Abstract
Classical methods for identification of Mycobacterium species rely on morphology and biochemical profiles. Speciation of a Mycobacterium isolate using these standard methods is a lengthy process based on subjective data interpretation. In this study, Mycobacterium species were characterized by utilizing matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). This technology is designed to provide a characteristic mass spectral fingerprint based on desorbed ions from the cell surface. Thirty-seven strains were analyzed; these represented thirteen species and five subspecies that included the Mycobacterium tuberculosis complex and the M. avium-M. intracellulare complex, as well as rapid- and slow-growing mycobacteria. All 37 strains were analyzed in triplicate, and a database was generated. This method produced species-specific patterns for all but 1 of the 37 isolates and provided reliable differentiation at the strain level. The data suggest that whole-cell MALDI-TOF MS has potential as a rapid and reproducible method for the identification and characterization of Mycobacterium species.
The genus Mycobacterium consists of approximately 100 heterogeneous species of rapid- and slow-growing acid-fast bacilli (9, 25). Most members of this genus are harmless microbes that live in diverse soil and aqueous environments; however, there are a number of pathogenic species that infect humans and animals (25, 42). The number of infections caused by mycobacteria has increased over the past few decades (2, 5, 10, 16, 44). The two most well-known human pathogens are Mycobacterium tuberculosis and M. leprae. M. tuberculosis belongs to one of two major groups within the genus called the Mycobacterium tuberculosis complex (MTC), which includes pathogens such as M. microti, M. africanum, and M. bovis. The MTC has become a major cause of death in many developing countries and continues to be a public health problem globally (8, 17). A number of the environmental Mycobacterium species, collectively known as nontuberculosis mycobacteria (NTM), are responsible for many opportunistic infections that are increasingly common among immunocompromised individuals. For example, the M. avium-M. intracellulare complex (MAC) is a leading cause of secondary infections in patients infected with human immunodeficiency virus (42). Another member of the NTM, M. marinum, is also emerging as a pathogen in humans, causing a variety of cutaneous and related illnesses in immunocompromised hosts (16, 24). Differentiation of the MTC and NTM has become important due to the rise of infections and antimicrobial resistance in this genus.
Despite their medical and environmental importance, mycobacteria have always proven difficult to identify. This is due to a combination of factors, principal among them being their low growth rate and fastidious growth requirements. Conventional methods for phenotypic identification rely on standard biochemical tests that may involve cultivation on specific growth media and extensive incubation time. One of the most commonly used identification tests for mycobacteria is analysis of the unique mycolic acids found in their cell wall (4, 22). This test requires harvesting cell material, followed by extracting and derivatizing mycolic acid esters and analyzing their contents by high-pressure liquid chromatography (HPLC). This method has proven reliable in identifying many Mycobacterium species (42) and is commonly used in clinical laboratories.
In the past several years, molecular methods, including PCR and hybridization assays using specific probes, have been developed for the identification of mycobacteria (6, 15, 34). These methods provided a fast, reproducible way of identifying specific Mycobacterium species, reducing the detection time from 6 weeks to approximately 3 weeks. A PCR-restriction endonuclease analysis based on hsp65, the gene encoding the 65-kilodalton heat shock protein, has been used extensively for differentiating M. tuberculosis from other NTM members (3, 7, 27, 33). Hybridization assays require initial species identification to specify probes for the hybridization, and there are only limited numbers of species that can be identified by these assays (6, 34). Currently, DNA probes are available to identify M. tuberculosis complex, MAC, M. kansasii, and M. gordonae (12). Other commercially available DNA probes have their limitations as well, which include misidentification, inability to differentiate among the MTC complex members, and the need to confirm results with alternate methods (23, 32, 38, 42).
The existing molecular typing methods concentrate on MTC strains. These include restriction fragment length polymorphism of the IS6110 repetitive sequence (36) and spacer oligotyping (spoligotyping) to look at genetic loci which contain a variable number of tandem repeats (42). For the other mycobacterial species, restriction fragment length polymorphism analysis and pulsed-field gel electrophoresis are the most widely used typing techniques, providing different levels of discrimination (10, 13, 26, 44, 45). In addition, multilocus enzyme electrophoresis has been used to subtype isolates of MAC and rapidly growing mycobacteria (RGM) (11, 46).
With the rise of opportunistic pathogens and with several new species being named annually (5, 28, 37, 39, 40), the need to have a rapid and reproducible identification method for a wider range of Mycobacterium species is evident. Recent developments in mass spectrometry (MS) have made it possible to use whole-cell matrix-assisted laser desorption ionization- time-of-flight MS (MALDI-TOF MS) or MALDI to aid in the identification of microorganisms. This technology is designed to provide a characteristic mass spectral fingerprint based on desorbed ions from the cell surface. Cells were picked from bacterial colonies, applied to a 96-well mass spectrometer plate, and overlaid with a matrix solution. The instrument software finished the process by automatically acquiring and analyzing data and using statistical algorithms to generate a profile (18, 29). A wide variety of prokaryotes have been characterized by using this approach, including clinically or environmentally relevant bacteria such as Escherichia coli, Staphylococcus aureus, Bacillus species, and Pseudomonas species (19, 21, 30, 31, 43). Here we present an evaluation of MALDI in the identification of Mycobacterium species and in its ability to distinguish between multiple strains within a species. A total of 37 strains, representing 13 species and four groups of Mycobacterium, were analyzed by using MALDI-TOF MS.
