Abstract
We developed an assay for rapid detection of rifampin resistance in Mycobacterium tuberculosis based on Pyrosequencing technology, involving a technique for real-time sequencing. A 180-bp region of the rpoB gene was amplified in clinical isolates of both rifampin-resistant and -susceptible M. tuberculosis. The PCR products were subjected to Pyrosequencing analysis using four different sequencing primers in four overlapping reactions. These four sequencing reactions covered the 81-bp region where >96% of the mutations associated with rifampin resistance are located. The results were compared to those obtained with two other molecular methods, the line probe assay and cycle sequencing, and the phenotypic BACTEC method. The genotypic determination methods all detected the mutations that previously have been correlated with rifampin resistance. In addition, Pyrosequencing analysis and the two other molecular methods found additional mutations within the rpoB gene in phenotypically susceptible strains. We found that Pyrosequencing technology, in particular, offers high accuracy, short turnaround time, and a potentially high throughput in detection of rifampin resistance in M. tuberculosis.
Mycobacterium tuberculosis lays a heavy burden in the global population, with an estimate of 8.3 million new cases and 1.8 million deaths in the year 2000 (5). The emergence of drug-resistant and multidrug-resistant (MDR) tuberculosis (TB), i.e., resistance to at least rifampin (RIF) and isoniazid makes the situation even worse. In the community, MDR TB originates from two possible sources, either from the spread of already resistant strains or by the selection of resistant bacteria due to suboptimal treatment (4, 15). The treatment of resistant TB is both difficult and expensive. It includes the prolonged use of expensive antibiotics, which are less effective and more prone to give adverse side effects than standard TB chemotherapy (6, 17). To cure the patient and eliminate the risk of spread within a population, a prompt detection of drug resistance in the infectious agent is crucial. Conventional drug susceptibility tests are either time-consuming, as the proportion method on solid media (Löwenstein-Jensen or Middlebrook 7H10 agar), or expensive, such as commercial broth-based culturing systems.
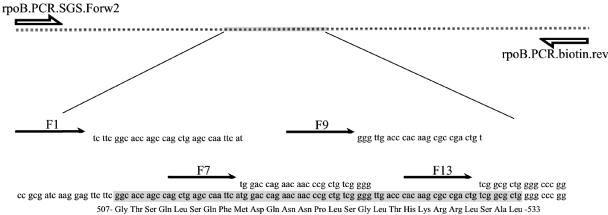
Rifampin is of one of the most important drugs for the treatment of tuberculosis. Mutations correlated to RIF resistance have been detected in the rpoB gene, encoding the β-subunit of the DNA-dependent RNA polymerase (12). Among the RIF-resistant clinical isolates of M. tuberculosis, approximately 96% have mutations within an 81-nucleotide rifampin resistance-determining region (RRDR) comprising codons 507 to 533 (Fig. 1) (20, 25).
FIG. 1.
Distribution of PCR primers and the four sequencing primers for analysis of the RRDR in the rpoB gene. A fraction of the gene was first amplified in a PCR, followed by Pyrosequencing analysis with four overlapping reactions.
Pyrosequencing technology is a rapid sequencing technology which has mostly been used on short read sequences in single-nucleotide polymorphism analyses of human DNA (8). The technology is also increasingly assessed for species identification or typing of various microbes (21), including mycobacteria (28). Quantitative Pyrosequencing analysis of linezolid resistance in enterococci, as well as macrolide resistance in different bacterial species, shows the potential for detection in samples of mixed-template populations (9, 24). Recently, Pyrosequencing technology has been used to detect resistant tuberculosis with special emphasis on screening (1, 33) or to specifically detect ethambutol resistance (11). We applied this technology to the detection of rifampin resistance in M. tuberculosis. The Pyrosequencing chemistry differs in several aspects from standard sequencing, e.g., no fluorochromes or radioactivity is used and it does not require any postreaction step (22). Furthermore, it generates shorter DNA sequences, typically 20 to 70 bp in length in a 96-well format. The technology allows fast prediction of mutations and can be used in screening of large numbers of samples simultaneously. Our goal was to develop a method for genotypic analysis of rpoB mutations correlated to rifampin resistance that is reliable, fast, and easy to perform. The results obtained in this analysis were compared with line probe assay (LiPA) (13), standard Sanger sequencing, and an internationally recognized reference standard for determination of rifampin resistance, the phenotypic radiometric BACTEC 460 method (10).
