Abstract
Pti4 is a tomato (Lycopersicon esculentum) transcription factor that belongs to the ERF (ethylene-responsive element binding factor) family of proteins. It interacts with the Pto kinase in tomato, which confers resistance to the Pseudomonas syringae pv tomato pathogen that causes bacterial speck disease. To study the function of Pti4, transgenic Arabidopsis plants were generated that expressed tomato Pti4 driven by the strong constitutive promoters, cauliflower mosaic virus 35S and tCUP. Global gene expression analysis by Affimetric GeneChip indicated that expression of Pti4 in transgenic Arabidopsis plants induced the expression of GCC box-containing PR genes. We also demonstrated that Pti4 enhanced GCC box-mediated transcription of a reporter gene. The data suggests that tomato Pti4 could act as a transcriptional activator to regulate expression of GCC box-containing genes. Furthermore, we show that the expression of tomato Pti4 in transgenic Arabidopsis plants produced a phenotype similar to that seen in plants treated with ethylene, thus providing evidence that the Pti4 gene is involved in the regulation of a subset of ethylene-responsive genes containing the GCC box.
The ERF (ethylene-responsive element binding factor) proteins (formerly known as EREBPs [ethylene-responsive element binding proteins]) were first isolated as GCC box binding proteins from tobacco (Nicotiana tabacum; Ohme-Takagi and Shinshi, 1995). The GCC box contains a conserved AGCCGCC sequence, which was first indentified from the promoters of ethylene-inducible genes in tobacco (Ohme-Takagi and Shinshi, 1990). ERF proteins contain a highly conserved DNA binding domain (ERF domain) consisting of 58 or 59 amino acids (Ohme-Takagi and Shinshi, 1995; Hao et al., 1998). Although the ERF domain shares some sequence homology with the AP2 domain found in the Arabidopsis protein APETALA2 (Jofuku et al., 1994; Riechmann and Meyerowitz, 1998), the ERF and AP2 domain proteins belong to distinct families (Fujimoto et al., 2000).
A number of ERF proteins have been identified from different plant species (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Stockinger et al., 1997; Zhou et al., 1997; Liu et al., 1998; Solano et al., 1998; Menke et al., 1999; Fujimoto et al., 2000; Ohta et al., 2000). For example, CBF1, DREBP1A, and DREB2A have been shown to bind to a C-repeat/dehydration-responsive element that is involved in drought and cold stress (Stockinger et al., 1997; Liu et al., 1998). The ERFs, Pti4/5/6, AtERP, Arabidopsis ERF1, AtERFs, and ORCA2 bind to the GCC box in the ethylene-responsive element that is essential for the responsiveness of some promoters to ethylene (Ohme-Takagi and Shinshi, 1995; Buttner and Singh, 1997; Zhou et al., 1997; Solano et al., 1998; Menke et al., 1999; Fujimoto et al., 2000; Gu et al., 2000; Ohta et al., 2000). In addition, a tobacco ERF protein, Tsi1, that could bind both the GCC and the C-repeat/dehydration-responsive element sequences was also identified (Park et al., 2001).
Studies on the tomato (Lycopersicon esculentum) resistance (R) gene, Pto, provided evidence that linked the ERF genes to the defense response. Pto is a protein kinase that confers resistance to Pseudomonas syringae pv tomato, a bacteria that expresses the avirulence gene avrPto. Pto was shown to directly interact in two-hybrid assays with the tomato ERF proteins, Pti4/5/6 (Zhou et al., 1997). Pti4/5/6 proteins have been shown to bind the GCC box cis-element, which is present in the promoter region of many ethylene-regulated pathogenesis-related (PR) genes (Ohme-Takagi and Shinshi, 1995; Zhou et al., 1997; Gu et al., 2000). It has been proposed that the Pti4/5/6 proteins may activate PR gene expression by binding to the GCC box of the PR gene promoters (Zhou et al., 1997; Gu et al., 2000). Ethylene has been implicated in the regulation of basic-type PR genes during the defense responses of plants attacked by pathogens. Infection by a pathogen and treatment with an elicitor both promote the synthesis of ethylene and ethylene activates the transcription of basic-type PR genes (Ecker, 1995; Yamamoto et al., 1999). The differential expression of Pti4, Pti5, and Pti6 in various tomato tissues implies that they may have distinct roles in plants (Thara et al., 1999; Gu et al., 2000). Pti4 is particularly interesting because its expression is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase (Gu et al., 2000).
We have generated transgenic Arabidopsis plants transformed with binary vectors carrying fusions of the tCUP or cauliflower mosaic virus (CaMV) 35S promoter to the tomato Pti4 cDNA. Overexpression of Pti4 in Arabidopsis induces the expression of the GCC box-containing PR genes. Etiolated Pti4 transgenic seedlings show inhibition of hypocotyl elongation, which is a typical characteristic of plants treated with ethylene (Ecker, 1995). In addition, Pti4 transgenic plants also display a dwarf phenotype similar to that of constitutive ethylene-responsive mutants. Our study provides direct evidence that the Pti4 gene product is involved in the regulation of the ethylene-responsive genes containing the GCC box.
