46). The DNA patterns obtained by PFGE were analyzed with BioNumerics software (Applied Maths, Saint-Martens-Latem, Belgium) using the Dice coefficient (34, 35). S. aureus N315 was used as a DNA reference standard because the genome of N315 has been determined (18).
Statistical analysis.
Differences in distribution of qacA/B in MRSA between patients with impetigo and SSSS and those with other diseases were tested by the χ2 test, with P values of <0.05 considered to be statistically significant.
RESULTS
Antimicrobial susceptibility.
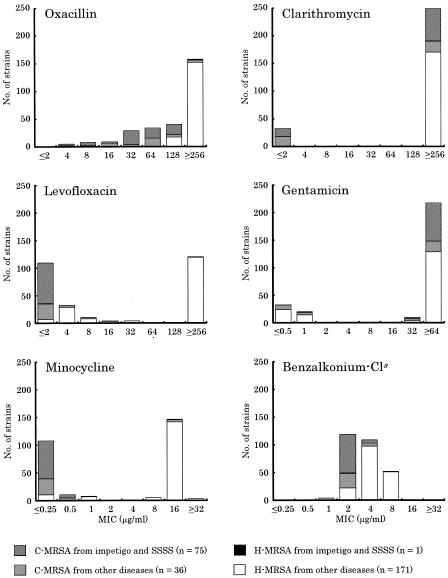
The MIC50s and MIC90s of the antimicrobial agents for the MRSA strains isolated from patients with impetigo and SSSS were compared with those of MRSA strains isolated from patients with other diseases, excluding impetigo and SSSS (Table 1). All MRSA strains isolated from patients with impetigo and SSSS, except for one stain, were susceptible to levofloxacin and minocycline, although 17 and 26% of MRSA strains isolated from patients with other diseases were susceptible to levofloxacin and minocycline, respectively. However, the rate of gentamicin-resistant MRSA strains isolated from impetigo and SSSS patients (95%) was higher than that of MRSA strains isolated from patients with other diseases. The rates of clarithromycin-resistant MRSA strains isolated from patients with impetigo and SSSS and from patients with other diseases were 80 and 91%, respectively. The MRSA strains isolated from impetigo and SSSS patients were more susceptible to the antiseptic agents tested than MRSA strains isolated from patients with other diseases (Table 1).
TABLE 1.
Antimicrobial susceptibility of MRSA strains in this study
| Antimicrobial agenta | MIC (μg/ml) for strains isolated from patients:
|
|||||
|---|---|---|---|---|---|---|
| With impetigo and SSSS
|
Without impetigo and SSSS
|
|||||
| Range | 50% | 90% | Range | 50% | 90% | |
| Oxacillin | 4-≥256 | 64 | 128 | 4-≥256 | ≥256 | ≥256 |
| Cefmetazole | 4-≥64 | 32 | ≥64 | 4-≥64 | ≥64 | ≥64 |
| Clarithromycin | ≤0.125-≥256 | ≥256 | ≥256 | ≤0.125-≥256 | ≥256 | ≥256 |
| Levofloxacin | ≤0.125-≥256 | ≤0.125 | ≤0.125 | ≤0.125-≥256 | ≥256 | ≥256 |
| Gentamicin | 0.5-≥64 | ≥64 | ≥64 | 0.25-≥64 | ≥64 | ≥64 |
| Arbekacin | 0.5-8 | 2 | 4 | 0.25-≥32 | 1 | 2 |
| Vancomycin | ≤0.5-2 | 1 | 1 | ≤0.5-2 | 1 | 1 |
| Minocycline | ≤0.125-16 | ≤0.125 | 0.25 | ≤0.125-32 | 16 | 16 |
| Benzalkonium-Cl | 2-4 | 2 | 2 | 1-8 | 4 | 8 |
| Benzethonium-Cl | 1-8 | 1 | 2 | 0.5-16 | 4 | 8 |
| Chlorhexidine-Glu | 1-4 | 2 | 2 | 1-8 | 4 | 4 |
| Cetyltrimethylammonium-Br | ≤0.5-16 | 4 | 8 | 2-16 | 8 | 8 |
| Ethidium-Br | 1-32 | 8 | 8 | 1-≥256 | 8 | ≥256 |
Benzalkonium-Cl, benzalkonium chloride; benzethonium-Cl, benzethonium chloride; chlorhexidine-Glu, chlorhexidine digluconate; cetyltrimethylammonium-Br, cetyltrimethylammonium bromide; ethidium-Br, ethidium bromide.
