Abstract
Recently, we demonstrated rapid dissemination of different methicillin-resistant Staphylococcus aureus (MRSA) clones at the Institute for Microbiology at the University of Magdeburg (B. Ghebremedhin, W. König, and B. König, Eur. J. Clin. Microbiol. Infect. Dis. 24:388-398, 2005). The majority of them harbored the readily transmissible mec cassette type IV. Thus, theoretically, methicillin-susceptible Staphylococcus aureus (MSSA) might capture the mecA gene from circulating MRSA, or MRSA strains might catch mobile toxin genes from MSSA. Therefore, we characterized MSSA strains circulating at the University Hospital in Magdeburg. Among a total of 84 MSSA strains under study, about 40% possessed the tst (toxic shock syndrome toxin) gene and up to four additional enterotoxin genes. tst-positive MSSA strains belonged to all known agr groups (I to IV) and to 14 different spa types (t008, t012, t015, t019, t024, t056, t065, t127, t133, t162, t271, t287, t399, and t400), and they were classified by multilocus sequence typing (MLST) as ST1, ST8, ST30, ST39, ST45, ST101, ST121, ST395, and ST426. In contrast, simultaneously circulating MRSA strains (n = 24) harbored in general two or three genes of the enterotoxin gene cluster, and the tst-positive MRSA isolates belonged to the well-known epidemic types ST22, ST45, and ST228 and were classified as spa types t001, t028, and t032. From our results, one may conclude that the pool of circulating MSSA strains is an important parameter with regard to the epidemiology of hospital- and community-acquired MRSA clones and their potential virulence.
The emergence of Staphylococcus aureus strains resistant to methicillin (MRSA) and other antimicrobial agents has become a major concern, especially in the hospital environment, because of the higher mortality due to systemic MRSA infections (45). Methicillin resistance is conferred by carriage of the mecA gene (3), which is located on a genetic element called the staphylococcal cassette chromosome (SCC) in S. aureus (16, 21). The mechanism(s) responsible for mecA transfer is not known, but evidence supports horizontal transfer of the mecA gene between different staphylococcal species (14) as well as different gram-positive bacteria (2).
Analysis of the natural population dynamics and expansion of pathogenic clones of S. aureus provided evidence that essentially any S. aureus genotype carried by humans can transform into a life-threatening human pathogen but that certain clones are more virulent than others (28).
Many S. aureus strains produce one or more specific staphylococcal exotoxins, including staphylococcal enterotoxins (SEs), staphylococcal exfoliative toxins, and toxic shock syndrome toxin 1 (TSST-1). These toxins cause infections ranging from relatively mild involvement of the skin and soft tissue to life-threatening sepsis, necrotizing pneumonia, and toxic shock syndrome (TSS) (13, 24, 26, 27, 33).
SEs have been classified as members of the pyrogenic toxin superantigen family because of their biological activities and structural relatedness. SEs have been divided into five serological types (sea through see) on the basis of their antigenicity. In recent years, the existence of new types of SE genes (seg, seh, sei, sej, sek, sel, sem, sen, and seo), which belong to the operon of the enterotoxin gene cluster (egc), has been reported (19, 20, 32, 34).
These toxins cause TSS and related illnesses through their capacity to induce massive cytokine release from both macrophages and T cells by direct binding rather than classical antigen presentation to the major histocompatibility complex class II molecules and the Vβ region of specific T cells (39, 40).
TSST-1 is a potent superantigen and the most common cause of TSS. It is produced exclusively by S. aureus, and approximately 20% of natural isolates are producers. Lindsay et al. suggested that tst is carried by a family of closely related pathogenicity islands that interact in a highly specific way with certain staphylococcal phages, and they stated that this interaction may be responsible for the spread of tst among staphylococcal strains (25).
The Panton-Valentine leukocidin (Luk-PVL) belongs to the family of bicomponent toxins (37). Luk-PVL is associated with skin and soft-tissue infections as well as with more serious infections, e.g., severe necrotizing pneumonia (8, 24).
The aim of this work was to characterize the methicillin-susceptible S. aureus (MSSA) strains at the University Hospital in Magdeburg and the nearby rehabilitation and chronic care facility (RCCF) as a potential source for newly emerging MRSA strains by horizontal genetic exchange and to compare these with the simultaneously circulating MRSA clones. To serve this purpose, we determined their antibiotic resistance phenotypes and used a combination of different molecular typing methods, including multilocus sequence typing (MLST), spa typing, agr specificity, and analysis of their pathogenicity profiles.
MATERIALS AND METHODS
Bacterial isolates, isolate identification, and antibiotic susceptibility testing.