RESULTS
Pti4 Protein Activates GCC Box-Mediated Transcription of a Reporter Gene
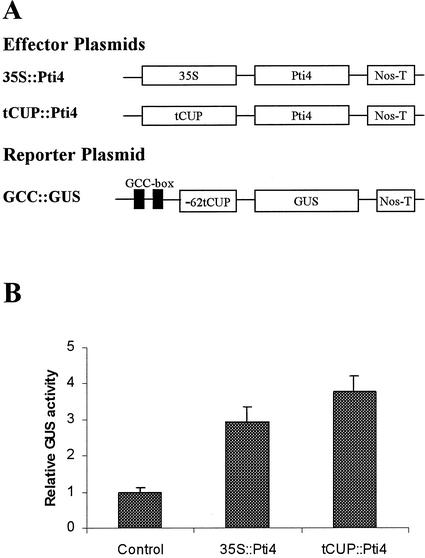
To test whether the tomato Pti4 protein can interact with the GCC box, Pti4 effector plasmids were constructed in which the Pti4 cDNA was driven by a strong constitutive promoter, CaMV 35S or tCUP (Fig. 1A). The reporter plasmids, GCC::GUS and mGCC::GUS, were constructed using a β-glucuronidase (GUS) reporter gene. Two GCC boxes or mutated GCC boxes (mGCC; Ohme-Takagi and Shinshi, 1995) were fused to a minimal promoter, -62tCUP (Wu et al., 2001) to drive the GUS reporter gene expression. The effector plasmids were cobombarded into tobacco leaves together with a reporter plasmid. As shown in Figure 1B, cotransfection of the reporter plasmid GCC::GUS with a effector plasmid resulted in a 3- to 4-fold increase in GUS expression, indicating that Pti4 protein can interact with the GCC boxes in the promoter of the reporter construct to activate transcription. Transcription of the reporter gene that had a mutated GCC box was not activated by Pti4 (data not shown).
Figure 1.
Activation of the GCC box-mediated transcription of GUS reporter gene by Pti4 protein in transient expression assays. A, Schematic diagram of the effector and reporter constructs used in cobombardment experiments. The effector constructs contain the Pti4 cDNA fused to the Nos terminator driven by the 35S or tCUP promoter. The reporter construct contains two GCC boxes fused to the -62tCUP minimal promoter-GUS construct. B, Activation of the GCC::GUS fusion gene by Pti4. The reporter plasmid GCC::GUS was cobombarded with each effector plasmid or the control plasmid pUC19. GUS activity was reported as picomoles of 4-methylumbelliferone per milligram of protein per minute. Bars indicate the se of three replicates.
Ectopic Expression of Tomato Pti4 Induces Resident Basic Chitinase Gene Expression
Transgenic Arabidopsis plants were generated that expressed Pti4 driven by a strong constitutive promoter, CaMV 35S or tCUP (Foster et al., 1999). Southern-blot analysis was performed to determine whether the genomic DNA of the putative transformants contained the transgenic DNA (data not shown). Four of the transgenic lines (tCUP::Pti4-1, tCUP::Pti4-3 and tCUP::Pti4-4, and tCUP::Pti4-5) contained the Pti4 transgene driven by tCUP promoter and two transgenic lines (35S::Pti4-3 and 35S::Pti4-6) contained Pti4 transgene driven by CaMV 35S promoter.
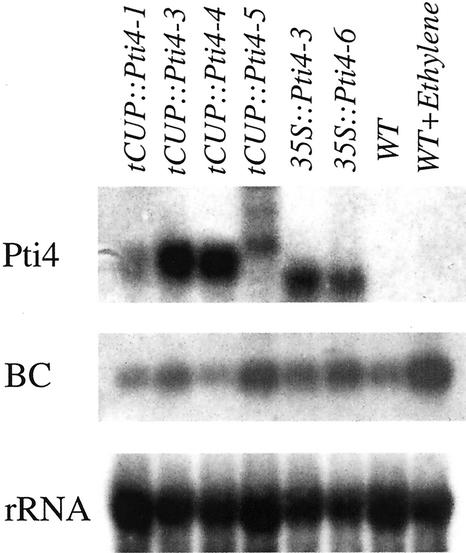
The expression of Pti4 RNA in the transgenic lines was determined by northern analysis. The predicted 1-kb transcript was detected in five transgenic lines, tCUP::Pti4-1, tCUP::Pti4-3, tCUP::Pti4-4, 35S::Pti4-3 and 35S::Pti4-6, using the Pti4 cDNA probe. It was absent from the wild-type plants as expected (Fig. 2). One transgenic line, tCUP::Pti4-5, showed bands that were larger in size than the bands in the other lanes of the transgenic plants. This is most likely due to the downstream termination of transcription. Different levels of Pti4 transcript accumulation were detected in the transgenic lines, with the transgenic line tCUP::Pti4-1 having the lowest level of Pti4 expression.