Distributions of qacA/B and smr.
Although the antiseptic resistance gene qacA/B was detected in 45.9% (95/207) of MRSA strains isolated from patients with diseases other than impetigo and SSSS, only one MRSA (1.3%) strain isolated from a patient with impetigo and SSSS carried qacA/B (Table 2). The smr gene was detected in only four strains of MRSA isolated from patients with impetigo but not found among MRSA isolates from patients with diseases other than impetigo and SSSS. The prevalence of qacA/B in MRSA isolates from patients with impetigo and SSSS was significantly lower than that in MRSA isolates from patients with other diseases (P < 0.0001).
TABLE 2.
Distributions of qacA/B and smr
| Gene(s) or parameter | No. (%) of strains isolated from patients
|
||
|---|---|---|---|
| With impetigo and SSSS (n = 76) | Without impetigo and SSSS (n = 207) | Total (n = 283) | |
| qacA/B | 1 (1.3) | 95 (45.9) | 96 (33.9) |
| smr | 4 (5.3) | 0 | 4 (1.4) |
| NDa | 71 (93.4) | 112 (54.1) | 183 (64.7) |
ND, not detected.
Molecular and epidemiological analysis.
To study the genotypic characteristics and genetic relatedness of MRSA isolates, 283 MRSA strains were analyzed by SCCmec and PFGE typings. The SCCmec type of MRSA strains isolated from patients with impetigo and SSSS was type IV, with the exception of one strain. In 207 MRSA strains isolated from patients with other diseases, types I, II, III, and IV were detected in 5, 164, 1, and 37 strains, respectively. The numbers of SCCmec types of MRSA strains carrying qacA/B were 1 of type I, 91 of type II, 0 of type III, and 4 of type IV. One strain carrying both qacA/B and etb had type IV SCCmec. The antimicrobial susceptibilities of H-MRSA and C-MRSA were compared (Fig. 1). Although the susceptibilities of H-MRSA to oxacillin, levofloxacin, and minocycline were different from those of C-MRSA, no distinction in the resistance profiles between C-MRSA strains isolated from patients with impetigo and SSSS and C-MRSA isolated from patients with other diseases was present.
FIG. 1.
Comparison of antimicrobial susceptibilities for H-MRSA and C-MRSA strains used in this study. a, benzalkonium chloride.
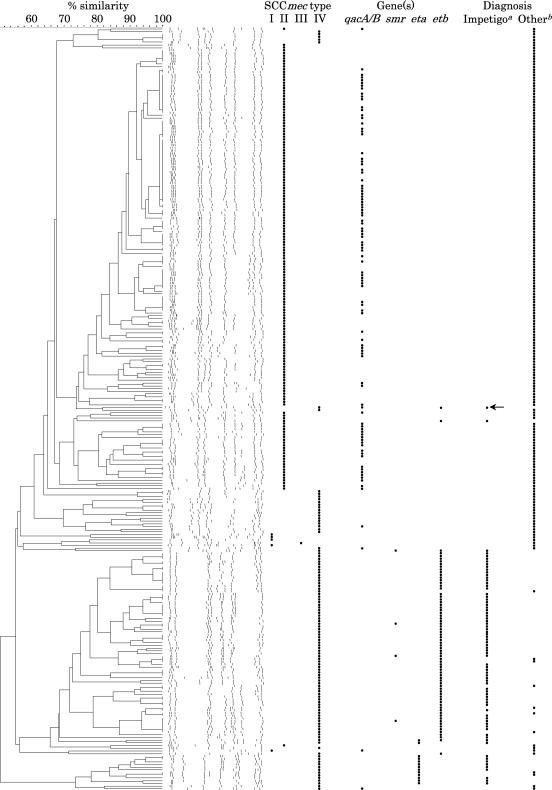
On the other hand, the PFGE type of MRSA carrying SCCmec type IV differed significantly from that of MRSA carrying SCCmec type II (Fig. 2). Seventy-six MRSA strains which had SCCmec type IV, except one strain, isolated from patients with impetigo and SSSS were classified into 49 PFGE types, and 96 MRSA isolates carrying qacA/B were classified into 44 PFGE types. When the qacA/B plasmids carrying six MRSA strains with different PFGE types were tested, the patterns of the restriction digestion of all of the qacA/B plasmids were identical (data not shown). These genetic findings suggest that a specific MRSA clone carrying qacA/B was not prevalent, but qacA/B was horizontally transferred among various MRSA clones.