S. aureus strains were isolated and identified from various clinical specimens sent to the Institute for Microbiology at the University of Magdeburg. In this regard, we obtained swabs (n = 70) from different locations (e.g., wound, skin, ear, throat, and peritoneum) as well as secretions from the lung, stomach, peritoneum, and abscesses (n = 12). Colonies that were isolated from the respective specimens and that were yellow on mannitol salt agar (mannitol fermenters) were plated to purity on blood agar and incubated at 37°C in air for 24 h. The isolates were identified as S. aureus on the basis of positive catalase, coagulase, and DNase tests. Final identification and antimicrobial resistance testing were performed with the Phoenix, an automated bacteriology system that performs bacterial identification and susceptibility testing analyses (Becton Dickinson). The results were interpreted in accordance with the Clinical and Laboratory Standards Institute (http://www.clsi.org). In addition, resistance to methicillin was detected on oxacillin resistance screening agar medium (Mueller-Hinton oxacillin [bioMérieux]) and was confirmed by screening for PBP2a (penicillin-binding protein 2a) (Slidex MRSA detection; Denka Seiken). The mecA gene was detected by PCR as described by Murakami et al. (31).
DNA extraction.
Strains were grown on brain heart infusion agar or in the same broth at 37°C overnight. Genomic DNA used as a target for all molecular methods was extracted by using the QIAGEN DNA extraction kit according to the manufacturer's suggestions, with the modification that 20 μl of lysostaphin (1 mg/ml) and 20 μl of lysozyme (100 mg/ml) were added at the cell lysis step. The concentration of DNA was estimated spectrophotometrically.
Multilocus sequence typing (MLST).
MLST was carried out by the methodology described by Enright et al. (10). The allelic profile of S. aureus isolates was obtained by sequencing (using BigDye fluorescent terminators) internal fragments of seven “housekeeping” genes (arcC [carbamate kinase], aroE [shikimate dehydrogenase], glpF [glycerol kinase], gmk [guanylate kinase], pta [phosphate acetyltransferase], tpi [triosephosphate isomerase], and yqiL [acetyl coenzyme A acetyltransferase]) and submitted to the MLST home page (http://www.mlst.net), where seven numbers depicting the allelic profile were assigned that defined the MLST type. For phylogenetic analysis, the relatedness of lineages was displayed as a dendrogram constructed from the matrix of pair-wise differences in allelic profiles by using the unweighted pair-group method with arithmetic averages.
spa typing.
The spa type was received by single-locus DNA sequencing (using BigDye fluorescent terminators) of repeat regions of the Staphylococcus protein A gene (spa) as described by Harmsen et al. (15). Repeats were assigned a numerical code, and the spa type was deduced from the order of specific repeats.
agr group-specific multiplex PCR and toxin gene detection.
Extracted genomic DNA was used as a template to amplify specific agr alleles (GenBank accession numbers X52543, AF001782, AF001783, AF288215, Z49220, AF346724, and AF346725). For multiplex PCR, one primer set was prepared to amplify the four specific S. aureus agr alleles using the primers described by Lina et al. (23). Amplification was carried out under the following conditions: an initial 5-min denaturation step at 95°C, then 25 stringent cycles (1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 72°C), and a final extension step at 72°C for 10 min.
Sequences specific for sea-see, seg-sej, tst, and lukS-lukF were detected by PCR on a PE-9600 thermocycler (Perkin-Elmer). The primers used to detect sea to see, seg to sej, and tst were described by Becker et al. (4, 5). luk-PV genes were detected as described by Lina et al. (24). PCR products were analyzed by electrophoresis through 1.5% agarose gels.
RESULTS
Distribution of MSSA isolates among the different departments.
In total, 82 consecutive MSSA strains were under study. The highest proportions of MSSA clones have been isolated from the department of dermatology (35.37%) and the intensive care units (50.01%). The remaining MSSA strains (14.62%) were distributed among eight different departments equally.
Antimicrobial resistance phenotypes of the MSSA strains.
We primarily examined the resistance of the MSSA isolates (n = 82) to antibiotics of different classes. Our data indicate that 58 (70.7%), 19 (23.2%), 13 (15.9%), 13 (15.9%), and 3 (3.7%) MSSA strains were resistant to penicillin (PEN), ciprofloxacin (CIP), clindamycin (CLI), erythromycin (ERY), and gentamicin (GEN), respectively (data not shown). Eighteen (23%) MSSA isolates showed no resistance to the tested antibiotics.