Figure 2.
Northern-blot analysis of the Pti4 transgenic plants. Total RNA was isolated from wild-type (WT) and transgenic lines (1–6). Lanes 1 to 6 correspond to transgenic lines tCUP::Pti4-1, tCUP::Pti4-3, tCUP::Pti4-4, tCUP::Pti4-5, 35S::Pti4-3, and 35S::Pti4-6, respectively. Five micrograms of total RNA was probed with a Pti4 cDNA, a basic chitinase (BC), and an rDNA probe, respectively.
Solano et al. (1998) reported that overexpression of another ERF protein, ERF1, in transgenic Arabidopsis plants induced basic chitinase gene expression. Basic chitinase is an ethylene-responsive gene, which contains the GCC box in its promoter (Samac et al., 1990). We therefore examined whether the expression of tomato Pti4 in Arabidopsis could induce the expression of the Arabidopsis basic chitinase gene. As shown in the Figure 2, the basic chitinase gene was expressed at a relative low level in the wild-type but was induced in the transgenic lines tCUP::Pti4-3, tCUP::Pti4-5, 35S::Pti4-3, and 35S::Pti4-6. The transgenic line tCUP::Pti4-1, which had the lowest level of Pti4 expression among the six transgenic lines, did not show the induction of chitinase expression. These data indicated that there was a general correlation between Pti4 expression and chitinase RNA accumulation, suggesting that Pti4 induced the expression of the basic chitinase gene in Arabidopsis. The expression of other GCC box-containing genes, PDF1.2, PR-1, and HOOKLESS1, however, was not induced by expression of Pti4 in transgenic plants (data not shown), suggesting that the expression of these genes and basic chitinase gene was regulated by different mechanisms.
Pti4 Induces Expression of the GCC Box-Containing Genes
To identify other genes regulated by Pti4, we compared global gene expression in tCUP::Pti4-3 and wild-type seedlings by cDNA hybridization to GeneChip (Affymetrix, Santa Clara, CA) containing 8,247 Arabidopsis genes. Of the 8,247 Arabidopsis genes, only 28 genes that exhibited greater than 2.5-fold expression in tCUP::Pti4-3 compared with the wild-type (Table I). In comparison, the expression of the control genes such as actin, GAPDH, and UBQ4 did not show significant difference. Search for cis-elements in promoter regions of these 28 induced genes revealed that 18 of them contain GCC box related sequences in 5′ upstream sequences. Some of them encode well-known pathogen-related proteins. These include chitinase and β-1,3-glucanase that have antifungal activities; xyloglucan endo-transglycosylase and β-glucosidase and monooxygenase 1 involve in cell wall modification; and peroxidases, basic blue protein, and protein disulfide isomerase involve in oxidative burst.
Table I.
Genes induced by Pti4 in Arabidopsis
| Accession No. | Gene Product | −Fold Induction | GCC Motifb |
|---|---|---|---|
| X98453 | Peroxidase ATPN | 20.7 | GCCGCC |
| AC005662 | Unknown | 18.6 | GCCGCC |
| AL022604 | Putative sugar transport protein | 13.9 | GCCGAC |
| X98809 | Peroxidase ATP5a | 13.2 | GCCACC |
| AC004138 | Putative basic blue protein | 9.6 | GCCACC |
| AC002333 | Putative endochitinase | 7.8 | GCCGCC |
| Y14070 | Heat shock protein 17.6A6.7 | 6.7 | — |
| AF088280 | PAP3 | 4.9 | GCCGNC |
| X92975 | Xyloglucan endo-transglycosylase | 4.3 | GCCACC |
| AC003033 | Putative protein disulfide isomerase | 3.8 | GCCACC |
| AF082157 | β-glucosidase | 3.7 | GCCGNC |
| Z97340 | β-1,3-glucanase precursor | 3.7 | GCCGCC |
| AJ001809 | Succinate dehydrogenase flavaprotein | 3.7 | GCCGAC |
| AB023448 | Basic endochitinase | 3.6 | GCCGCC |
| AC006340 | Unknown protein | 3.6 | — |
| AC002335 | Putative trypsin inhibitor | 3.6 | GCCACC |
| AF002109 | Putative anthocyanin 5-aromatic acyltransferase | 3.4 | — |
| AF082299 | AGP2 | 3.4 | GCCGNC |
| X79052 | SRG1 | 3.3 | — |
| AB003280 | Phosphoglycerate dehydrogenase | 3.2 | — |
| L04637 | Lipoxygenase | 3.1 | — |
| AL034567 | Putative ubiquinol-cytochrome c reductase | 3.0 | GCCGNC |
| AC004561 | Putative tropinone reductase | 2.9 | — |
| AJ007587 | Monooxygenase 1 | 2.8 | GCCGNC |
| AC002343 | HSP90-like protein | 2.7 | GCCGCC |
| AC003114 | Calreticulin | 2.6 | — |
| X75365 | Sucrose-proton symporter | 2.6 | — |
| AC005662 | Putative embryo-abundant protein | 2.6 | GCCGNC |
| U41998 | Actin | 1.0 | — |
| M64115 | GAPDH | 1.3 | — |
| U33014 | UBQ4 | 1.1 | — |
28 genes that exhibited greater than 2.5-fold expression in tCUP∷Pti4-3 compared with the wild type were shown. In addition, three control genes (actin, GAPDH, and UBQ4) that did not show significant difference in expression were also listed for comparison.