MATERIALS AND METHODS
Bacteria, media, and growth conditions.
The 37 Mycobacterium strains representing 13 species used in the present study were obtained from the American Type Culture Collection (ATCC) (Table 1) . The freeze-dried cultures were revived by resuspending them in Middlebrook broth 7H9 (Becton Dickinson, MD) with albumin-dextrose-catalase. M. avium subsp. paratuberculosis and M. avium subsp. silvaticum were resuspended in Middlebrook 7H9 supplemented with Tween 80 and Mycobactin J. A total of 0.1 ml of this suspension was streaked for isolation onto Middlebrook 7H10 with oleic acid-albumin-dextrose-catalase, with the exception of M. microti, which was grown in 7H9 broth. RGM were incubated for 3 to 5 days at 37°C in an aerobic atmosphere. Slow-growing mycobacteria were incubated for 2 to 6 weeks at 37°C in an aerobic atmosphere, with the exception of M. tusciae and M. doricum, which were incubated at 30°C.
TABLE 1.
Mycobacterium strains used in this study
| Group | Organisma | ATCC no. | Isolation source |
|---|---|---|---|
| MAC | M. avium subsp. aviumT | 25291 | Diseased hen's liver |
| M. avium subsp. avium | 35717 | Human sputum | |
| M. avium subsp. avium | 35718 | Human cervical lymph node | |
| M. avium subsp. paratuberculosis | 43544 | Human intestinal tissue | |
| M. avium subsp. paratuberculosis | 43545 | Human intestinal tissue | |
| M. avium subsp. silvaticumT | 49884 | Wood pigeon's liver and spleen | |
| M. intracellulareT | 13950 | NAb | |
| M. intracellulare | 700662 | Human sputum | |
| M. intracellulare | 700664 | Derived from ATCC 700662 | |
| MTC | M. africanumT | 25420 | Sputum |
| M. microtiT | 19422 | NA | |
| M. microti | 11152 | Vole | |
| M. microti | 35781 | Field vole | |
| M. microti | 35782 | Field vole | |
| M. tuberculosis | 25177 | NA | |
| RGM | M. abscessusT | 19977 | NA |
| M. abscessus | 23016 | Human | |
| M. abscessus | 700869 | Joint aspiration | |
| M. chelonaeT | 35752 | Tortoise | |
| M. chelonae | 14472 | Sputum | |
| M. chelonae | 35751 | Eye infection | |
| M. fortuitum subsp. acetamidolyticumT | 35931 | Human sputum | |
| M. fortuitum subsp. acetamidolyticum | 43266 | Sputum | |
| M. fortuitum subsp. fortuitumT | 6841 | Cold abscess | |
| M. fortuitum subsp. fortuitum | 49403 | Facial abscess | |
| M. fortuitum subsp. fortuitum | 49935 | Leg wound | |
| M. mucogenicumT | 49650 | Infected thyroglossal duct cyst | |
| M. mucogenicum | 49649 | Peritoneal fluid | |
| M. mucogenicum | 49651 | Postinjection site abscess | |
| Slow growing | M. doricumT | BAA-565 | Cerebrospinal fluid |
| M. kansasiiT | 12478 | From a fatal case | |
| M. kansasii | 25100 | NA | |
| M. kansasii | 25101 | NA | |
| M. kansasii | 25414 | NA | |
| M. marinumT | 927 | Fish | |
| M. marinum | 11566 | Swimming pool | |
| M. tusciaeT | BAA-564 | Lymph node |
A superscript “T” signifies a type strain of the species.
NA, not available.
Streptomyces melanosporofaciens, ATCC BAA-668, was grown in ISP medium 1 (ATCC medium 1877) at 30°C. It was used as an outgroup for the mycobacterial dendrogram.
Bacterial preparation for MALDI-TOF analysis.
Isolated colonies from agar were applied to a MALDI plate well (12 wells per strain) of a 96-well target plate and allowed to dry for 1 h. Isolates growing in broth were harvested by centrifugation, washed with HPLC-grade water, and resuspended in 30 μl of HPLC water. Then, 1 μl of the final suspension was spotted onto a MALDI plate well and allowed to dry for 1 h. Next, 1 μl of matrix was overlaid on each sample well and allowed to dry for 15 min. The matrix used was a saturated solution of α-cyano-4-hydroxycinnamic acid (Sigma) dissolved in 1:1:1 acetonitrile, water, and methanol with 0.1% (vol/vol) formic acid and 0.01 M 18-Crown 6. A 1-μl portion of the standard was applied to lock mass wells for mass calibration using average molecular weights. The standard mixture consisted of 1 pmol of bradykinin/μl, angiotensin I, glu-fibrinopeptide B, rennin substrate tetradecapeptide, adrenocorticotropin (ACTH), 2 pmol of bovine insulin/μl, and 10 pmol of ubiquitin (Sigma)/μl.