MATERIALS AND METHODS
M. tuberculosis strains.
The M. tuberculosis reference strain H37Rv (ATCC 25618) as well as 27 rifampin-resistant and 25 rifampin-susceptible clinical isolates of M. tuberculosis from different parts of the world were selected from the national strain collection at the Swedish Institute for Infectious Disease Control. The included strains were verified not to originate from the same patient. Of these 27 isolates, 24 were MDR TB, of which 10 were resistant to one additional drug and 9 were resistant to two or more additional drugs. All strains were stored at −70°C and cultured on standard solid Löwenstein-Jensen medium in a biosafety level III laboratory.
Phenotypic drug susceptibility testing.
Phenotypic drug susceptibility was determined using the BACTEC 460 assay (Becton Dickinson, Sparks, MD) (23), with the following critical concentrations: rifampin, 2.0 μg/ml; isoniazid, 0.2 μg/ml; streptomycin, 4.0 μg/ml; ethambutol, 5.0 μg/ml.
Isolation of DNA.
A loopful (approximately 10 μl) of bacterial cells was harvested from Löwenstein-Jensen medium and mixed with 1 ml of 50 mM Tris-HCl (pH 8.0). The bacteria were inactivated and lysed for 30 min at 85°C. The suspension was then chilled to room temperature and centrifuged at a relative centrifugal force of 12,000 × g for 5 min, and the supernatant was subsequently removed. The pellet was suspended in 50 μl chloroform and 50 μl water, mixed by vortexing for 5 min, and centrifuged at a relative centrifugal force of 12,000 × g for another 5 min. The aqueous phase containing DNA was removed and quantified by optical density measurement at 260 nm. Each sample was diluted (typically 100-fold) to 5 ng/μl, renumbered to meet the blinded criteria, and stored at −20°C.
Genotypic drug susceptibility testing.
All strains were previously analyzed with the INNO-LiPA Rif. TB kit (Innogenetics, Ghent, Belgium) (13). Apart from an M. tuberculosis complex species-specific probe, the line probe assay comprises 5 wild-type overlapping hybridization probes (∼20 bp/each) covering nearly the whole 81-bp RRDR in rpoB as well as four probes exclusively detecting the four most common mutations conferring rifampin resistance. According to the kit manual, even if none of these four probes hybridizes, a strain should still be regarded as resistant whenever at least one of the wild-type probes does not hybridize. In such a case, one can only determine that the mutation is present within the region for the hybridization probe (approximately 20 bp) and no discrimination between silent, nonsense, and missense mutations can be obtained. To determine the precise mutation, the strains were also analyzed by cycle sequencing.
Pyrosequencing analysis.
The rifampin resistance-determining region was amplified in a Gene-Amp PCR system 2400 (Perkin Elmer, Norwalk, CT) by using primers rpoB.PCR.SGS.Forw2 and rpoB.biotin.Rev2 (Table 1), the latter being biotinylated at its 5′ end. Each 50-μl reaction mixture contained 1 mM Mg2+, 200 nM concentrations of each primer, 100 μM concentrations of each deoxynucleoside triphosphate, 10 ng template, 5 μl of 10× PCR buffer (Perkin Elmer), and 1 U AmpliTaq Gold (Perkin Elmer). The reaction was initiated at 95°C for 10 min, followed by 45 cycles of (94°C for 30 s, 63°C for 20 s, 72°C for 20 s), and finally, elongated at 72°C for 7 min. The approximately 180-bp product was analyzed on a 2% agarose gel.
TABLE 1.