GCC-motifs were found between 100 and 1,500 bp upstream of the translation start site for a subset of genes up-regulated in tCUP∷Pti4-3 transgenic plants.
Pti4 Transgenic Plants Display an Ethylene-Responsive Phenotype
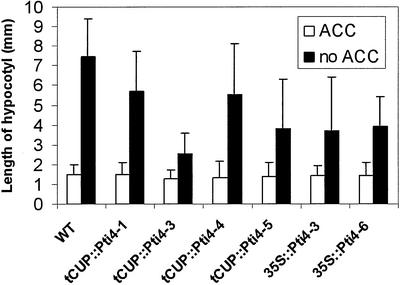
To evaluate the involvement of Pti4 in the ethylene signaling pathway, Pti4 transgenic plant lines were examined for the ethylene-responsive phenotype. This is characterized by a triple response in Arabidopsis, which includes inhibition of root and hypocotyl elongation, radial swelling of the hypocotyl and root, and exaggeration in the curvature of the apical hook (Ecker, 1995; Chang and Shockey, 1999). The hypocotyls of the etiolated transgenic seedlings were measured 72 h after germination. As shown in Figures 3 and 4, the transgenic lines showed inhibition of hypocotyl elongation, a phenotype similar to those observed in the constitutive ethylene response-mutants or in wild-type plants exposed ethylene (Solano et al., 1998). The seedlings from the transgenic line tCUP::Pti4-3, which had a high Pti4 expression, displayed strong inhibition of hypocotyl elongation. The seedlings from the transgenic line tCUP::Pti4-1, which had a lower level of Pti4 transgene expression, showed weak inhibition of hypocotyl elongation (Fig. 3). These data indicated that there was a correlation between the Pti4 expression and the inhibition of hyopcotyl elongation.
Figure 3.
Length of hypocotyl of transgenic Arabidopsis seedlings. Surface-sterilized seeds from wild-type (WT) and transgenic lines were planted in growth medium and cold treated at 4°C for 4 d before germination and growth in the dark at 23°C for 72 h in the presence (with ACC) or absence (without ACC) of 1-aminocyclopropane-1-carboxylic acid. The lengths of seedling hypocotyls were measured to the closest millimeter. Fourteen to 20 seedlings from each line were measured. Error bars correspond to the se.
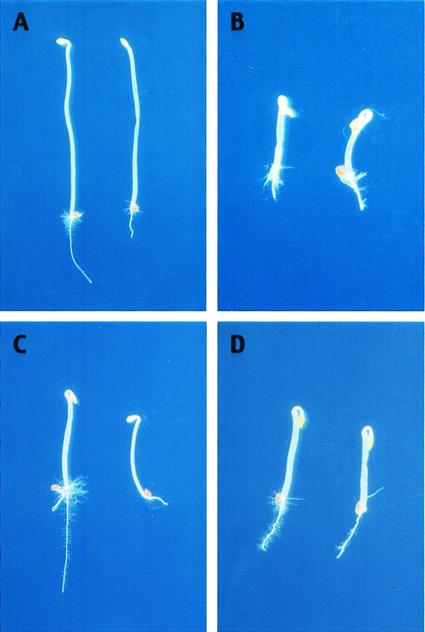
Figure 4.
Phenotype of Pti4 overexpression in transgenic seedlings. Each panel is composed of two etiolated Arabidopsis seedling. Surface-sterilized seeds were planted in growth medium and cold treated at 4°C for 4 d before germination and growth in the dark at 23°C for 72 h. A, Wild type incubated without aminocyclopropane carboxylic acid (ACC); B, wild type displaying the triple response in the presence of 10 μm ACC; C, tCUP::Pti4-3 transgenic seedlings incubated without ACC; and D, tCUP::Pti4-3 transgenic seedlings incubated in the presence of 10 μm ACC.
The seedlings from the transgenic lines did not show strong curvature of the apical hook (Fig. 4), suggesting a partial seedling triple response phenotype. This is consistent with the observation that the HOOKLESS1 gene, a gene required for apical hook curvature (Lehman et al., 1996), was not expressed in the transgenic plants (data not shown). The adult plants from the transgenic lines also displayed reduced leaf size when compared with the wild-type plants (Fig. 5). This is a phenotype similar to that of the constitutive ethylene-responsive mutants such as ctr1 (Ecker, 1995).
Figure 5.
