Abstract
The genetic diversity of group A streptococci (GAS) throughout much of the world has not been adequately explored. To assess genetic variation among GAS in western Nepal, 120 noninvasive GAS, collected from eight different villages, were genetically characterized using emm typing, sof sequencing, and multilocus sequence typing (MLST). A high level of genetic diversity was observed among these isolates, with 51 genotypes based upon 51 multilocus sequence types (STs), 45 emm sequence types, and 28 sof sequence types. On the basis of shared ST-emm and sof-emm associations, 40 of the 51 genotypes were identical or highly related to genotypes characterized from locations outside of Nepal, even though most of the emm sequence and clonal types are rare among GAS within the United States. When analyzing all known STs highly related to Nepal STs, only one example of similar STs shared between a sof PCR-positive strain and a sof PCR-negative strain was observed. Since previous data indicate free exchange of MLST loci between sof-positive and sof-negative strains, there is possibly selection against the expansion of subclones resulting from horizontal transfers of sof or emm genes between sof-positive and sof-negative strains. All 45 emm types encountered in Nepal have also been documented from other countries. These data, together with data encompassing the past decade of emm type surveillance, support the possibility that most existing GAS emm types have been discovered. Similarly, since most (40/51) strain types were highly related to strains found elsewhere, it is likely that a major fraction of the existing GAS clonal complexes have been discovered.
For the eventual introduction of vaccines, especially those including multiple M protein protective epitopes, the global distribution of M protein types and genotypes from group A streptococci (GAS) should be explored. During the past 10 years the number of documented M protein gene (emm) types has expanded markedly due to the ease of characterizing emm sequence types of newly encountered strains. Currently, there are more than 170 emm types and 750 emm subtypes known from group A streptococci (4). Although emm types are good predictors of clonal types within a given geographic location (8), there is considerable genetic diversity within certain emm types, especially when comparing strains from different countries (1, 18).
GAS are subdivided into two large divisions by virtue of either harboring or lacking the sof gene, which maps approximately 10 kb upstream of the emm gene and encodes a hypervariable cell surface-bound virulence factor that is usually associated with one principle emm type (1, 5, 15, 19). In earlier work we demonstrated that sof gene-positive strains of a given emm type can be associated with multiple other emm-sof gene associations, indicating that sof-positive strains exchange emm and sof loci (1). Interestingly, while there appears to be free exchange of multilocus sequence typing (MLST) alleles between sof-positive and sof-negative strains (11, 16), we are not aware of evidence indicating past horizontal exchange of sof or emm loci between sof-negative and sof-positive GAS.
sof-negative strains are largely divided into pattern A to C and pattern D strains, based upon the organization of emm and similar emm-like genes at the mga locus (2, 3). Pattern A to C strains are common within the United States and relatively rare in tropical countries. Pattern D strains are rare within the United States and common within tropical countries (2). Most known sof-positive strains are mga locus pattern E strains (18). There are previous data suggesting that pattern A to C strains and pattern D strains primarily colonize at the throat and skin sites, respectively (2, 3, 7). All or most pattern E isolates are sof positive, and data suggest no site preferences for colonization (2, 3, 7). Here we describe 120 noninvasive isolates collected from skin impetigo lesions and the oropharynx of asymptomatic children in far-western Nepal. We did not see compelling site associations for the different mga locus patterns.
Although we have observed a great range of genetic diversity during analysis of Streptococcus pyogenes over the past 10 years recovered from many different geographic locations (1, 4, 18), there are indications that we have already sampled a significant portion of this diversity. For example, while we routinely encountered new emm types 5 to 10 years ago, new emm types are now only rarely discovered (4; personal observations). In this study of noninvasive GAS isolates recovered in remote villages in Nepal, we did not encounter a single new emm type that we have not also found in other countries. Furthermore, we found that the majority of strains characterized in this study were highly genetically related to strains encountered elsewhere.
MATERIALS AND METHODS
Isolates.
This work was performed as a follow-up to a previous study (12, 13) involving eight villages in the districts of Kailali and Kanchanpur, in the Seti and Mahakali zones in far-western Nepal. The study included the administration of a single dose of azithromycin (20 mg/kg) in November 1998 and also in November 1999 to all village children aged 1 to 10 years.