FIG. 2.
Comparison of PFGE patterns, SCCmec types, and antiseptic resistance genes of MRSA isolated from patients with impetigo and SSSS and those of MRSA strains isolated from patients with other diseases. The arrow indicates the strain carrying both qacA/B and etb isolated from patients with SSSS. a, strain isolated from patients with impetigo and SSSS; b, strain isolated from patients without impetigo and SSSS.
DISCUSSION
In recent studies, the incidence rate of C-MRSA among patients with skin infections, including impetigo and SSSS, has been increasing, and most C-MRSA strains carried type IV SCCmec. Unlike H-MRSA, C-MRSA is frequently susceptible to non-β-lactam antibiotics, such as aminoglycosides, tetracyclines, and fluoroquinolones (3, 31). In this study, 99% of the MRSA strains isolated from patients with impetigo and SSSS had type IV SCCmec. However, most C-MRSA strains isolated from patients with impetigo and SSSS were resistant to clarithromycin and gentamicin, although those strains were susceptible to levofloxacin and minocycline. The results suggest that the C-MRSA strains isolated from patients with impetigo and SSSS have evolved not only β-lactam resistance but also multidrug resistance. Recently, skin and soft tissue infections and severe necrotizing pneumonia caused by C-MRSA strains carrying the Panton-Valentine leukocidin gene (pvl) have become a serious problem (6, 21, 44, 51). Although screening for pvl was carried out by PCR, no pvl was detected among the C-MRSA strains used in this study (data not shown).
Cationic antiseptic agents such as quaternary ammonium compounds and chlorhexidine digluconate and iodine compounds, such as povidon iodine, are commonly used for the disinfection of MRSA on skin and hands (5). The qacA/B gene that confers resistance to cationic antiseptic agents was detected in 46% of MRSA isolates cultured from patients with diseases other than impetigo and SSSS. This frequency (44%) of qacA/B was very similar to that previously reported in Japan (35). Almost all MRSA isolates with qacA/B had SCCmec type II (91/96). This seems to be due to the significant exposure of H-MRSA isolates to antiseptics in hospital environments. Therefore, our data predict that the qacA/B plasmid has been further dispersed among various H-MRSA by horizontal transfer.
SCCmec type IV MRSA strains have been frequently isolated from patients with skin infections. In SCCmec type IV MRSA isolates, qacA/B was detected in 8.1% (3/37) of strains isolated from patients with other diseases. These data showed that qacA/B was able to maintain SCCmec type IV MRSA, whereas the genes encoding exfoliative toxin are the causative genes of impetigo and SSSS and the etb gene is located on a plasmid (19, 39). Although the ET genes were found in nine C-MRSA strains (SCCmec type IV) isolated from patients without impetigo and SSSS, no qacA/B was detected in the nine strains. Only one strain carrying both qacA/B and etb was found in patients with SSSS. This SCCmec type IV strain had a PFGE pattern which resembled that of the SCCmec type II MRSA group (Fig. 2) and was resistant to levofloxacin, minocycline, and gentamicin. Therefore, the C-MRSA carrying both qacA/B and etb seemed to be a rare MRSA clone among the MRSA carrying the ET genes. When the plasmid was purified and analyzed by using restriction enzymes, the strain with qacA/B and etb carried a single plasmid that had the same restriction pattern as the qacA/B plasmid (data not shown). Therefore, the qacA/B plasmid seemed to be maintained in the strain with etb by integration of etb into the chromosome. These data suggest that the qacA/B plasmid may be incompatible with the etb plasmid. Further studies of the plasmids encoding qacA/B and etb are necessary to demonstrate the incompatibility between the qacA/B and etb plasmids.
On the contrary, the smr gene was found in 5% (4/76) of the MRSA strains isolated from patients with impetigo and SSSS but not detected in MRSA strains isolated from patients with other diseases. This frequency of this gene in patients with impetigo and SSSS (3.4%) was similar to that of smr previously reported in Japan (35). The smr plasmid was also detected in MRSA strains carrying the etb plasmid from patients with impetigo. It might be possible to maintain smr plasmid in the cells carrying the etb plasmid, because the smr plasmid is compatible with the qacA/B plasmid (24, 35).
In summary, the qacA/B gene, which is a dominant plasmid-borne antiseptic resistance gene in MRSA, was not prevalent in C-MRSA strains isolated from patients with impetigo and SSSS, although qacA/B was likely transferred horizontally among various H-MRSA clones. Consequently, our results suggest that antiseptic agents have a higher potential to prevent the infection of impetigo and SSSS caused by C-MRSA.