Phenotype of Pti4 overexpression in transgenic plants. The transgenic tCUP::Pti4-3 plants (middle and right) displayed a reduced leaf size when compared with the wild-type plant (left). The photo was taken after plants were grown for 5 weeks in a growth chamber (16 h of light and 8 h of darkness at 23°C).
DISCUSSION
The Pti4 protein belongs to the ERF-type proteins, which is a large family of plant transcription factors (Ohme-Takagi and Shinshi, 1995; Riechmann and Meyerowitz, 1998; Fujimoto et al., 2000). The binding of some ERF proteins to the GCC box in the ethylene-responsive element suggests a role for these proteins in the regulation of ethylene-responsive gene expression. In tomato, Pti4 transcripts rapidly accumulated in response to ethylene, before expression of the GCC box-containing GluB and Osm genes (Thara et al., 1999; Gu et al., 2000), further supporting a role in ethylene-regulated PR gene expression. Using a transient expression system, we have shown that Pti4 can function as a transcriptional activator of a GCC box-containing reporter gene. Furthermore, expression of Pti4 in Arabidopsis induced the expression of GCC box-containing genes and conferred a constitutive ethylene-responsive phenotype. These data suggested that Pti4 is involved in the regulation of ethylene-responsive genes containing the GCC box.
The GCC box contains a conserved AGCCGCC sequence, which was first identified from the promoters of ethylene-inducible PR genes in tobacco (Ohme-Takagi and Shinshi, 1990; Eyal et al., 1993; Hart et al., 1993). It has been suggested that this sequence is a target in the ethylene signal transduction pathway because deletion of the GCC box eliminates ethylene responsiveness (Broglie et al., 1989; Meller et al., 1993; Vogeli-Lange et al., 1994; Shinshi et al., 1995). A search for plant promoter sequences containing the GCC box sequence uncovered a number of predominantly basic PR genes from bean (Phaesoleus vulgarus), tobacco, potato (Solanum tuberosum), Arabidopsis, Brassica sp., and tomato (Zhou et al., 1997; Jia and Martin, 1999), suggesting that these PR genes might be regulated by related ERF transcriptional factors. In Arabidopsis, the GCC box-containing genes include basic chitinase gene, PDF1.2, PR-1, and HOOKLESS1 (Samac et al., 1990; Lehman et al., 1996; Lebel et al., 1998; Manners et al., 1998). Overexpression of Pti4 in Arabidopsis induces the expression of the basic chitinase gene and other GCC box-containing genes. The expression of PDF1.2, PR-1, and HOOKLESS1, however, was not induced, suggesting that the expression of these genes might be regulated by different ERF proteins.
The Arabidopsis ethylene-responsive factor 1 (ERF1) was induced rapidly by ethylene, and its role in regulating ethylene-inducible genes was demonstrated (Solano et al., 1998). The Arabidopsis ERF1 activates GCC box-containing PR genes such as the basic chitinase gene and PDF1.2 and confers constitutive ethylene response when overexpressed in Arabidopsis. Pti4 could be a tomato functional homolog of ERF1 in mediating ethylene-regulated expression of PR genes containing a GCC box. In Arabidopsis, expression of ERF1 is controlled by a novel DNA-binding protein encoded by the EIN3 gene, indicating that ERF1 acts downstream of EIN3 in ethylene signaling (Solano et al., 1998). It remains to be determined whether Pti4 gene expression is regulated by a transcriptional factor similar to EIN3 in tomato.
In tobacco, at least four different ERF proteins, ERF1 through -4, have been identified (Ohme-Takagi and Shinshi, 1995). ERF2 and ERF4 enhance the GCC box-mediated transcription of a reporter gene in tobacco protoplasts, suggesting that they act as transcriptional activators (Ohta et al., 2000). In contrast to ERF2 and ERF 4, ERF3 reduces the transcription of the reporter gene in tobacco protoplasts, indicating that ERF3 functions as a repressor. Several Arabidopsis ERF-like genes, AtERF1 to -5, were also isolated from an Arabidopsis cDNA library by using tobacco ERFs as probes (Fujimoto et al., 2000). It has been shown that AtERF1, AtERF2, and AtERF5 act as transcriptional activators for GCC box-dependent transcription. AtERF3 and AtERF4, however, act as transcriptional repressors (Fujimoto et al., 2000). These studies indicate that GCC box-dependent transcription is controlled by a dynamic system utilizing antagonistic mechanisms in plants. Different ERF proteins also possess distinct DNA binding preferences, suggesting they could play different roles in the differential control of GCC box-containing gene expression (Hao et al., 1998; Fujimoto et al., 2000).