Sixty children from each village were randomly selected. During November 2000 oropharyngeal swabs were collected from all children and skin swabs were collected from those with clinically diagnosed impetigo lesions. Impetigo lesions were cleaned and left intact for swabbing. Swabs were transported in silica packets at room temperature to the Centers for Disease Control and Prevention (CDC), where they were cultured and subjected to serologic and phenotypic testing by established methods (23). GAS were isolated from 67 (17%) of 399 oropharyngeal swabs and 53 (58%) of 92 impetigo lesions. In total, 120 throat and skin lesion GAS isolates were recovered from 111 individuals and included in this analysis. Five children yielded isolates from both skin and throat that differed in emm type, and four children yielded isolates from both sites with identical emm subtypes. All nine of these isolates were included in the analysis. One additional child yielded two different emm subtypes from two throat culture isolates which were also included. Seven children yielded two isolates with identical emm subtypes recovered from different individual impetigo lesions. The seven duplicate lesion isolates were not included in the analysis.
Antibiotic susceptibility testing.
MICs to erythromycin and tetracycline were determined by standard disk diffusion and E test techniques.
Genotyping.
Multilocus, emm, and sof sequence typing was performed as previously described (1, 4, 8, 21). All isolates were subjected to emm and sof sequence typing when amplicon positive. sof PCR-negative isolates sharing the same emm subtype and T agglutination pattern were grouped within the same multilocus sequence type (ST) obtained from one or two representative isolates. sof-positive isolates sharing the same emm subtype, T agglutination pattern, and sof sequence type were assigned the same ST, which was obtained from one to three representative isolates. To verify the probability that such patterns represented the same ST within all isolates of a given subtype and T agglutination profile, one to seven MLST target sequences were obtained from additional independent isolates. Multilocus sequence data were analyzed using eBURST (10) and the 319 STs in the database as of 16 August 2005 (21).
M protein sequence dendrogram.
Sequences consisting of 22 signal sequence residues plus 66 N-terminal residues that encompass the 50-residue emm type/subtype-determining region according to the CDC emm typing scheme (4) were aligned using the Wisconsin Package version 10.3 PileUp program (gap creation penalty of 8 and gap extension penalty of 2) followed by the Distances program (with the uncorrected distance option; 1,103 of 1,128 pairs had distances between 30.0 and 100.0.) and finally the GrowTree program (neighbor-joining method). These data were entered into the TreeView program downloaded from http://taxonomy.zoology.gla.ac.uk/rod/treeview.html to generate an unrooted tree.
RESULTS
Genetic diversity of Nepal isolates and comparison with GAS recovered in the United States.
We found a remarkable degree of genetic diversity among the 120 isolates characterized in this study, including 51 multilocus sequence types and 45 distinct emm types (Table 1). Emphasizing this diversity is the observation that in general the genotypes that were the same did not cluster by village. This markedly contrasts with pharyngitis surveillance carried out in the United States, where only 35 emm types accounted for 1,975 isolates (20). Similarly, in recent invasive surveillance, only 41 emm types were found among 1,061 isolates (17). Twenty-one of the 45 emm types found among Nepal isolates were not found among the combined 3,036 invasive and noninvasive U.S. GAS isolates found in recent invasive and noninvasive GAS disease surveillance, and 10 additional types were found within less than 1% of both invasive and noninvasive U.S. GAS surveillance isolates (Table 2).
TABLE 1.