Instrumentation and data analysis.
A MALDI-linear TOF mass spectrometer (Micromass UK, Ltd., Manchester, United Kingdom) was used (19). The instrument was operated, under high vacuum, in the positive ion mode with an acceleration voltage of 15 kV and was set to acquire mass spectral peaks with mass/charge (m/z) ratios from 500 to 10,000 Da. The nitrogen laser output was 337 nm with a 3-ns pulse width, and the laser fluence was set just above the threshold for ion production. The laser fires randomly (spot size, ∼2 μm) within each well until 15 spectra of sufficient intensity are recorded. For this analysis, data from at least eight replicate wells (i.e., a minimum of 120 spectra) were used for each organism.
The Microbelynx software included with the instrument processed the data by comparing all collected spectra and determining a standard deviation for the peak intensity and placement. From these data a consensus profile is created using all spectra whose peak intensities and placements fall within threshold values that are deemed statistically significant. This reference profile becomes part of an instrument database. For identification, duplicate samples of each organism were then analyzed and compared to the database. For comparisons of samples within the database, a root mean square (RMS) value is reported for the comparison of different profiles; the closer the value is to zero, the better the match. Dendrograms based on the RMS value could also be used to show the relationship of groups of organisms based on their spectral profiles.
Streptomyces melanosporofaciens (ATCC BAA-668) served as an outgroup.
16S rRNA and hsp65 species sequencing.
The universal primers 27F (AGAGTTTGATCMTGGCTCAG) and 519R (GTATTACCGCGGCTGCTG) were used to amplify the first 500 bp of the 16S rRNA gene from the genomic DNA template (20). For the hsp65 (heat shock protein) gene, universal primers Tb11 (ACCAACGATGGTGTGTCCAT) and Tb12 (CTTGTCGAACCGCATACCCT) were used to amplify the 439-bp region from the genomic DNA template (35). The PCR product was run on a 1% precast SeaKem Gold Agarose gel (1× Tris-borate-EDTA buffer plus ethidium bromide from Cambrex BioScience, Maine). The specific band was excised from the gel and purified using a QIAquick gel extraction kit (QIAGEN, Maryland). The purified DNA was sequenced by using a CEQ 8000 genetic analyzer (Beckman Coulter, California). The results were analyzed by the Codon Code Aligner software (CodonCode Corp., Massachusetts) and compared against the NCBI GenBank database by BLAST analysis (1).
RESULTS
A total of 37 strains, representing 13 species belonging to four groups of Mycobacterium, were analyzed (Table 1). Strains were selected to yield a representative data set of both clinical and environmental isolates, as well as 14 type cultures. Whole cells of each isolate were subjected to MALDI-TOF MS analysis. A unique mass spectral fingerprint was produced for each isolate in a mass range between 500 and 4,000 Da (see Fig. 1 to 5). The majority of ions detected were less than 1,000 Da.
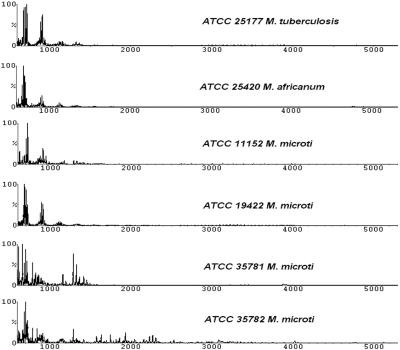
FIG. 1.
Spectral profiles generated from the MTC which comprised several slow-growing species. All except one strain (ATCC 35781) showed similar profiles. ATCC 19422 is the type strain of M. microti.
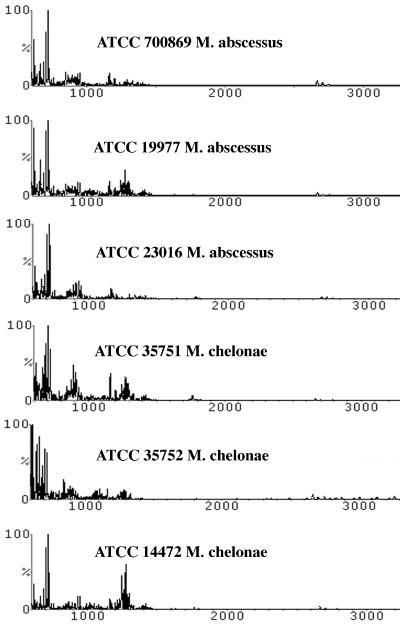
FIG. 5.
M. chelonae and M. abscessus, which belong to the RGM, are difficult to distinguish by phenotypic and molecular testing. MALDI analysis indicated that each strain among these species had some unique biomarkers, although the major peaks were very similar.
A database which contained the mass spectra of all 37 isolates was created at the ATCC. In addition, a separate database containing more than 400 spectra generated from many different clinically and environmentally important species was also utilized. Both databases were searched simultaneously as a single library to demonstrate specificity of this technique to identify Mycobacterium species. Comparison of all mass spectra to the search library resulted in unambiguous identification of the isolates to the genus and species level, with the exception of M. tusciae (ATCC BAA-564), which could not be resolved to the species level. There were eight strains that were misidentified as a very closely related strain at times (i.e., M. kansasii and M. marinum; see Fig. 3). In replicates where the correct strain did not result with the highest identity, it was always identified as the second-closest strain.