In tomato, three ERF-like genes, Pti4/5/6, were identified by their interaction with Pto kinase in yeast two-hybrid screening (Zhou et al., 1997). Pti4/5/6 show differential expression patterns in various tomato tissues, implying that they may play distinct roles (Thara et al., 1999; Gu et al., 2000). Our study demonstrates that expression of the Pti4 gene in Arabidopsis induces the expression of GCC box-containing genes. In tomato, several GCC box-containing genes, such as GluB, Osm, and one 1-aminocyclopropane-1-carboxylic acid oxidase gene, have been identified (Jia and Martin, 1999). Pti4 transcripts rapidly accumulated in response to ethylene, before expression of the GCC box-containing GluB and Osm genes (Thara et al., 1999; Gu et al., 2000), suggesting that Pti4 may control the expression of a subset of GCC box-containing genes in tomato. In Arabidopsis, an ERF-like protein, AtEBP, was shown to interact with a basic Leu zipper transcription factor (Buttner and Singh, 1997), indicating that ERF proteins may interact with other transcriptional factors to regulate gene expression. Further research is required to investigate how subsets of GCC box-containing genes are regulated by different EFR proteins.
In summary, we have demonstrated that Pti4 can act as a transcriptional activator to enhance GCC box-mediated gene transcription. Expression of Pti4 in transgenic Arabidopsis plants confers a constitutive ethylene phenotype and induces the GCC box-containing gene expression. Our study provides evidence that Pti4 gene product is involved in the regulation of a subset of ethylene-responsive genes containing the GCC box.
MATERIALS AND METHODS
Plant Material
Arabidopsis (ecotype Columbia) was grown in a growth chamber (16 h of light and 8 h of darkness at 23°C) after a 2- to 4-d vernalization period. For growth under sterile conditions, seeds were surface sterilized (15-min incubation in 5% [v/v] sodium hypochlorite, and a three-time rinse in sterile distilled water) and sown on one-half-strength Murashige and Skoog salts (Sigma, St. Louis; Murashige and Skoog, 1962) supplemented with 1% (w/v) Suc, pH 5.7, and 0.8% (w/v) agar in petri dishes.
To test the triple response of seedlings, surface-sterilized seeds were planted in Murashige and Skoog growth medium and cold treated at 4°C for 4 d. Seeds were then grown in the dark at 23°C for 72 h in the presence or absence of 1-aminocyclopropane-1-carboxylic acid, and the hypocotyl lengths of seedlings were measured.
Southern- and Northern-Blot Analysis
Total genomic DNA from Arabidopsis was extracted as described (Dellaporta et al., 1983). For Southern blots, Arabidopsis genomic DNA was digested with restriction enzymes, separated by agarose gel electrophoresis, and transferred to nylon membranes (Sambrook et al., 1989). For northern analysis, total RNA was isolated from 100 to 200 mg of Arabidopsis tissues using TriPure Reagent as described by the manufacturer (Boehringer Mannheim, Basel). Northern blots were prepared by electrophoresis of 5- to 10-μg samples of total RNA through agarose gels in the presence of formaldehyde (Strommer et al., 1993), followed by transfer to nylon membranes. Southern and northern blots were probed with 32P-labeled probes. Prehybridization and hybridization were performed at 65°C in 0.5 m Na2HPO4 (pH 7.2), 7% (w/v) SDS, and 1 mm EDTA. Filters were washed once for 15 min in 2× SSC with 0.1% (w/v) SDS at room temperature, then twice for 20 min in 0.1× SSC, 0.1% (w/v) SDS at 65°C. The damp filters were autoradiographed at −80°C using two intensifying screens. Filters were stripped in 5 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 0.05% (w/v) SDS at 100°C for 2 min when reprobing was required.
Synthesis of Biotin-Labeled cRNA
The methods for preparation of cRNA directly from total RNA and subsequent steps leading to hybridization and scanning of the U95 GeneChip Arrays were provided by the manufacturer (Affymetrix). Briefly, first-stranded cDNA was synthesized from 20 μg of total RNA with a special oligo(dT)24 primer containing a T7 RNA polymerase promoter at its 5′ end in 20 μL of first-strand reaction mix at 42°C for 1 h. The second-strand was synthesized in second-strand reaction mix for 2 h at 16°C. After second-strand synthesis, biotin-labeled cRNA was generated from the cDNA sample by an in vitro transcription reaction using BioArray RNA Transcript Labeling Kit (Enzo Diagnostics, New York) with biotin-labeled CTP and UTP. The labeled cRNA was purified by using RNeasy spin columns (Qiagen USA, Valencia, CA). Fifteen micrograms of each cRNA sample was fragmented at 94°C for 35 min in fragmentation buffer (40 mm Tris-acetate, pH 8.1, 100 mm potassium acetate, and 30 mm magnesium acetate) and then used to prepare 300 μL of mixture. A biotinylated oligonucleotide, B2, was added that hybridizes to unique features at the center and four corners of each chip to map the probe sets on the chip.