Primers used for amplification and Pyrosequencing analysis of the RRDR of rpoB in M. tuberculosis
| Primer name | Sequence (5′-3′) | Source |
|---|---|---|
| rpoB.PCR.SGS.Forw2 | TTTCGATCACACCGCAGACGTTa | SGS ABb |
| rpoB.biotin.Rev2 | Biotin-GGCACGCTCACGTGACAGAC | Interactivac |
| F1 | GCGATCAAGGAGT | Life Technologiesd |
| F7 | CAGCCAGCTGAGCCAATTCA | Life Technologies |
| F9 | ACCAGAACAACCCGCTGTCG | Life Technologies |
| F13 | TGACCCACAAGCGCCGACTG | Life Technologies |
A 3-nucleotide 5′ poly(T) tail was added.
Scandinavian Gene Synthesis AB, Köping, Sweden.
Thermo Hybaid, Ulm, Germany.
Invitrogen, Paisley, Scotland.
Preparation of template and sequencing was performed with a slightly modified version of the PSQ 96 SQA reagent kit 1X96 manual (Biotage AB, Uppsala, Sweden). In brief, the single-stranded biotinylated PCR products were separated from the nonbiotinylated strand by denaturation and vacuum filtration in a 96-well format. Annealing of sequencing primers was done by heating to 90°C for 2 min, followed by cooling to room temperature. The combination of the sequencing primers, F1, F7, F9, and F13 (Table 1), was designed to detect mutations in the entire 81-bp RRDR (20) in four overlapping reactions (Fig. 1). A cyclic dispensation of deoxynucleoside triphosphates was used for primer F1 [22(CTAG)], for primers F7 and F9 [22(CTGA)], and for primer F13 [17(TCGA)]. Each strain was analyzed at least twice with each sequencing primer. Due to the limited accuracy of the early beta version of the software, all pyrograms were manually interpreted. Each sequencing reaction was analyzed from the first downstream nucleotide until the first read nucleotide of the following downstream sequencing primer, except for primer F13, which was analyzed to include codon 534 (Fig. 1). All sequences obtained from each sequencing primer were gathered and compared in multiple ClustalW alignments with the H37Rv wild-type rpoB sequence (Rv0667) as the master sequence. A strain was considered potentially resistant if a mutation that would alter the amino acid sequence was present. All runs were reanalyzed and aligned when the new PSQ96MA2.1 sequencing software was released, confirming the previously obtained results.
RESULTS
We have searched for mutations conferring rifampin resistance in 53 strains of M. tuberculosis with Pyrosequencing technology. Each strain was sequenced at least twice and aligned with the H37Rv wild-type sequence. The turnaround time from harvest of cells to the interpreted sequence was estimated to be approximately 1 workday.
Of the analyzed strains, 24 were found to contain the wild-type DNA sequence in the RRDR, as determined by all three genetic methods. In the remaining 29 strains, Pyrosequencing analysis and standard cycle sequencing detected mutations in the RRDR. These results were in agreement with the results obtained with the LiPA method. When the genotypic results were compared to the phenotypic reference standard BACTEC, 27 of the 53 strains were found to be resistant to rifampin. Thus, 2 of the 53 samples were susceptible to rifampin, although they contained a mutation in the RRDR as determined by the genotypic methods. Both Pyrosequencing analysis and Sanger sequencing revealed a Leu511Pro mutation (Fig. 2), whereas in LiPA these mutations were indicated by the lack of hybridization to one of the wild-type probes. The Leu511Pro substitution was also found together with an Asp516Gly substitution in another strain, which was phenotypically resistant (Table 2). The Leu511Pro mutation has been reported to be present in resistant, borderline resistant, and susceptible strains (2, 16, 30, 31). To rule out borderline resistance in these strains, the MIC for the two strains with the single Leu511Pro mutation were determined both in BACTEC 460 and on Middlebrook 7H10 agar. The two strains were confirmed to be susceptible to rifampin with MIC of 0.5 μg/ml and 0.25 to 0.5 μg/ml, respectively, by both methods. In contrast, one of the strains with two mutations in the RRDR (Leu511Pro plus Asp516Gly) was highly resistant with a MIC of >256 μg/ml in BACTEC 460.