We next analyzed the resistance phenotypes of the respective MSSA strains (n = 82). As is apparent from Table 1, the majority of MSSA strains (n = 32; 39%) possessed resistance to only PEN. Besides resistance to penicillin, 12 (14.6%) strains were resistant to only one additional antibiotic, 8 (9.75%) strains were resistant to two additional antibiotics, and 5 strains (6.1%) were resistant to three additional antibiotics. One MSSA strain showed a wide resistance pattern to PEN, CIP, CLI, ERY, and GEN. Only six strains were resistant to antibiotics (GEN, CIP, TET-CIP, ERY-CLI, or ERY-CIP-CLI) other than PEN.
TABLE 1.
Resistance phenotypes of MSSA strains during the study period at the Otto-von-Guericke University and the RCCF in Flechtingen, Germanya
| Resistance phenotype | No. (%) of MSSA strains positive for phenotype |
|---|---|
| No resistance | 18 (23) |
| PEN | 32 (39) |
| GEN | 1 (1.22) |
| CIP | 1 (1.22) |
| PEN-CIP | 7 (8.54) |
| PEN-GEN | 1 (1.22) |
| TET-CIP | 2 (2.44) |
| PEN-TET | 4 (4.88) |
| ERY-CLI | 1 (1.22) |
| PEN-TET-CIP | 3 (3.66) |
| PEN-ERY-CLI | 5 (6.1) |
| ERY-CIP-CLI | 1 (1.22) |
| PEN-TET-ERY-CLI | 1 (1.22) |
| PEN-ERY-CIP-CLI | 4 (3.7) |
| PEN-ERY-CIP-CLI-GEN | 1 (1.22) |
| Total | 82 (100) |
Antibiotics given: penicillin (PEN), oxacillin (OXA), ciprofloxacin (CIP), erythromycin (ERY), clindamycin (CLI), gentamicin (GEN), and tetracycline (TET).
Analysis of toxin genes.
The ability of S. aureus to cause a variety of diseases in humans and animals may be attributed to its ability to produce a plethora of virulence factors. Therefore, we analyzed the MSSA strains for the presence of tst, sea to sej, and the Panton-Valentine leukocidin gene (luk-PV). Among the 82 MSSA strains, 8 expressed no enterotoxin gene under study. Eighteen strains were positive for only a single gene of the staphylococcal enterotoxin cluster. Thirty strains harbored two genes simultaneously, 19 strains harbored three genes simultaneously, and 6 strains harbored four genes simultaneously. Only one strain possessed five genes of the enterotoxin gene cluster (Table 2). The latter strain was isolated from an outpatient at the department of dermatology and possessed the luk-PV gene as well. With regard to the enterotoxin genes, the sea gene was detected nine times and the seb gene was detected four times. The sea and seb genes were never detected together. The individual results of the PCR analysis are summarized in Table 2. We next analyzed the presence of another classical superantigen, the tst gene, among the MSSA strains under study. About 40% of MSSA strains (n = 33) possessed the tst gene. All tst-positive MSSA strains were also positive for up to four additional genes of the staphylococcal enterotoxin gene cluster (Table 3). Twenty-one of the 33 tst-positive MSSA strains were predominantly found in the departments of dermatology and in the intensive care units. Due to the fact that most tst-positive MSSA strains were isolated from departments at high risk for MRSA, we performed a detailed analysis of the tst-positive S. aureus strains.
TABLE 2.
Pathogenicity profiles of MSSA strains under study at the Otto-von-Guericke University and the RCCF in Flechtingen, Germany
| Toxin gene(s) | No. of MSSA (n = 82) strains positive for toxin gene(s) |
|---|---|
| None | 8 |
| sea | 3 |
| sed | 1 |
| sei | 13 |
| sej | 1 |
| sec, sei | 4 |
| sed, sei | 4 |
| seg, sei | 18 |
| seh, sei | 1 |
| sei, sej | 3 |
| sea, seg, sei | 4 |
| seb, seg, sei | 1 |
| sec, seg, sei | 5 |
| sed, sei, sej | 5 |
| seg, sei, sej | 4 |
| sea, seg, sei, sej | 2 |
| seb, sed, sei, sej | 2 |
| sec, seg, sei, sej | 1 |
| sec, seh, sei, sej | 1 |
| seb, sed, seg, sei, sej | 1 |
TABLE 3.
Pathogenicity profiles of tst-positive MSSA strains under study at the Otto-von-Guericke University and the RCCF in Flechtingen, Germany
| Toxin genes | No. of MSSA (n = 33) positive for toxin genes |
|---|---|
| sei, tst | 5 |
| sec, sei, tst | 4 |
| sed, sei, tst | 1 |
| seg, sei, tst | 6 |
| seh, sei, tst | 1 |
| sei, sej, tst | 1 |
| sea, seg, sei, tst | 4 |
| sec, seg, sei, tst | 1 |
| sec, seg, sej, tst | 1 |
| sed, sei, sej, tst | 4 |
| seg, sei, sej, tst | 1 |
| sea, seg, sei, sej, tst | 1 |
| seb, sed, sei, sej, tst | 2 |
| sec, seg, sei, sej, tst | 1 |
Clonal relatedness of tst-positive S. aureus isolates. (i) Multilocus sequence typing (MLST).