Acknowledgments
We thank T. Ito and K. Hiramatsu for providing us the SCCmec type strains of MRSA. We also thank K. Narui, K. Sato, K. Goto, R. Tamura, and T. Watanabe for their technical assistance.
This work was supported by High-Tech Research Centre Project for Private Universities provided by the Ministry of Education, Culture, Sports, Science and Technology and by the Matching Fund Subsidy for Private Schools of Japan.
REFERENCES
- 1.Alam, M. M., M. Ishino, and N. Kobayashi. 2003. Analysis of genomic diversity and evolution of the low-level antiseptic resistance gene smr in Staphylococcus aureus. Microb. Drug Resist. 9(Suppl. 1):S1-S7. [DOI] [PubMed] [Google Scholar]
- 2.Alam, M. M., N. Kobayashi, N. Uehara, and N. Watanabe. 2003. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb. Drug Resist. 9:109-121. [DOI] [PubMed] [Google Scholar]
- 3.Almer, L. S., V. D. Shortridge, A. M. Nilius, J. M. Beyer, N. B. Soni, M. H. Bui, G. G. Stone, and R. K. Flamm. 2002. Antimicrobial susceptibility and molecular characterization of community-acquired methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 43:225-232. [DOI] [PubMed] [Google Scholar]
- 4.Bjorland, J., T. Steinum, M. Sunde, S. Waage, and E. Heir. 2003. Novel plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob. Agents Chemother. 47:3046-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce, J. M., D. Pittet, et al. 2002. Guideline for hand hygiene in health-care settings. Recommendations of the Healthcare Infection Control Practices Advisory Committee and the HICPAC/SHEA/APIC/IDSA Hand Hygiene Task Force. Morb. Mortal. Wkly. Rep. Recomm. Rep. 51:1-45, CE1-CE4. [Google Scholar]
- 6.Diep, B. A., G. F. Sensabaugh, N. S. Somboona, H. A. Carleton, and F. Perdreau-Remington. 2004. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J. Clin. Microbiol. 42:2080-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fournier, B., Q. C. Truong-Bolduc, X. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grinius, L., G. Dreguniene, E. B. Goldberg, C.-H. Liao, and S. J. Projan. 1992. A staphylococcal multidrug resistance gene product is a member of a new protein family. Plasmid 27:119-129. [DOI] [PubMed] [Google Scholar]
- 9.Grinius, L. L., and E. B. Goldberg. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269:29998-30004. [PubMed] [Google Scholar]
- 10.Groom, A. V., D. H. Wolsey, T. S. Naimi, K. Smith, S. Johnson, D. Boxrud, K. A. Moore, and J. E. Cheek. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 11.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heir, E., G. Sundheim, and A. L. Holck. 1999. The qacG gene on plasmid pST94 confers resistance to quaternary ammonium compounds in staphylococci isolated from the food industry. J. Appl. Microbiol. 86:378-388. [DOI] [PubMed] [Google Scholar]
- 13.Heir, E., G. Sundheim, and A. L. Holck. 1998. The Staphylococcus qacH gene product: a new member of the SMR family encoding multidrug resistance. FEMS Microbiol. Lett. 163:49-56. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., and K. Hiramatsu. 1998. Acquisition of methicillin resistance and progression of multiantibiotic resistance in methicillin-resistant Staphylococcus aureus. Yonsei Med. J. 39:526-533. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaatz, G. W., S. M. Seo, L. O'Brien, M. Wahiduzzaman, and T. J. Foster. 2000. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1404-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Ladhani, S., C. L. Joannou, D. P. Lochrie, R. W. Evans, and S. M. Poston. 1999. Clinical, microbial, and biochemical aspects of the exfoliative toxins causing staphylococcal scalded-skin syndrome. Clin. Microbiol. Rev. 12:224-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leelaporn, A., N. Firth, I. T. Paulsen, A. Hettiaratchi, and R. A. Skurray. 1995. Multidrug resistance plasmid pSK108 from coagulase-negative staphylococci; relationships to Staphylococcus aureus qacC plasmids. Plasmid 34:62-67. [DOI] [PubMed] [Google Scholar]
- 21.Liassine, N., R. Auckenthaler, M. C. Descombes, M. Bes, F. Vandenesch, and J. Etienne. 2004. Community-acquired methicillin-resistant Staphylococcus aureus isolated in Switzerland contains the Panton-Valentine leukocidin or exfoliative toxin genes. J. Clin. Microbiol. 42:825-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Littlejohn, T. G., D. DiBerardino, L. J. Messerotti, S. J. Spiers, and R. A. Skurray. 1991. Structure and evolution of a family of genes encoding antiseptic and disinfectant resistance in Sthphylococcus aureus. Gene 101:59-66. [DOI] [PubMed] [Google Scholar]