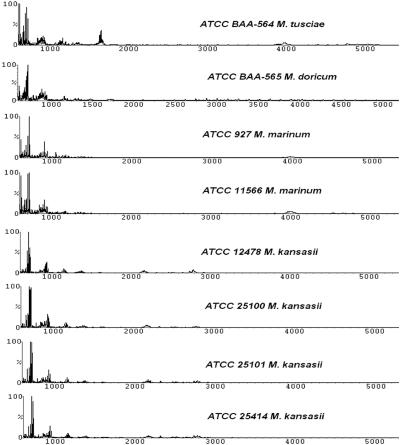
FIG. 3.
The slow-growing mycobacteria comprised four species, with M. doricum and M. tusciae as recently described new species. The profiles for four strains of M. kansasii were almost identical, and the two strains of M. marinum were very similar to each other. At times the software was not able to correctly identify M. kansasii and M. marinum at the strain level; however, the correct species was always consistently determined.
In the present study, six strains belonging to the three species of the MTC—M. tuberculosis (one strain), M. africanum (one strain), and M. microti (four strains)—were analyzed. Figure 1 shows that five of the six strains exhibited spectra that were highly similar to each other. This is further demonstrated by the dendrogram generated in Fig. 6. The exception within this group was one strain of M. microti (ATCC 35781) that clustered with M. marinum instead. A recently described species, M. doricum (ATCC BAA-565), in spite of its scotochromogenic property (39), clustered in the MTC clade where all members were nonchromogenic (42) (see Fig. 6).
FIG. 6.
A dendrogram of MALDI profiles was generated by using a separate algorithm based on the RMS value of each isolate done in triplicate. Type strains for each species were noted. Streptomyces melanosporofaciens (ATCC BAA-668) served as an outgroup.
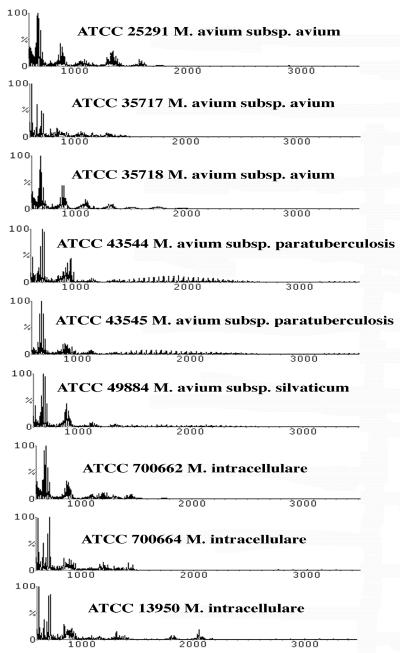
Nine strains from the MAC, which also included M. intracellulare species, were analyzed, and a similarity matrix of RMS values was prepared (Table 2). RMS values are used to determine reproducibility, and a lower RMS value signifies greater similarity. (Briefly, the RMS values were calculated by comparison of the averaged profile of a strain [based on 12 replicates] in the database with the 12 replicate profiles of a test strain.) Consistently, the lowest RMS value was obtained when each strain was compared against itself. These results indicated that although the spectral patterns were often very similar (Fig. 2) the software was capable of reliably distinguishing differences in the spectral peak positions and intensities to provide a correct identification. The spectral patterns fell into two groups (see Fig. 6), with M. avium strains (ATCC 35718, ATCC 49884, ATCC 43544, and ATCC 43545) and M. intracellulare (ATCC 13950) in one clade and M. avium (ATCC 35717), along with two strains of M. intracellulare (ATCC 700662 and ATCC 700664), forming the other. One exception was M. avium ATCC 25291, which clustered with M. tusciae (ATCC BAA-564) and was more related to the M. fortuitum clade. Interestingly, ATCC 25291 is the type strain of M. avium subsp. avium.
TABLE 2.
RMS values of MALDI profiles for MACa
| Strain | RMS value for strain:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 1 | 0.42 | ||||||||
| 2 | 0.87 | 0.34 | |||||||
| 3 | 0.92 | 2.43 | 0.35 | ||||||
| 4 | 2.57 | 5.37 | 2.67 | 0.54 | |||||
| 5 | 2.53 | 5.87 | 2.59 | 0.88 | 0.35 | ||||
| 6 | 1.12 | 2.76 | 1.04 | 1.53 | 1.41 | 0.65 | |||
| 7 | 1.21 | 1.46 | 1.58 | 2.36 | 2.55 | 2 | 0.44 | ||
| 8 | 1.23 | 1.1 | 2 | 1.96 | 2.52 | 2.04 | 1.38 | 0.28 | |
| 9 | 2.07 | 3.53 | 2.39 | 1.68 | 2.08 | 1.56 | 2.76 | 2.79 | 0.29 |
Strains: 1, ATCC 25291 (M. avium subsp. avium); 2, ATCC 35717 (M. avium subsp. avium); 3, ATCC 35718 (M. avium subsp. avium); 4, ATCC 43544 (M. avium subsp. paratuberculosis); 5, ATCC 43545 (M. avium subsp. paratuberculosis); 6, ATCC 49884 (M. avium subsp. silvaticum); 7, ATCC 700662 (M. intracellulare); 8, ATCC 700664 (M. intracellulare); 9, ATCC 13950 (M. intracellulare). Each value in boldface type represents comparison of a strain against itself (see text for explanation).