We determined the genetic backgrounds of tst-positive MSSA strains (n = 21) by multilocus sequence typing (MLST). MLST revealed nine different sequence types (STs) among the 21 MSSA isolates. In this regard, we detected 1, 6, 1, 2, 5, 2, 1, 2, and 1 strain of ST1, ST8, ST30, ST39, ST45, ST101, ST121, ST395, and ST426, respectively. These results indicate a high heterogeneity among the MSSA isolates with regard to their MLST types.
(ii) Phylogenetic analysis of and relationship between strains with different ST patterns.
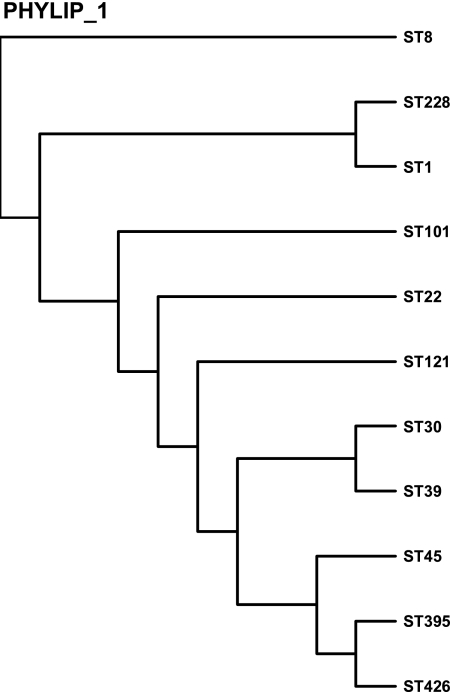
In order to analyze the relationship among strains with different ST patterns, a dendrogram (Fig. 1) was constructed; zero genetic distance corresponds to identical ST patterns. Each terminal branch of the dendrogram represents a sequence type (ST). For comparison, we added MRSA clones of MLST ST22, ST45, and ST228, which are the major MRSA clones at the University Hospital in Magdeburg (12). As the S. aureus isolates were compared in a dendrogram tree, three major clusters were identified. ST8 belongs to cluster I, while ST1 and ST228 belong to cluster II. The third cluster is heterogenous and accounts for ST101, ST22, ST121, ST30, ST39, ST45, ST395, and ST426. According to the denodrogram, ST426, ST395, and ST45 clones of cluster III are more closely related to each other than to other cluster III clones, whereas ST30 and ST39 clones, cluster more closely. ST121 clones seem to be not directly related to the other members of cluster III.
FIG. 1.
Dendrogram constructed to analyze the clonal relatedness of the tst-positive MSSA strains.
(iii) spa typing.
The spa gene of S. aureus encodes protein A and was used for the characterization of MSSA isolates. The analysis of the spa gene sequences revealed different repeats (Ridom StaphType). Among the 21 MSSA isolates we detected, 14 different spa types were detected: t008, t012, t015, t019, t024, t056, t065, t127, t133, t162, t271, t287, t399, and t400. The results are presented in Table 4. It is apparent from Table 4 that a distinct MLST type could also cover different spa types. In this regard, ST8 covered spa types t008, t024, and t400, and ST45 belonged to spa types t015, t065, and t133. The remaining MLST types—ST1, ST30, ST39, ST101, ST121, ST395, and ST426—were each assigned to one spa type only: t127, t012, t399, t056, t162, t287, and t271, respectively.
TABLE 4.
Analysis of spa types and the corresponding MLST types for the different MSSA strains
| spa type (n) | No. of S. aureus strains of MLST type:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ST1 | ST8 | ST30 | ST39 | ST45 | ST101 | ST121 | ST395 | ST426 | |
| t008 (2) | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| t012 (1) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| t015 (2) | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| t019 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| t024 (2) | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| t056 (2) | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| t065 (2) | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| t127 (1) | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| t133 (1) | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| t162 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| t271 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| t287 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| t399 (2) | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| t400 (2) | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
(iv) Analysis of agr.
Variation in agr specificity type has been proposed as a possible influence on population dynamics of S. aureus (23). Therefore, we investigated the agr specificity groups of the different tst-positive MSSA strains by PCR using agr group-specific primers.
As is apparent from Table 5, the tst-positive MSSA strains belonged to agr types I, II, III, and IV. All ST8, ST101, and ST395 strains expressed agr type I. Among the five strains of ST45, the agr type of one stain was detected as type IV, while the other four strains belonged to agr type I. The agr type of the ST121 isolate was detected as type IV, and the ST426 isolate expressed agr type II. ST1 and ST30 isolates belonged to the new emerging American clones, which were detected among community-acquired MRSA (CA-MRSA) strains, harboring agr type III. The ST39 strains also expressed agr type III.