Oligonucleotide Array Hybridization and Scanning
cRNA hybridization mix was heated to 94°C for 5 min, equilibrated to 45°C for 5 min, and clarified by centrifugation (14,000g) at room temperature for 6 min. Aliquots of each sample (10 mg of cRNA in 200 mL of the mixture) were hybridized GeneChip arrays at 45°C for 16 h in a rotisserie oven set at 60 rpm (GeneChip Hybridization Oven 640, Affymetrix). After this, the arrays were washed with SSPE, stained with streptavidin-phycoerythrin (Molecular Probes, Eugene, OR) and washed again. The whole procedure of washing and staining was carried out in GeneChip Fluidics Station 400 (Affymetrix). Then the chip was scanned by GeneArray Scanner (HP and Affymetrix). Average difference and expression call for each features on the chip was computed using Affymetrix GeneChip Analysis Suite version with a default parameters.
Plasmid Construction
To prepare reporter constructs, the CaMV 35S promoter of pBI221 (CLONTECH, Palo Alto, CA) was replaced with a truncated tCUP promoter, -394tCUP (Wu et al., 2001), to generate the pBI-BtCUP vector. The -394tCUP promoter of pBI-BtCUP was replaced with a minimal tCUP promoter, -62tCUP. DNA fragments containing two GCC boxes or mutated GCC boxes (Ohme-Takagi and Shinshi, 1995) were ligated into the PstI site located upstream of the -62tCUP promoter.
To construct the effector plasmids, we replaced the 35S promoter of pBI221 with the tCUP promoter (Foster et al., 1999) to generate the pBI-tCUP vector. The GUS gene in the pBI221 and pBI-tCUP was replaced with the Pti4 coding region to generate 35S::Pti4 and tCUP::Pti4, respectively.
To generate plasmid for Arabidopsis transformation, the 35S::Pti4 and tCUP::Pti4 plasmids were digested with EcoRI and HindIII, and the resulting fragment containing the promoters and the Pti4 gene were then subcloned into the multicloning sites of pCAMBIA2300 binary vector (Cambia, Canberra, Australia).
Plant Transformation and Selection
Plant transformation plasmids were electroporated into Agrobacterium tumefaciens GV3101 as described by Shaw (1995). The A. tumefaciens-mediated transformation of Arabidopsis was performed as described (Clough and Bent, 1998), with the following modifications. Plants with immature floral buds and few siliques were dipped into a solution containing A. tumefaciens, 2.3 g L−1 Murashige and Skoog salts (Sigma), 5% (w/v) Suc, and 0.03% (w/v) Silwet L-77 (Lehle Seeds, Round Rock, TX) for 0.5 min. T1 seeds were collected, dried at 25°C, and sown on sterile media containing 40 μg mL−1 kanamycin to select the transformants. Surviving T1 plantlets were transferred to soil to set seeds (T2).
Particle Gun Delivery Assays
Tobacco (SR1; Nicotiana tabacum) plants were maintained in vitro in one-half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) in Magenta containers (Magenta Corp., Chicago) in a growth chamber at 25°C. After transfer to fresh medium for 2 to 3 weeks, uniform-sized leaves (about 3 cm in width) were cut off from the plants and placed on a medium consisting of Murashige and Skoog salts, B5 vitamins (Gamborg et al., 1968), 1 mg L−1 6-benzyladenine, 0.1 mg L−1 naphthalene acetic acid, 3% (w/v) Suc, and 0.25% (w/v) Gelrite in a 20- × 15-mm petri dish. The leaves were preconditioned on this medium for 1 d before gene delivery.
Plasmid DNA was isolated using the Qiagen Plasmid Midi Kit. The reporter plasmid was mixed with an effector plasmid at a 1:5 ratio (w/v). In the control, the reporter was mixed with the pUC19 plasmid. A modified particle inflow gun (Brown et al., 1994) was used for DNA delivery. DNA was precipitated onto tungsten particles using following protocol: 5 μg of DNA was added to 25 μL of tungsten particles (100 mg mL−1) and followed by the addition of 25 μL of 2.5 m CaCl2 and 5 μL of 0.1 m spermidine. The leaves were bombarded at a distance 16 cm from the screen and under a pressure of 1,000 kPa of He gas. Bombarded leaves were maintained on the same medium for 24 h before assay for GUS activity. Gene expression was determined by histochemical and fluorometric assays (Jefferson, 1988). GUS activity was reported as picomoles of 4-methylumbelliferone per milligram of protein per minute.
ACKNOWLEDGMENTS
We are grateful to Dr. Gregory Martin (Cornell University) for providing us with the Pti4 cDNA clone. We thank Ming Hu, Teresa Martin, Marysia Latoszek-Green, and Susan Sibbold for technical assistance, Dr. Tim Xing and Linda Harris for critical reading of the manuscript.
Footnotes
This work was supported in part by the Matching Investment Initiative Program at Agriculture and Agri-Food Canada. This paper is Eastern Cereal and Oilseed Research Centre contribution no. 001554.
©Minister of Public Works and Government Services Canada 2002. For the Department of Agriculture and Agri-Food, Government of Canada.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010696.