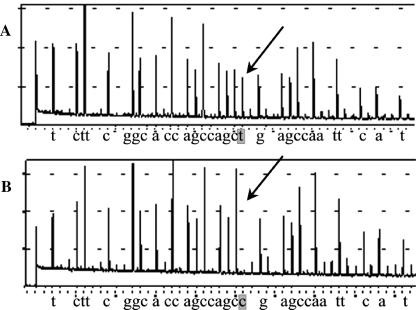
FIG. 2.
Mutation detection using the F1 Pyrosequencing primer. (A) Wild type; (B) CTG511CCG substitution. The Pyrosequencing chemistry differs from Sanger sequencing. In the former method, each nucleotide is dispensed automatically one at a time. Whenever a nucleotide is incorporated, a flash is produced and is monitored as a peak in the pyrogram. The height of the peak is proportional to the number of nucleotides inserted. Mutations are detected both at the peak level and at the aligned sequence output.
TABLE 2.
Mutations detected by Pyrosequencing analysis in comparison to the phenotypic BACTEC 460 and the genotypic LiPA
| No. of strains | BACTEC 460 resulta | Codon alteration(s) detected by sequencing primer:
|
LiPA resultb | |||
|---|---|---|---|---|---|---|
| F1 | F7 | F9 | F13 | |||
| 24 | S | WT | ||||
| 16 | R | TCG531TTG | R5 | |||
| 2 | R | GAC516GTC | R2 | |||
| 2 | R | CAC526GAC | R4b | |||
| 2 | S | CTG511CCG | ΔS1 | |||
| 1 | R | CTG511CCG | GAC516GGC | ΔS1 | ||
| 1 | R | CAC526TAC | R4a | |||
| 1 | R | CAC526CTC | ΔS4 | |||
| 1 | R | TCG531CAG, GGG534GGC | ΔS5 | |||
| 1 | R | AAC518deletion | NAc | ΔS2 | ||
| 1 | R | CAC526CGC | ΔS4 | |||
| 1 | R | GAC516TAC | GCG532GTG | ΔS2, ΔS5, R4a, R4b | ||
R denotes resistance and S denotes sensitivity to RIF at 2 μg/ml.
Four specific probes define the four most commonly detected rifampin mutations: R2, GAC516GTC; R4a, CAC526TAC; R4b, CAC526GAC; R5, TCG531TTG. Other mutations are detected by lack of hybridization to any of the five overlapping wild-type (WT) probes, which are denoted as ΔS1 to ΔS5 according to the location of the mutation (13).
NA, no data obtained (see text for further explanation).
The most frequently detected mutation in these samples was Ser531Leu, which was present in 59% (16/27) of all resistant strains. Each of the F1, F7, F9, and F13 sequencing primers detected 3, 5, 5, and 19 mutations, respectively, including three strains with double mutations. One of these double mutants revealed a mutation that altered the amino acid sequence (Ser531Gln), while the other mutation was a silent mutation (GGG534GGC), i.e., not altering the glutamine at position 534.
DISCUSSION
An early and more detailed detection of antibiotic resistance in M. tuberculosis can improve the treatment outcome and reduce the spread of resistant tuberculosis. A rapid diagnosis allowing initiation of correct therapy is not only of major importance for the individual patient but also from the public health perspective, since it can decrease the period of infectiousness and thus reduce the spread of disease in society (7).