Characteristics of GAS isolates from eight villages in Nepal
| emm subtype (T pattern) | No. of isolates
|
mga locus patterna | Village(s) | ST | sof type (other sof type[s] linked with emm typeb) | Other locations with related strainsc | |
|---|---|---|---|---|---|---|---|
| Throat | Skin | ||||||
| 1.0 (1) | 0 | 9 | A-C | e, f | 28 | Negative | United States, Chile, Brazil, India, Poland, Czech Republic, United Kingdom |
| 4.0 (4) | 0 | 7 | E | d, f | 39 | 4 (2920) | United States, Germany, Czech Republic, United Kingdom |
| 4.2 (8/25) | 1 | 0 | E | a | 289 | 2920 (4) | Australia, Brazil |
| 11.0 (11) | 3 | 1 | E | b, e | 999 | 25 (11) | United States (DLV), United Kingdom (DLV) |
| 22.8 (2) | 2 | 0 | E | h | 360 | 174901 (22) | None known |
| 25.2 (14) | 2 | 1 | E | e | 350 | 4470 (25, 4958) | United States |
| 43.3 (3/13) | 0 | 1 | D | b | 343 | Negative | None known |
| 44.0 (11/12) | 3 | 3 | E | b, e, g, h | 351 | 3930 (44, 61) | Australia, Brazil |
| 49.4 (12) | 1 | 0 | E | h | 228 | 66 (49) | India |
| 53.0 (3/13/B) | 1 | 0 | D | a | 344 | Negative | Trinidad (DLV), United States (DLV) |
| 53.0 (13/1) | 0 | 2 | D | d | 363 | Negative | None known |
| 55.0 (NT) | 1 | 0 | A-C | f | 100 | Negative | Trinidad, United Kingdom |
| 56.0 (3/13) | 1 | 1 | D | g, h | 355 | Negative | Australia |
| 57.0 (3) | 2 | 1 | A-C | a, b | 348 | Negative | None known |
| 63.3 (6) | 0 | 1 | E | h | 297 | 76301 | Australia, United Kingdom |
| 63.4 (3/2/5/4) | 1 | 0 | E | E | 359 | 107 | Malaysia |
| 65.0 (3/13) | 1 | 1 | D | g | 129 | Negative | Egypt |
| 66.0 (4) | 2 | 0 | E | d, f | 249 | 137001 (66) | India, United Arab Emirates |
| 67.0 (3/13) | 2 | 0 | D | e, g | 267 | Negative | United Arab Emirates |
| 68.0 (1/13) | 0 | 1 | E | h | 361 | 68 (4438, 4470) | Egypt, United States |
| 70.0 (3/13) | 0 | 1 | D | a | 24 | Negative | Japan |
| 71.0 (14) | 3 | 3 | D | b, f, g, h | 130 | Negative | Egypt |
| 73.0 (3/13) | 4 | 0 | E | c | 331 | 73 | United Kingdom, United States |
| 74.0 (13) | 2 | 0 | D | e | 120 | Negative | Australia |
| 75.1 (NT) | 1 | 0 | E | e | 230 | 170901 (75, 4958) | United Arab Emirates |
| 75.1 (2/8/14) | 1 | 0 | E | g | 357 | 94 (94) | None known |
| 77.0 (2) | 3 | 1 | E | a, c, d, f | 347 | 131501 (77, 27L) | Poland (SLV) |
| 78.0 (3/5/27/44) | 1 | 1 | E | b, e | 181 | 170201 | Australia |
| 79.1 (12) | 5 | 0 | E | a, f | 362 | 130501 (79) | None known |
| 82.1 (3/13) | 4 | 1 | E | b, f, h | 320 | 74801 (82) | Egypt |
| 85.0 (3/13/B) | 0 | 2 | E | c, h | 109 | 76 | Australia, Trinidad, United States |
| 88.1 (NT) | 1 | 0 | E | c | 352 | 87 (88) | None known |
| 89.0 (13) | 1 | 2 | E | d | 356 | 136501 (89, 4835) | None known |
| 97.1 (NT) | 1 | 0 | D | b | 345 | Negative | None known |
| 100.1 (6/28) | 1 | 3 | D | a, f, h | 119 | Negative | Australia |
| 102.2 (11/12) | 3 | 1 | E | g, h | 349 | 102 | Australia, United States |
| 103.0 (13/B) | 0 | 1 | E | b | 201 | 103 | New Guinea, Malaysia, Brazil, United States |
| 104.0 (3/B) | 1 | 0 | E | c | 353 | 104 | Egypt |
| 105.0 (NT) | 1 | 0 | D | g | 151 | Negative | Malaysia |
| 106.0 (3/5/27/44) | 0 | 1 | E | b | 140 | 106 | Malaysia |
| 106.1 (3/5/27/44) | 1 | 0 | E | b | ND | 106 | Malaysia |
| 111.1 (NT) | 2 | 0 | D | f, g | 214, 346e | Negative | United Arab Emirates, Japan (DLV) |
| 116.1 (NT) | 1 | 0 | D | c | 227 | Negative | United Arab Emirates |
| 118.0 (12) | 2 | 0 | E | d, e | 354 | 118 | United Kingdom (DLV), United States (DLV) |
| 119.2 (NT) | 1 | 0 | D | f | 237 | Negative | United Arab Emirates |
| 122.0 (12) | 0 | 2 | D | c | 342 | Negative | None known |
| st1389.0 (13/B) | 1 | 1 | E | a, e | 358 | 1389 | United States |
| st1731.0 (12) | 2 | 0 | E | f, g | 303 | 1731 | None known |
| st2147.0 (8/25/I) | 0 | 2 | E | a | 169 | 2147 | Egypt |
| st6030.0 | 0 | 2 | D | a, c | 294 | Negative | United States |
| 1 | 0 | D | e | 224 | Negative | United Arab Emirates, Egypt (SLV) | |
| Total | 67 | 53 | |||||
See reference 4 for mga locus pattern information.