FIG. 2.
Mass spectral profiles from the MAC. Although the patterns were very similar among the strains of M. avium and M. intracellulare, the software was able to distinguish individual isolates based on the RMS values (Table 2). Note that the two profiles for M. avium subsp. paratuberculosis are almost identical.
The four strains of M. kansasii and two strains of M. marinum represented the slow-growing mycobacteria in our study. All four strains of M. kansasii yielded similar spectra (Fig. 3) and formed a tight cluster on the dendrogram (see Fig. 6). The M. marinum strains also showed nearly identical patterns and clustered together, although based on the mass spectral profiles they did not belong to M. kansasii clade (see Fig. 6).
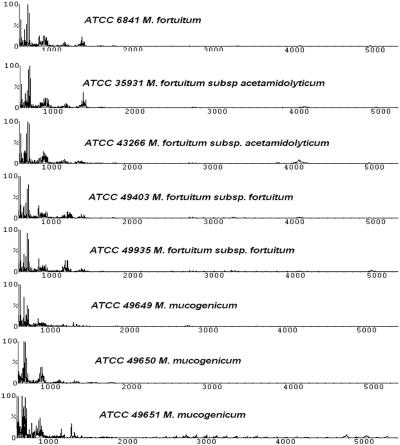
The RGM consisted of 14 strains. As was the case for the MAC, a similarity matrix indicated each strain identified itself as the best match (Table 3), although the spectra for a number of strains of M. fortuitum were highly similar (Fig. 4) and they clustered closely together (see Fig. 6). The three strains of M. mucogenicum were a conundrum since all of these strains ended up in different clades. ATCC 49649 grouped with M. abscessus-M. chelonae, which was reported previously (25). However, based on a DNA sequence comparison of the 16S rRNA gene, it was confirmed to be most closely related to M. mucogenium (results not shown). The type strain (ATCC 49650) ended up in the MAC, probably reflecting its earlier designation as an “M. chelonei-like” organism (45). The third strain, ATCC 49651, clustered with the slow-growing MTC members.
TABLE 3.
RMS values of MALDI profiles for RGMa
| Strain | RMS value for strain:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| 1 | 0.34 | |||||||||||||
| 2 | 1.29 | 0.42 | ||||||||||||
| 3 | 0.64 | 0.82 | 0.45 | |||||||||||
| 4 | 0.88 | 0.8 | 1.85 | 0.33 | ||||||||||
| 5 | 2.03 | 1.59 | 2.05 | 2.22 | 0.37 | |||||||||
| 6 | 1.77 | 0.66 | 5 | 1.32 | 1.73 | 0.29 | ||||||||
| 7 | 1.16 | 1.42 | 1.09 | 2.13 | 1.92 | 1.94 | 0.33 | |||||||
| 8 | 1.47 | 3.98 | 1.15 | 2.63 | 3.79 | 7.07 | 0.73 | 0.38 | ||||||
| 9 | 1.13 | 1.54 | 1.07 | 2 | 1.9 | 2.38 | 1.18 | 0.88 | 0.5 | |||||
| 10 | 1.25 | 1.3 | 1.73 | 1.93 | 1.81 | 1.78 | 1.7 | 1.96 | 1.85 | 0.27 | ||||
| 11 | 1.45 | 1.83 | 1.7 | 2.36 | 2.67 | 2.33 | 1.99 | 1.76 | 1.77 | 1.98 | 0.38 | |||
| 12 | 0.71 | 0.95 | 0.8 | 1.25 | 1.01 | 1.74 | 1.09 | 1.05 | 1.04 | 1.59 | 1.34 | 0.43 | ||
| 13 | 0.94 | 1.5 | 1.14 | 1.24 | 1.16 | 2.6 | 1.53 | 1.31 | 0.98 | 2.56 | 2.09 | 1.5 | 0.32 | |
| 14 | 1.25 | 1.64 | 1.6 | 2.23 | 1.27 | 2.6 | 1.93 | 1.91 | 1.83 | 2.82 | 2.16 | 1.65 | 1.69 | 0.3 |
Strains: 1, ATCC 700869 (M. abscessus); 2, ATCC 19977 (M. abscessus); 3, ATCC 23016 (M. abscessus); 4, ATCC 35751 (M. chelonae); 5, ATCC 35752 (M. chelonae); 6, ATCC 14472 (M. chelonae); 7, ATCC 6841 (M. fortuitum); 8, ATCC 35931 (M. fortuitum subsp. acetamidolyticum); 9, ATCC 43266 (M. fortuitum subsp. acetamidolyticum); 10, ATCC 49403 (M. fortuitum subsp. fortuitum); 11, ATCC 49935 (M. fortuitum subsp. fortuitum); 12, ATCC 49649 (M. mucogenicum); 13, ATCC 49650 (M. mucogenicum); 14, ATCC 49651 (M. mucogenicum). Each value in boldface type represents comparison of a strain against itself (see text for explanation).