To reduce the turnaround time, several PCR-based methods have been evaluated for the detection of mutations conferring RIF resistance in M. tuberculosis. Among these are LiPA and the single-stranded conformation polymorphism (SSCP). Besides the specific identification of the four most common rpoB mutations in LiPA, both of these methods are an indirect detection of mutations, either by lack of hybridization to wild-type probes (LiPA) or different electrophoretic migration in comparison to a wild-type fragment (SSCP). They do not have the discriminatory power to define all different types of mutations that can be expected to be found. This factor is, however, essential, as shown in this investigation, as silent mutations do occur, both at the genetic level (i.e., no change in the amino acid chain) as well as the phenotypic level (i.e., altered amino acid but no change in the drug susceptibility pattern) (14). The latter would be the case with the Leu511Pro mutation we found in two strains susceptible to rifampin. These types of mutations cannot be discriminated from other mutations affecting the same detection probe in LiPA and would thereby be defined as resistant with this method. Defining the exact mutation might also be of importance when having to select a rifamycin for treatment. Due to the possible pharmacological interactions of rifampin and certain antiretroviral drugs, rifabutin has been suggested as an agent for the treatment of dually infected TB and human immunodeficiency virus patients (3). Rifampin and rifabutin cross-resistance does not always seem to be the case but might be dependent on the actual resistance mutation, where some mutations appear to give cross-resistance while others seem to give resistance to rifampin but not to rifabutin (31, 32). The significance of electrophoretic separation methods such as SSCP still needs to be evaluated. It has been reported that the most frequent mutation conferring RIF resistance could not be discriminated from the susceptible wild-type strain (27). This indicates that methods such as these, when they are used to screen for RIF resistance, might need to be confirmed with sequencing to determine the characteristics of the mutation, especially in the case of novel mutations. Cycle sequencing has frequently been used as a gold standard for mutation detection, but for large-scale routine diagnostics, this method's usefulness can be questioned, since it is costly and time-consuming and requires technical expertise (26).
Thus, our aim was to create a robust system with the same resolution as Sanger sequencing but easier to handle and, thus, more suitable for screening high numbers of samples. A comparison of Pyrosequencing technology to other molecular methods for RIF resistance detection showed advantages in its short turnaround time, easiness to automate, high capacity, and finally, accuracy in the determination of the exact mutations.
Longer sequences of the 3′ end of the RRDR than those we used can be analyzed by Pyrosequencing (1, 33). Arnold et al. showed that longer sequences (26 bp) can reduce the reproducibility. Zhao et al. used long sequences (about 40 bp) and did not comment on any problems with this layout. However, they report a frameshift mutation in the essential rpoB gene, which can be an indication of problems with the interpretation of the sequence data. We have found that running longer sequences reduces the reproducibility in our method, i.e., acceptable sequences were not received in all runs. This was true for various primer designs as well as whether or not different single-stranded protein concentrations were applied (data not shown). As a measure of the stability of this assay, we observed that 7 different single-nucleotide substitutions were found to be located at the annealing sites of the F7, F9, and F13 sequencing primers together. Although present at the annealing sites, these mutations have not affected the quality of the sequences, presumably because low sequencing temperature will allow for single mismatching in sequencing primers. On the other hand, a 3-nucleotide deletion at the annealing site for the F9 primer did negatively affect the sequencing reaction for this primer. Although not satisfactorily sequenced by the F9 primer, this sample was regarded as resistant, since the AAC518 deletion was detected by sequencing with the F7 primer. During optimization of the assay, we found evidence for extensive looping and self-priming (data not shown). This was predicted by the Pyrosequencing primer design program and confirmed in experiments when running the template in the absence of the sequencing primer. This is a recognized potential problem with Pyrosequencing technology, and various publications have dealt with this either by adding a dideoxynucleoside triphosphate to the nonbiotinylated end of the template or using blocking primers (18, 29). We successfully circumvented the self-priming by using highly purified primers (SGS, Köping, Sweden) in combination with the addition of a poly(T) tail to the 5′ end of the forward PCR primer, i.e., the sequencing template. The reason for looping is probably a reflection of the fact that the sequence is prone to secondary structure, which may be correlated to the high (65%) GC content and to the fact that the Pyrosequencing analysis is performed at a low reaction temperature.