TABLE 5.
Analysis of MLST types and the corresponding accessory gene regulator (agr) types for the different MSSA strains
| MLST (n) | No. of S. aureus isolates of agr type:
|
|||
|---|---|---|---|---|
| I | II | III | IV | |
| ST1 (1) | 0 | 0 | 1 | 0 |
| ST8 (6) | 6 | 0 | 0 | 0 |
| ST30 (1) | 0 | 0 | 1 | 0 |
| ST39 (2) | 0 | 0 | 2 | 0 |
| ST45 (5) | 4 | 0 | 0 | 1 |
| ST101 (2) | 2 | 0 | 0 | 0 |
| ST121 (1) | 0 | 0 | 0 | 1 |
| ST395 (2) | 2 | 0 | 0 | 0 |
| ST426 (1) | 0 | 1 | 0 | 0 |
DISCUSSION
MRSA is still a dominant hospital-associated pathogen (h-MRSA). However, there are ongoing changes in the epidemiology of MRSA. In former times, MRSA strains were clonal and there were only a few epidemic strains; MRSA strains are now more heterogenous. In addition, there is an evolution of so-called community-acquired MRSA (CA-MRSA) with characteristics distinct from those of the traditional h-MRSA.
Genetically, methicillin-resistant S. aureus (MRSA) is produced when methicillin-susceptible S. aureus (MSSA) acquires a mobile genetic element, staphylococcal cassette chromosome mec (SCCmec). Toxin-producing MSSA may also alter the pathogenicity of established MRSA by the transfer of virulence factors via plasmids or mobile elements. It is hypothesized that the evolution of CA-MRSA is a recent event due to the acquisition of mec DNA by previously methicillin-susceptible strains that circulated in the community. Thus, besides the tracking of MRSA dissemination, we need precise knowledge about the circulating MSSA strains and have to monitor the pathogenicity profiles of MRSA and MSSA strains. This study investigated the heterogeneity of MSSA at the University Hospital in Magdeburg, Germany.
It is well known that MRSA prevalence varies almost 100-fold around the world. Quite recently we described that among the S. aureus isolates collected over a 1-year period at the University Hospital of Magdeburg, Germany, approximately 7.3% were classified as MRSA by laboratory analysis (12). The highest prevalence of MRSA has been in the departments of dermatology and the intensive care units of anesthesiology and surgery (12). In accordance with this finding, in this study the highest levels of MSSA isolated from clinical specimens were from the respective departments as well.
Until 1995, most MRSA strains from around the world, including Germany, exhibited multiresistance phenotypes (45). Meanwhile, the resistance phenotypes of MRSA strains have changed, and we also described a narrowing of the resistance pattern in epidemic MRSA strains quite recently at the University Hospital in Magdeburg (12, 43). In this regard, we also detected Barnim (ST22) and southern German (ST228) epidemic MRSA strains which were resistant to only one, two, or no further antibiotics beside penicillin and oxacillin. The Berlin (ST45) epidemic MRSA showed a minor resistance pattern as well (12). A rising number of CA-MRSA strains show low-level resistances as well (44). Among the 82 MSSA strains in this study, most isolates possessed resistance to only penicillin, followed by strains with resistance to one or two more antimicrobial substances. Thus, the capture of the mecA gene, probably through SCCmec cassette type IV, will lead to MRSA strains with narrowed susceptibility profile. In our study, we show a penicillin resistance of 70.7%. These data are in good concordance with the data from the GENARS (German Network for Antimicrobial Resistance Surveillance) project (http://www.genars.de). From 2002 up to now, approximately 73% of all clinical Staphylococcus aureus isolates analyzed during the GENARS project (n = 4,200) exhibited penicillin resistance. However, surveillance data may differ in one country. In this regard, the data from the Antimicrobial Surveillance Study of the Paul Ehrlich Society for Chemotherapy (http://www.p-e-g.org) from 2004 showed a resistance of 76.7% against penicillin. However, only 841 Staphylococcus aureus strains were under study. With regard to the levels of susceptibility to gentamicin, erythromycin, clindamycin, and ciprofloxacin, our data are similar to those obtained by the Paul Ehrlich Society for Chemotherapy in 2004. In summary, with the exception of methicillin resistance, both MRSA and MSSA strains are heterogenous in their susceptibility patterns.