LITERATURE CITED
- Broglie KE, Biddle P, Cressman R, Broglie R. Functional analysis of DNA sequences responsible for ethylene regulation of a bean chitinase gene in transgenic tobacco. Plant Cell. 1989;1:599–607. doi: 10.1105/tpc.1.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DCW, Tian L-N, Buckley DJ, Lefebvre M, McGrath A, Webb J. Development of a simple particle bombardment device for gene delivery into plant cells. Plant Cell Tiss Org Cult. 1994;37:47–53. [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/s1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation, version II. Plant Mol Rep. 1983;1:19–21. [Google Scholar]
- Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Lev-Yadun S, Fluhr R. A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J. 1993;4:225–234. doi: 10.1046/j.1365-313x.1993.04020225.x. [DOI] [PubMed] [Google Scholar]
- Foster E, Hattori J, Labbe H, Ouellet T, Fobert P, James L, Miki B. A tobacco cryptic constitutive promoter, tCUP, revealed by T-DNA tagging. Plant Mol Biol. 1999;41:45–55. doi: 10.1023/a:1006229501860. [DOI] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirement suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gu Y, Yang C, Thara YK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–786. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hart CM, Nagy F, Meins F., Jr A 61 bp enhancer element of the tobacco beta-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol Biol. 1993;21:121–131. doi: 10.1007/BF00039623. [DOI] [PubMed] [Google Scholar]
- Jefferson R. Plant reporter genes: the GUS gene fusion system. In: Setlow JK, Hollaender A, editors. Genetic Engineering: Principles and Methods. New York: Plenum Press; 1988. pp. 247–263. [Google Scholar]
- Jia Y, Martin GB. Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol Biol. 1999;40:455–465. doi: 10.1023/a:1006213324555. [DOI] [PubMed] [Google Scholar]
- Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel E, Heifetz P, Thorne L, Uknes S, Ryals J, Ward E. Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 1998;16:223–233. doi: 10.1046/j.1365-313x.1998.00288.x. [DOI] [PubMed] [Google Scholar]
- Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Penninckx IA, Vermaere K, Kazan K, Brown RL, Morgan A, Maclean DJ, Curtis MD, Cammue BP, Broekaert WF. The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol Biol. 1998;38:1071–1080. doi: 10.1023/a:1006070413843. [DOI] [PubMed] [Google Scholar]
- Meller Y, Sessa G, Eyal Y, Fluhr R. DNA-protein interactions on a cis-DNA element essential for ethylene regulation. Plant Mol Biol. 1993;23:453–463. doi: 10.1007/BF00019294. [DOI] [PubMed] [Google Scholar]
- Menke FLH, Champion A, Kijne JW, Memelink J. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 1999;18:4455–4463. doi: 10.1093/emboj/18.16.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Ohme-Takagi M, Shinshi H. Structure and expression of a tobacco beta-1,3-glucanase gene. Plant Mol Biol. 1990;15:941–946. doi: 10.1007/BF00039434. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Ohme-Takagi M, Shinshi H. Three ethylene-responsive transcriptional factors in tobacco with distinct transactivation functions. Plant J. 2000;22:29–38. doi: 10.1046/j.1365-313x.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- Park JM, Park C, Lee S, Ham B, Shin R, Pack K. Overexpression of the tobacco Isi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Samac DA, Hironaka CM, Yallaly PE, Shah DM. Isolation and characterization of the genes encoding basic and acidic chitinase in Arabidopsis thaliana. Plant Physiol. 1990;93:907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shaw CH. Introduction of cloning plasmids into Agrobacterium tumefaciens. Methods Mol Biol. 1995;49:33–37. doi: 10.1385/0-89603-321-X:33. [DOI] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M. Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol. 1995;27:923–932. doi: 10.1007/BF00037020. [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strommer J, Gregerson R, Vayda M. Isolation and characterization of plant mRNA. In: Glik BR, Thompson JE, editors. Methods in Plant Molecular Biology and Biotechnology. Boca Raton, FL: CRC Press; 1993. pp. 49–66. [Google Scholar]
- Thara VK, Tang X, Gu YQ, Martin GB, Zhou JM. Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 1999;20:475–483. doi: 10.1046/j.1365-313x.1999.00619.x. [DOI] [PubMed] [Google Scholar]
- Vogeli-Lange R, Frundt C, Hart CM, Nagy F, Meins F., Jr Developmental, hormonal, and pathogenesis-related regulation of the tobacco class I beta-1,3-glucanase B promoter. Plant Mol Biol. 1994;25:299–311. doi: 10.1007/BF00023245. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Suzuki K, Shinshi H. Elicitor-responsive, ethylene-independent activation of GCC box-mediated transcription that is regulated by both protein phosphorylation and dephosphorylation in cultured tobacco cells. Plant J. 1999;20:571–579. doi: 10.1046/j.1365-313x.1999.00634.x. [DOI] [PubMed] [Google Scholar]
- Wu K, Malik K, Tian L, Hu M, Martin T, Foster L, Brown D, Miki B. Enhancers and core promoter elements are essential for the activity of a cryptic gene activation sequence from tobacco, tCUP. Mol Genet Genom. 2001;265:763–770. doi: 10.1007/s004380100478. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]