- 23.Littlejohn, T. G., I. T. Paulsen, M. T. Gillespie, J. M. Tennent, M. Midgley, I. G. Jones, A. S. Purewal, and R. A. Skurray. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 74:259-265. [DOI] [PubMed] [Google Scholar]
- 24.Mayer, S., M. Boos, A. Beyer, A. C. Fluit, and F. J. Schmitz. 2001. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 47:896-897. [DOI] [PubMed] [Google Scholar]
- 25.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 28.NCCLS. 2001. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed., approved standard M7-M4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Neyfakh, A. A., C. M. Borsch, and G. W. Kaatz. 1993. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 37:128-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng, E. Y. W., M. Trucksis, and D. C. Hooper. 1994. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob. Agents Chemother. 38:1345-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishijima, S., and I. Kurokawa. 2002. Antimicrobial resistance of Staphylococcus aureus isolated from skin infections. Int. J. Antimicrob. Agents 19:241-243. [DOI] [PubMed] [Google Scholar]
- 32.Noguchi, N., M. Hase, M. Kitta, M. Sasatsu, K. Deguchi, and M. Kono. 1999. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 172:247-253. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi, N., H. Okada, K. Narui, and M. Sasatsu. 2004. Comparison of the nucleotide sequence and expression of norA genes and microbial susceptibility in 21 strains of Staphylococcus aureus. Microb. Drug Resist. 10:197-203. [DOI] [PubMed] [Google Scholar]
- 34.Noguchi, N., T. Okihara, Y. Namiki, Y. Kumaki, Y. Yamanaka, M. Koyama, K. Wakasugi, and M. Sasatsu. 2005. Susceptibility and resistance genes to fluoroquinolones in methicillin-resistant Staphylococcus aureus isolated in 2002. Int. J. Antimicrob. Agents 25:374-379. [DOI] [PubMed] [Google Scholar]
- 35.Noguchi, N., J. Suwa, K. Narui, M. Sasatsu, T. Ito, K. Hiramatsu, and J. H. Song. 2005. Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J. Med. Microbiol. 54:557-565. [DOI] [PubMed] [Google Scholar]
- 36.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plano, L. R. 2004. Staphylococcus aureus exfoliative toxins: how they cause disease. J. Investig. Dermatol. 122:1070-1077. [DOI] [PubMed] [Google Scholar]
- 40.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, P. E., and D. F. Brown. 1985. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett. 192:28-32. [DOI] [PubMed] [Google Scholar]
- 42.Rogolsky, M., B. W. Beall, and B. B. Wiley. 1986. Transfer of the plasmid for exfoliative toxin B synthesis in mixed cultures on nitrocellulose membranes. Infect. Immun. 54:265-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4:2051-2062. [DOI] [PubMed] [Google Scholar]
- 44.Takizawa, Y., I. Taneike, S. Nakagawa, T. Oishi, Y. Nitahara, N. Iwakura, K. Ozaki, M. Takano, T. Nakayama, and T. Yamamoto. 2005. A Panton-Valentine leucocidin (PVL)-positive community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J. Clin. Microbiol. 43:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tennent, J. M., B. R. Lyon, M. Midgley, I. G. Jones, A. S. Purewal, and R. A. Skurray. 1989. Physical and biochemical characterization of the qacA gene encoding antiseptic and disinfectant resistance in Staphylococcus aureus. J Gen. Microbiol. 135:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ubukata, K., S. Nakagami, A. Nitta, A. Yamane, S. Kawakami, M. Sugiura, and M. Konno. 1992. Rapid detection of the mecA gene in methicillin-resistant staphylococci by enzymatic detection of polymerase chain reaction products. J. Clin. Microbiol. 30:1728-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamaguchi, T., Y. Yokota, J. Terajima, T. Hayashi, M. Aepfelbacher, M. Ohara, H. Komatsuzawa, H. Watanabe, and M. Sugai. 2002. Clonal association of Staphylococcus aureus causing bullous impetigo and the emergence of new methicillin-resistant clonal groups in Kansai district in Japan. J. Infect. Dis. 185:1511-1516. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]