FIG. 4.
MALDI profiles for the RGM. Patterns for the different strains of M. fortuitum were highly similar to each other, but each culture could be distinguished by the RMS values. Strains for M. mucogenicum, however, ended up in different clades (see Fig. 6).
The spectral profiles for the last two members of the RGM, M. chelonae and M. abscessus, appeared to be ambiguous (Fig. 5). They formed two clades: with M. chelonae (ATCC 14472) clustering with two strains of M. abscessus (ATCC 19977 and ATCC 35751) and the other M. chelonae (ATCC 35752) closely related to two different strains of M. abscessus (ATCC 700869 and ATCC 23016). The fact that ATCC 35752 is the type strain of M. chelonae made the interpretation more complicated. Verifying the identities of these isolates by hsp65 gene sequencing confirmed ATCC 19977, ATCC 23016, and ATCC 700869 as M. abscessus (results not shown). Interestingly, ATCC 14472 was also identified as M. abscessus by the same sequencing method. Needless to say, the identity of ATCC 14472 needs to be investigated further to resolve whether it is M. chelonae as named.
DISCUSSION
The results of this study show that identification of diverse species of Mycobacterium, and strains within those species, is possible using whole-cell MALDI. All of the 37 strains that were analyzed yielded reproducible, unique mass spectral profiles. In general, the identities revealed by the MALDI approach were consistent with those revealed by other methods that have been used to identify mycobacteria, although there were exceptions (see below).
The results in our study extend those of Hettick et al., who used MALDI-TOF to perform proteomic profiling of mycobacteria (14). These authors performed extensive comparisons between single strains of each of six different Mycobacterium species and demonstrated good reproducibility. These investigators also compared whole cells versus cells extracted with the organic solvent trifluoroacetic acid and found that either preparation yielded informative results, although trifluoroacetic acid-extracted cells were used for most of their analyses. Furthermore, these authors utilized a different mass spectrometer with a laser power capable of observing ions with mass/charge (m/z) ratios of up to 20 kDa; therefore, direct comparison between the profiles in that study and the ones described here is not possible.
In the present study, eight additional species of Mycobacterium were analyzed, along with multiple strains of individual species. These constituted clinical and environmental isolates, some with fastidious requirements and others with less-complex nutrition requirements. The isolates included representatives from the four major physiological groups: the MTC, the MAC, the RGM, and the slow-growing mycobacteria. Two newer species, M. tusciae and M. doricum, were also included to assess the technology, since they represented less-well-known members of this genus (37, 39).
Relationships among mycobacteria.
Compared to the members of Bacteria domain as a whole, the Mycobacterium spp. produced spectral patterns that were relatively consistent. In Fig. 6, Streptomyces melanosporofaciens was included as an outgroup in the dendrogram, and it showed little similarity to the mycobacteria. Another study (19) that utilized the same MALDI techniques to investigate a broad group of Bacteria and Archaea found much more variability, especially among disparate groups of Bacteria, than is seen for the Mycobacterium species tested here. Furthermore, when the organisms used in the present study were compared to a database of over 400 strains of bacterial species, their closest matches were all mycobacteria (results not shown).
Among Mycobacterium spp., some species—M. kansasii, M. fortuitum, and M. marinum—formed similar patterns and clustered together in the dendrogram. Others form two clades (MAC complex, M. chelonae, and abscessus). It is interesting that members of MTC formed a unique cluster distinct from the nontuberculosis mycobacteria. This is significant since it suggests that MALDI has the potential to effectively identify these important pathogens and unambiguously distinguish them from their less-harmful relatives. However, this work will need to be confirmed by testing many more virulent strains of M. tuberculosis and building a robust database. This in itself is a challenge due to biosafety concerns of vaporized molecules from these pathogens which are transmitted by air. Hettick et al. (14) reported that incubating the strains in acetonitrile-trifluoroacetic acid solvent resulted in inactivation of the cells; this suggests the use of cell extracts rather than whole cells as a possible method for safely working with these highly pathogenic bacteria.
The biomarkers detected by MALDI were not distinguished in the present study. Most of the ions detected by the system were relatively small, between 500 and 2,000 Da. Based on their molecular masses, it is reasonable to assume that these biomarkers are lipid molecules including mycolic acids, as well as small polypeptides that are constituents of the cell wall. The phenotypic characteristics identified by MALDI are known to vary both with culture age and with the propagation medium upon which the cells are grown (41). By using standard mycobacterium medium, the effects of medium variability can be kept to a minimum. We have not yet systematically investigated the effects of culture age on MALDI patterns; however, we have not noticed significant differences between preparations of the same strain harvested within a 24-h period.