The relative virulence of MRSA and fully methicillin-susceptible S. aureus has been scrutinized. The majority of studies support the concept that MSSA and MRSA strains have equivalent potentials for colonization and causing disease. In general, bacteremia isolates of S. aureus often contain these classical members of the superantigen family, isolates from patients with diarrhea carry seb, and isolates from wound infections harbor the sec gene (40). A large number of S. aureus strains isolated from furuncles and carbuncles produce Panton-Valentine leukocidin. In addition to the enterotoxins, toxic shock syndrome toxin 1 (TSST-1) of S. aureus is associated with septic shock and toxic shock syndrome (39).
In the present study, we detected only one luk-PV-positive MSSA strain. Only a small percentage of MSSA strains harbored the gene sea, seb, or sec, the classical staphylococcal enterotoxin genes. In contrast, we detected in nearly all MSSA strains the sei gene, which belongs to the egc cluster. Surprisingly, about 40% of the MSSA strains under study possessed the tst gene. In contrast to other reports, we detected two tst-positive strains (n = 22) which possessed the seb gene as well (6). Interestingly, the tst-positive MSSA strains were recovered from the departments with the highest MRSA rates (dermatology and intensive care units of anesthesiology and surgery).
Up to now, we rarely detected tst-positive MRSA strains, mainly ST22 and ST228, at the University Hospital in Magdeburg. tst-positive MRSA strains belonged to the well-known epidemic MLST types ST22, ST45, and ST228 and were classified as spa types t001, t028, and t032 (data not shown). From the literature, it is known that in Europe the epidemic MRSA strain EMSRA-16 sometimes harbored the tst gene. Other reports about TSST-1 production by MRSA strains exist as well (38), although there are no available MLST data for the respective strains.
In this context, understanding the epidemiology of TSST-1-producing MSSA is clinically important because of the rare but potentially devastating symptoms caused by toxic shock syndrome toxin 1 (TSST-1). Thus, one has to keep in mind the emergence of CA-MRSA harboring the tst gene. There has been considerable speculation about the origin and evolution of the MRSA strains. According to Fitzgerald et al., MRSA strains have arisen multiple independent times by lateral transfer of the mec elements into methicillin-susceptible precursors (11). Up to now, the mec gene has been found to be present in up to eight distinct S. aureus lineages that are highly differentiated in terms of overall chromosomal gene content. Presently there are further changes occurring in the emergence and spread of epidemic MRSA in German hospitals (22, 44, 45, 46). MLST has been used to study the evolution of pandemic clones of MRSA (9, 10, 36). We recently described for the University Hospital in Magdeburg that the most abundant types were two of the newly emerging MRSA clones, the Barnim epidemic MRSA (ST22) and the southern German epidemic MRSA (ST228). In contrast to other parts of Germany, the Berlin epidemic MRSA (ST45) was less abundant in Magdeburg (12).
Thus, overall changes in the prevalence and spread of different epidemic MRSA strains are observed, and thus the emergence of new MRSA clones may be expected. In this study, the tst-positive MSSA strains were further characterized according to their MLST and spa types. In our study, the TSST-1-producing MSSA isolates represent a heterogenous group covering different STs and spa types. STs (sequence types) of the dominant MRSA clones at the University Hospital in Magdeburg during the last 3 years (ST22, ST45, and ST228) were scarcely found or were not found among the present MSSA collection. These results are in agreement with the findings described by Aires de Sousa et al. (1). In contrast to Aires de Sousa et al., we do not conclude from our data that the introduction of SCCmec into susceptible clones is most likely a relatively infrequent event. In our study, we detected MLST types ST1, ST8, and ST30 among the tst-positive MSSA strains. MLST analysis indicated distinct genetic backgrounds for the arising CA-MRSA strains associated with each geographic origin, namely ST80 in Germany and France (17, 42), ST1, ST8, and ST59 in the United States (35, 29), and ST30 in Australia (7). Among the MSSA strains in this study, we have not detected any MSSA sharing the background of the major European CA-MRSA clone, ST80. We detected MSSA isolates of ST1, ST8, ST30, and ST39. ST30 was recently reported to have spread in the community in Europe. ST39 was associated with MSSA and MRSA in Australia (http://www.mlst.net). Thus, we conclude from our results that similar to luk-PV-positive MSSA strains, tst-positive MSSA strains, at least of MLST types ST1, ST8, and ST30, have the potential to capture the mecA gene and are thus a potential source of tst-positive CA-MRSA strains. Whether the additional MLST types may acquire the mecA gene is not known. Moreover, many S. aureus accessory genes carry virulence factors such as tst. These genes are often carried on mobile elements, such as phages and pathogenicity islands, which transfer horizontally between strains (sea, tst, and eta) (30). Thus, it is assumed that the transfer of the tst gene can occur at an extremely high frequency (25).