In comparison to phenotypic methods, detection of rifampin resistance using Pyrosequencing technology is an accurate and time-efficient method, permitting genotype determination within approximately one working day. It would further be a major advantage if the DNA could be isolated directly from clinical specimens or at least from early broth cultures. This would circumvent the lengthy isolation of bacterial cultures, which can take several weeks. With Pyrosequencing technology, it is also possible to include quantitative analysis of clinical samples, i.e., to determine the ratio of genes containing the resistance mutation, as has been done with human immunodeficiency virus and bacteria (9, 19, 24). It would allow for detection of the emergence of a mutation within a population of cells. In conclusion, Pyrosequencing technology offers a high-throughput assay for rapid and specific determination of rifampin resistance in M. tuberculosis.
REFERENCES
- 1.Arnold, C., L. Westland, G. Mowat, A. Underwood, J. Magee, and S. Gharbia. 2005. Single-nucleotide polymorphism-based differentiation and drug resistance detection in Mycobacterium tuberculosis from isolates or directly from sputum. Clin. Microbiol. Infect. 11:122-130. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer, T., G. Zurcher, P. Imboden, and A. Telenti. 1995. Mutation position and type of substitution in the beta-subunit of the RNA polymerase influence in-vitro activity of rifamycins in rifampicin-resistant Mycobacterium tuberculosis. J. Antimicrob. Chemother. 35:345-348. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2000. Updated guidelines for the use of rifabutin or rifampin for the treatment and prevention of tuberculosis among HIV-infected patients taking protease inhibitors or nonnucleoside reverse transcriptase inhibitors. Morb. Mortal. Wkly. Rep. 49:185-189. [PubMed] [Google Scholar]
- 4.Cohn, D. L., F. Bustreo, M. C. Raviglione, et al. 1997. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD Global Surveillance Project. Clin. Infect. Dis. 24(Suppl. 1):S121-S130. [DOI] [PubMed] [Google Scholar]
- 5.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 6.Douglas, J. G., and M. J. McLeod. 1999. Pharmacokinetic factors in the modern drug treatment of tuberculosis. Clin. Pharmacokinet. 37:127-146. [DOI] [PubMed] [Google Scholar]
- 7.Drobniewski, F. A., M. Caws, A. Gibson, and D. Young. 2003. Modern laboratory diagnosis of tuberculosis. Lancet Infect. Dis. 3:141-147. [DOI] [PubMed] [Google Scholar]
- 8.Fakhrai-Rad, H., N. Pourmand, and M. Ronaghi. 2002. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum. Mutat. 19:479-485. [DOI] [PubMed] [Google Scholar]
- 9.Haanpera, M., P. Huovinen, and J. Jalava. 2005. Detection and quantification of macrolide resistance mutations at positions 2058 and 2059 of the 23S rRNA gene by pyrosequencing. Antimicrob. Agents Chemother. 49:457-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inderleied, C. B., and G. E. Pfyffer. 2003. Susceptibility test methods: mycobacteria, p. 1149-1177. In P. R. Murray et al. (ed.), Manual of clinical microbiology, 8th ed., vol. 1. American Society for Microbiology, Washington D.C. [Google Scholar]
- 11.Isola, D., M. Pardini, F. Varaine, S. Niemann, S. Rusch-Gerdes, L. Fattorini, G. Orefici, F. Meacci, C. Trappetti, M. Rinaldo Oggioni, and G. Orru. 2005. A Pyrosequencing assay for rapid recognition of SNPs in Mycobacterium tuberculosis embB306 region. J. Microbiol. Methods 62:113-120. [DOI] [PubMed] [Google Scholar]
- 12.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 13.Jureen, P., J. Werngren, and S. E. Hoffner. 2004. Evaluation of the line probe assay (LiPA) for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 84:311-316. [DOI] [PubMed] [Google Scholar]