In conclusion, our findings indicate that the identification of diverse Mycobacterium species is a tractable task by using MALDI. We also demonstrated that it is possible to resolve closely related strains of mycobacteria, and whole-cell MALDI-TOF MS can serve as an effective identification system for Mycobacterium species.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Brown-Elliott, B. A., and R. J. Wallace, Jr. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15:716-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunello, F., M. Ligozzi, E. Cristelli, S. Bonora, E. Tortoli, and R. Fontana. 2001. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 39:2799-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, W. R., K. C. Jost, and J. O. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooksey, R. C., J. H. de Waard, M. A. Yakrus, I. Rivera, M. Chopite, S. R. Toney, G. P. Morlock, and W. R. Butler. 2004. Mycobacterium cosmeticum sp. nov., a novel rapidly growing species isolated from a cosmetic infection and from a nail salon. Int. J. Syst. Evol. Microbiol. 54:2385-2391. [DOI] [PubMed] [Google Scholar]
- 6.De Beenhouwer, H., Z. Liang, P. De Rijk, C. Van Eekeren, and F. Portaels. 1995. Detection and identification of mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 33:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devallois, A., K. S. Goh, and M. Rastogi. 1997. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J. Clin. Microbiol. 35:2969-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, mortality by country. WHO global surveillance and monitoring project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 9.Euzeby, J. P., 13 July 2005 posting date. List of bacterial names with standing in nomenclature. [Online.] http://www.bacterio.cict.fr/ [DOI] [PubMed]
- 10.Falkinham, J. O. I. 1999. Molecular epidemiology: other mycobacteria, p. 136-160. In C. D. Ratledge (ed.), Mycobacteria: molecular biology and virulence. Blackwell Science, Oxford, United Kingdom.
- 11.Feizabadi, M. M., I. D. Robertson, D. V. Cousins, K. J. Dawson, and D. J. Hampson. 1997. Use of multilocus enzyme electrophoresis to examine genetic relationships amongst isolates of Mycobacterium intracellulare and related species. Microbiology 143:1461-1469. [DOI] [PubMed] [Google Scholar]
- 12.Hance, A. J., B. Grandchamp, V. Levy-Frebault, D. Lecossier, J. Rauzier, D. Bocart, and B. Gicquel. 1989. Detection and identification of mycobacteria by amplification of mycobacterial DNA. Mol. Microbiol. 3:843-849. [DOI] [PubMed] [Google Scholar]
- 13.Hector, J. S. R., Y. Pang, G. H. Mazurek, Y. Zhang, B. A. Brown-Elliott, and R. J. Wallace, Jr. 1992. Large restriction fragment patterns of genomic Mycobacterium fortuitum DNA as strain-specific markers and their use in epidemiologic investigation of four nosocomial outbreaks. J. Clin. Microbiol. 30:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hettick, J. M., M. L. Kashon, J. P. Simpson, P. D. Siegel, G. H. Mazurek, and D. N. Weissman. 2004. Proteomic profiling of intact mybocateria by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 76:5769-5776. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch, C. F., and J. M. Sigmund. 1995. Use of polymerase chain reaction (PCR) fingerprinting to differentiate bacteria for microbial products screening. J. Ind. Microbiol. 15:85-93. [DOI] [PubMed] [Google Scholar]
- 16.Holmes, G. F., S. M. Harrington, M. J. Romagnoli, and W. G. Merz. 1999. Recurrent, disseminated Mycobacterium marinum infection caused by the same genotypically defined strain in an immunocompromised patient. J. Clin. Microbiol. 37:3059-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iademarco, M. F., and K. G. Castro. 2003. Epidemiology of tuberculosis. Semin. Respir. Infect. 18:225-240. [DOI] [PubMed] [Google Scholar]
- 18.Jarman, K. H., S. T. Cebula, A. J. Saenz, C. E. Petersen, N. B. Valentine, M. T. Kingsley, and K. L. Wahl. 2000. An algorithm for automated bacterial identification using matrix-assisted laser desorption/ionization mass spectrometry. Anal. Chem. 72:1217-1223. [DOI] [PubMed] [Google Scholar]
- 19.Krader, P., and D. Emerson. 2004. Identification of Archaea and some extremophilic bacteria using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Extremophiles 8:259-268. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. G. Stackebrandt (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 21.Lay, J. O., Jr. 2001. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20:172-194. [DOI] [PubMed] [Google Scholar]
- 22.Levy-Frebault, V. V., and F. Portaels. 1992. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int. J. Syst. Bacteriol. 42:315-323. [DOI] [PubMed] [Google Scholar]
- 23.Miller, N., S. Infante, and T. Cleary. 2000. Evaluation of the LiPA MYCOBACTERIA assay for identification of mycobacterial species from BACTEC 12B bottles. J. Clin. Microbiol. 38:1915-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parent, L. J., M. M. Salam, P. C. Appelbaum, and J. H. Dossett. 1995. Disseminated Mycobacterium marinum infection and bacteremia in a child with severe combined immunodeficiency. Clin. Infect. Dis. 21:1325-1327. [DOI] [PubMed] [Google Scholar]