agr specificity type has been proposed as a possible influence on population dynamics in S. aureus (23). In agreement with Witte (41), we recently determined agr group II for the epidemic MRSA ST228 clone (southern German) and agr group I for the newly emerging ones, ST22 and ST45 strains, all circulating at the University Hospital in Magdeburg (12). We analyzed the agr specificity groups of the different tst-positive MSSA strains as a further contribution to the genotypic characterization of the MSSA isolates. Ji et al. argue that the presence of the tst gene in S. aureus is coupled to agr type III (18). In contrast, in our study all agr types were distributed among the different tst-positive MSSA strains. These results support the findings which were observed by Moore and Lindsay (30). Thus, we provide evidence that the presence of the tst gene is not coupled to a specific agr type, at least in MSSA. However, one feature of the described newly emerging CA-MRSA ST1 and ST30 clones is the presence of agr type III. Indeed, in our study the tst-positive MSSA strains of MLST types ST1, ST30, and ST39 belonged to agr group III. In contrast to the CA-MRSA strains of MLST type ST8 described in the literature, we detected only agr group II in our ST8 isolates (n = 6).
Through the multitude of applied methods, our data contribute to a more precise knowledge of the heterogeneity of MSSA in a clinical setting. In summary, we observed a heterogeneity of tst-positive MSSA clones with regard to MLST, to spa typing, to toxin profiles, and to antibiotic resistance patterns. Interestingly, tst-positive MSSA strains were predominantly found in the areas with a high incidence of MRSA, the department of dermatology and the intensive care units. Although it is not known if horizontal transfer of the tst gene occurs in clinical settings, care must be taken, and further investigations on virulence gene transfer must be conducted.
REFERENCES
- 1.Aires de Sousa, M., T. Conceicao, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 43:5150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer, G. L., and D. M. Niemeyer. 1994. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 2:343-347. [DOI] [PubMed] [Google Scholar]
- 3.Beck, W. D., B. Berger-Bachi, and F. H. Kayser. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J. Bacteriol. 165:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, K., A. W. Friedrich, G. Lubritz, M. Weilert, G. Peters, and C. von Eiff. 2003. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J. Clin. Microbiol. 41:1434-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohach, G. A., D. J. Fast, R. D. Nelson, and P. M. Schlievert. 1990. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit. Rev. Microbiol. 17:251-272. [DOI] [PubMed] [Google Scholar]
- 7.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, and P. M. Giffard. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diep, B. A., G. F. Sensabaugh, N. S. Somboona, H. A. Carleton, and F. Perdreau-Remington. 2004. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leukocidin. J. Clin. Microbiol. 42:2080-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghebremedhin, B., W. König, and B. König. 2005. Heterogeneity of methicillin-resistant Staphylococcus aureus strains at a German university hospital during a 1-year period. Eur. J. Clin. Microbiol. Infect. Dis. 24:388-398. [DOI] [PubMed] [Google Scholar]
- 13.Gillet, Y., B. Issartel, P. Vanhems, G. Lina, F. Vandenesch, J. Etienne, and D. Floret. 2001. Severe staphylococcal pneumonia in children. Arch. Pediatr. 8(Suppl. 4):742s-746s. [DOI] [PubMed] [Google Scholar]
- 14.Hanssen, A. M., G. Kjeldsen, and J. U. Sollid. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiramatsu, K., T. Ito, and H. Hanaki. 1999. Evolution of methicillin and glycopeptide resistance in Staphylococcus aureus, p. 221-242. In R. G. Finch and R. J. Williams (ed.), Bailliere′s clinical infectious disease. Bailliere Tindall, London, United Kingdom.