- 14.Kim, B. J., S. Y. Kim, B. H. Park, M. A. Lyu, I. K. Park, G. H. Bai, S. J. Kim, C. Y. Cha, and Y. H. Kook. 1997. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR-single-strand conformation polymorphism analysis for rifampin susceptibility testing. J. Clin. Microbiol. 35:492-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruuner, A., L. Pehme, S. Ghebremichael, T. Koivula, S. E. Hoffner, and M. Mikelsaar. 2002. Use of molecular techniques to distinguish between treatment failure and exogenous reinfection with Mycobacterium tuberculosis. Clin. Infect. Dis. 35:146-155. [DOI] [PubMed] [Google Scholar]
- 16.McNerney, R., P. Kiepiela, K. S. Bishop, P. M. Nye, and N. G. Stoker. 2000. Rapid screening of Mycobacterium tuberculosis for susceptibility to rifampicin and streptomycin. Int. J. Tuberc. Lung Dis. 4:69-75. [PubMed] [Google Scholar]
- 17.Moore-Gillon, J. 2001. Multidrug-resistant tuberculosis: this is the cost. Ann. N. Y. Acad. Sci. 953:233-240. [DOI] [PubMed] [Google Scholar]
- 18.Nordstrom, T., A. Alderborn, and P. Nyren. 2002. Method for one-step preparation of double-stranded DNA template applicable for use with Pyrosequencing technology. J. Biochem. Biophys. Methods 52:71-82. [DOI] [PubMed] [Google Scholar]
- 19.O'Meara, D., K. Wilbe, T. Leitner, B. Hejdeman, J. Albert, and J. Lundeberg. 2001. Monitoring resistance to human immunodeficiency virus type 1 protease inhibitors by pyrosequencing. J. Clin. Microbiol. 39:464-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 21.Ronaghi, M., and E. Elahi. 2002. Pyrosequencing for microbial typing. J. Chromatogr. B 782:67-72. [DOI] [PubMed] [Google Scholar]
- 22.Ronaghi, M., M. Uhlen, and P. Nyren. 1998. A sequencing method based on real-time pyrophosphate. Science 281:363-365. [DOI] [PubMed] [Google Scholar]
- 23.Siddiqi, S. H., J. P. Libonati, and G. Middlebrook. 1981. Evaluation of rapid radiometric method for drug susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 13:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinclair, A., C. Arnold, and N. Woodford. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in Enterococci. Antimicrob. Agents Chemother. 47:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 26.Telenti, A., and D. H. Persing. 1996. Novel strategies for the detection of drug resistance in Mycobacterium tuberculosis. Res. Microbiol. 147:73-79. [DOI] [PubMed] [Google Scholar]
- 27.Tracevska, T., I. Jansone, L. Broka, O. Marga, and V. Baumanis. 2002. Mutations in the rpoB and katG genes leading to drug resistance in Mycobacterium tuberculosis in Latvia. J. Clin. Microbiol. 40:3789-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuohy, M. J., G. S. Hall, M. Sholtis, and G. W. Procop. 2005. Pyrosequencing as a tool for the identification of common isolates of Mycobacterium sp. Diagn. Microbiol. Infect. Dis. 51:245-250. [DOI] [PubMed] [Google Scholar]
- 29.Utting, M., J. Hampe, M. Platzer, and K. Huse. 2004. Locking of 3′ ends of single-stranded DNA templates for improved Pyrosequencing performance. BioTechniques 37:66-67, 70-73. [DOI] [PubMed] [Google Scholar]
- 30.Watterson, S. A., S. M. Wilson, M. D. Yates, and F. A. Drobniewski. 1998. Comparison of three molecular assays for rapid detection of rifampin resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 36:1969-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams, D. L., L. Spring, L. Collins, L. P. Miller, L. B. Heifets, P. R. Gangadharam, and T. P. Gillis. 1998. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1853-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, B., H. Koga, H. Ohno, K. Ogawa, M. Fukuda, Y. Hirakata, S. Maesaki, K. Tomono, T. Tashiro, and S. Kohno. 1998. Relationship between antimycobacterial activities of rifampicin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621-628. (Erratum, 43:613, 1999.) [DOI] [PubMed] [Google Scholar]
- 33.Zhao, J. R., Y. J. Bai, Q. H. Zhang, Y. Wang, M. Luo, and X. J. Yan. 2005. Pyrosequencing-based approach for rapid detection of rifampin-resistant Mycobacterium tuberculosis. Diagn. Microbiol. Infect. Dis. 51:135-137. [DOI] [PubMed] [Google Scholar]