- 25.Pfyffer, G. E., B. A. Brown-Elliott, and R. J. Wallace, Jr. 2003. Mycobacterium: general characteristics, isolation, and staining procedures, p. 532-559. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 26.Picardeau, M., G. Prod'hom, L. Raskine, M. P. LePennec, and V. Vincent. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 35:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plikaytis, B. B., B. D. Plikaytis, M. A. Yakrus, W. R. Butler, C. L. Wookley, V. A. Silcox, and T. M. Shinnick. 1992. Differentiation of slowly growing Mycobacterium species, including Mycobacterium tuberculosis, by gene amplification and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 30:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritcher, E., S. B. Niemann, G. Rusch-Gerdes, and D. Harmsen. 2001. Description of Mycobacterium heckeshornense sp. nov. Int. J. Syst. Evol. Microbiol. 51:3023-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryzhov, V., and C. Fenselau. 2001. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73:746-750. [DOI] [PubMed] [Google Scholar]
- 30.Ryzhov, V., Y. Hathout, and C. Fenselau. 2000. Rapid characterization of spores of Bacillus cereus group bacteria by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Appl. Environ. Microbiol. 66:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smole, S. C., L. A. King, P. E. Leopold, and R. D. Arbeit. 2002. Sample preparation of gram-positive bacteria for identification by matrix-assisted laser desorption/ionization time-of-flight. J. Microbiol. Methods 48:107-115. [DOI] [PubMed] [Google Scholar]
- 32.Somoskovi, A., J. E. Hotaling, M. Fitzgerald, V. Jonas, D. Stasik, L. M. Parsons, and M. Salfinger. 2000. False-positive results for Mycobacterium celatum with the AccuProbe Mycobacterium tuberculosis complex assay. J. Clin. Microbiol. 38:2743-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steingrube, V. A., J. L. Gibson, B. A. Brown-Elliott, Y. Zhang, R. W. Wilson, M. Rajagopalan, and R. J. Wallace, Jr. 1995. PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33:149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suffys, P. N., A. da Silva Rocha, M. de Oliveira, C. E. Campos, A. M. Barreto, F. Portaels, L. Rigouts, G. Wouters, G. Jannes, G. van Reybroeck, W. Mijs, and B. Vanderborght. 2001. Rapid identification of mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J. Clin. Microbiol. 39:4477-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thierry, D., M. D. Cave, K. D. Eisenach, C. J. T., J. H. Bates, B. Gicquel, and J. L. Guesdon. 1990. IS-6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 18:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tortoli, E., R. M. Kroppenstedt, A. Bartoloni, G. Caroli, I. Jan, J. Pawlowski, and S. Emler. 1999. Mycobacterium tusciae sp. nov. Int. J. Syst. Evol. Microbiol. 49:1839-1844. [DOI] [PubMed] [Google Scholar]
- 38.Tortoli, E., A. Nanetti, C. Piersimoni, C. Cichero, G. Farina, C. Mucignat, L. Scarparo, L. Bartolini, R. Valentini, D. Nista, G. Gesu, C. P. Tosi, M. Crovatto, and G. Brusarosco. 2001. Performance assessment of new multiplex probe assay for identification of mycobacteria. J. Clin. Microbiol. 39:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortoli, E., C. Piersimoni, R. M. Kroppenstedt, J. I. Montoya-Burgos, U. Reischl, A. Giacometti, and S. Emler. 2001. Mycobacterium doricum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2007-2012. [DOI] [PubMed] [Google Scholar]
- 40.Turenne, C. Y., L. Thibert, K. Williams, T. V. Burdz, V. J. Cook, J. N. Wolfe, D. W. Cockcroft, and A. Kabani. 2004. Mycobacterium saskatchewanense sp. nov., a novel slowly growing scotochromogenic species from human clinical isolates related to Mycobacterium interjectum and Accuprobe-positive for Mycobacterium avium complex. Int. J. Syst. Evol. Microbiol. 54:659-667. [DOI] [PubMed] [Google Scholar]
- 41.Valentine, N., S. Wunschel, D. Wunschel, C. Petersen, and K. Wahl. 2005. Effect of culture conditions on microorganism identification by matrix-assisted laser desorption ionization mass spectrometry. Appl. Environ. Microbiol. 71:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincent, V., B. A. Brown-Elliott, K. C. Jost, and R. J. Wallace, Jr. 2003. Mycobacterium: phenotypic and genotypic identification, p. 560-584. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 43.Walker, J., A. J. Fox, V. Edwards-Jones, and D. B. Gordon. 2002. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: media effects and inter-laboratory reproducibility. J. Microbiol. Methods 48:117-126. [DOI] [PubMed] [Google Scholar]
- 44.Wallace, R. J., Jr., B. A. Brown-Elliott, and D. E. Griffith. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu. Rev. Microbiol. 52:453-490. [DOI] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., V. A. Silcomx, M. Tsukamura, B. A. Brown-Elliott, J. O. Kilburn, W. R. Butler, and G. O. Onyi. 1993. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J. Clin. Microbiol. 31:3231-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yakrus, M. A., M. W. Reeves, and S. B. Hunter. 1992. Characterization of isolates of Mycobacterium avium serotypes 4 and 8 from patients with AIDS by multilocus enzyme electrophoresis. J. Clin. Microbiol. 30:1474-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]