- 17.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 19.Jarraud, S., G. Cozon, F. Vandenesch, M. Bes, J. Etienne, and G. Lina. 1999. Involvement of enterotoxins G and I in staphylococcal toxic shock syndrome and staphylococcal scarlet fever. J. Clin. Microbiol. 37:2446-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166:669-677. [DOI] [PubMed] [Google Scholar]
- 21.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lelièvre, H., G. Lina, M. E. Jones, C. Olive, F. Forey, M. Roussel-Delvallez, M.-H. Nicolas-Chanoine, C. M. Bébéar, V. Jarlier, A. Andremont, F. Vandenesch, and J. Etienne. 1999. Emergence and spread in French hospitals of methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J. Clin. Microbiol. 37:3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay, J. A., A. Ruzin, H. F. Ross, N. Kurepina, and R. P. Novick. 1998. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29:527-543. [DOI] [PubMed] [Google Scholar]
- 26.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-522. [DOI] [PubMed] [Google Scholar]
- 27.McCormick, J. M., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 55:77-104. [DOI] [PubMed] [Google Scholar]
- 28.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van der Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishaan, A. M., E. O. Mason, Jr., G. Martinez-Aguilar, W. Hammerman, J. J. Propst, J. R. Lupski, P. Stankiewicz, S. L. Kaplan, and K. Hulten. 2005. Emergence of a predominant clone of community-acquired Staphylococcus aureus among children in Houston, Texas. Pediatr. Infect. Dis. J. 24:201-206. [DOI] [PubMed] [Google Scholar]
- 30.Moore, P. C. L., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-susceptible Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39:2760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami, K., W. Minamide, K. Wada, E. Nakamura, H. Teraoka, and S. Watanabe. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nilsson, H., P. Bjork, M. Dohlsten, and P. Antonsson. 1999. Enterotoxin H displays unique MHC class II-binding properties. J. Immunol. 163:6686-6693. [PubMed] [Google Scholar]
- 33.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 34.Omoe, K., M. Ishikawa, Y. Shimoda, D. L. Hu, S. Ueda, and K. Shinagawa. 2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates harboring seg, seh, or sei genes. J. Clin. Microbiol. 40:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan, E. S., B. A. Diep, E. D. Charlebois, C. Auerswald, H. A. Carleton, G. F. Sensabaugh, and F. Perdreau-Remington. 2005. Population dynamics of nasal strains of methicillin-resistant Staphylococcus aureus and their relation to community-associated disease activity. J. Infect. Dis. 192:811-818. [DOI] [PubMed] [Google Scholar]
- 36.Peacock, S. J., G. D. I. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. J. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prevost, G., P. Couppie, P. Prevost, S. Gayet, P. Petiau, B. Cribier, H. Monteil, and Y. Piemont. 1995. Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J. Med. Microbiol. 42:237-245. [DOI] [PubMed] [Google Scholar]
- 38.Schmitz, F. J., C. R. MacKenzie, R. Geisel, S. Wagner, H. Idel, J. Verhoef, U. Hadding, and H. P. Heinz. 1997. Enterotoxin and toxic shock syndrome toxin-1 production of methicillin resistant and methicillin susceptible Staphylococcus aureus strains. Eur. J. Epidemiol. 13:699-708. [DOI] [PubMed] [Google Scholar]
- 39.Uchiyama, T., T. Tadakuma, K. Imanishi, M. Araake, S. Saito, X. J. Yan, H. Fujikawa, H. Igarashi, and N. Yamaura. 1989. Activation of murine T cells by toxic shock syndrome toxin-1. The toxin-binding structures expressed on murine accessory cells are MHC class II molecules. J. Immunol. 143:3175-3182. [PubMed] [Google Scholar]
- 40.Uchiyama, T., X. J. Yan, K. Imanishi, and J. Yagi. 1994. Bacterial superantigens: mechanism of T cell activation by the superantigens and their role in the pathogenesis of infectious diseases. Microbiol. Immunol. 38:245-256. [DOI] [PubMed] [Google Scholar]
- 41.Witte, W. 2004. International dissemination of antibiotic resistant strains of bacterial pathogens. Infect. Genet. Evol. 4:187-191. [DOI] [PubMed] [Google Scholar]
- 42.Witte, W., C. Braulke, C. Cuny, B. Strommenger, G. Werner, D. Heuck, U. Jappe, C. Wendt, H. J. Linde, and D. Harmsen. 2005. Emergence of methicillin-resistant Staphylococcus aureus with Panton-Valentine leukocidin genes in central Europe. Eur. J. Clin. Microbiol. Infect. Dis. 24:1-5. [DOI] [PubMed] [Google Scholar]
- 43.Witte, W., C. Braulke, D. Heuck, and C. Cuny. 2000. Methicillin resistant Staphylococcus aureus in German hospitals develop narrower patterns of antimicrobial resistance. Eur. Surveill. 5:31-34. [DOI] [PubMed] [Google Scholar]
- 44.Witte, W., C. Cuny, B. Strommenger, C. Braulke, and D. Heuck. 2004. Emergence of a new community acquired MRSA strain in Germany. Eur. Surveill. 9:1-2. [DOI] [PubMed] [Google Scholar]
- 45.Witte, W., C. Cuny, C. Braulke, D. Heuck, and I. Klare. 1997. Widespread dissemination of epidemic MRSA in German hospitals. Eur. Surveill. 2:25-28. [DOI] [PubMed] [Google Scholar]
- 46.Witte, W., M. Enright, F. J. Schmitz, C. Cuny, C. Braulke, and D. Heuck. 2001. Characteristics of a new epidemic MRSA in Germany ancestral to United Kingdom EMRSA 15. Int. J. Med. Microbiol. 290:677-682. [DOI] [PubMed] [Google Scholar]