Strains with the same emm type combined with a related ST and/or the same sof type were considered related. STs are considered related if they share five to seven identical alleles; STs from the indicated locations are identical unless indicated in parentheses. SLV, single-locus variant; DLV, double-locus variant.
Entries in bold indicate there is the same emm-sof association as for the indicated Nepal isolates without accompanying MLST data.
STs 214 and 346 are SLVs associated with the emm111.1 isolates.
TABLE 2.
Proportion of Nepal emm types among recent U.S. invasive and pharyngitis GAS surveillance isolates
| Nepal emm type | % of 1,061 U.S. invasive isolatesa | % of 1,975 U.S. pharyngitis isolatesb |
|---|---|---|
| 1 | 18.2 | 19.0 |
| 4 | 2.4 | 8.2 |
| 11 | 3.4 | 1.0 |
| 22 | 2.4 | 2.6 |
| 25, 55, 56, 63, 67, 70, 74, 79, 85, 88, 97, 100, 105, 106, 111, 116, 119, 122, st1389, st1731 | 0 | 0 |
| 43, 65, 66, 71, 104 | <1 | 0 |
| 44 | 1.0 | 2.1 |
| 49 | 1.5 | 0 |
| 53, 68, 78, 118 | <1 | <1 |
| 57 | 0 | <1 |
| 73 | 2.2 | <1 |
| 77 | 3.6 | 3.8 |
| 82 | 5.9 | <1 |
| 89 | 5.5 | 4.6 |
| 102 | 1.1 | <1 |
According to existing MLST and sof sequence data, several clonal types found in this study are rare in the United States, even though they were associated with emm types routinely found within the United States (1, 4; B. Beall, unpublished observations). These clonal types included ST289 (emm4), ST351 (emm44), STs 230 and 357 (emm75), ST347 (emm77), ST320 (emm82), and ST356 (emm89). As is common in other tropical or subtropical locations (2), there were many more projected pattern D strains than there were pattern A to C strains (Table 3).
TABLE 3.
Association of projected mga locus patterns of Nepal isolates with skin and throat recovery sites
| mga locus pattern | No. of emm types | No. of throat isolates | No. of skin isolates | Total no. of isolates |
|---|---|---|---|---|
| A to C | 3 | 3 | 10 | 13 |
| D | 17 | 18 | 16 | 34 |
| E | 25 | 46 | 27 | 73 |
| Total | 45 | 67 | 53 | 120 |
emm types found among skin lesion and asymptomatic throat isolates recovered in Nepal.
Despite the rarity of the majority of emm types found in this study among GAS isolates recovered in the United States (Table 2), 44 of the 45 distinct emm types found in Nepal had been previously documented (4). The emm sequence type st1731 was documented from isolates recovered in other countries subsequent to this study (4).
Overlapping clonal relationships of Nepal strains with strains recovered elsewhere.
Thirty-four of the 51 STs derived from noninvasive isolates in Nepal (Tables 1 and 2) were highly related to STs from strains recovered elsewhere that share the same emm types and/or sof types (Table 1). Six additional genotypes displayed the same emm type-sof type associations as strains recovered elsewhere, indicating that 40 of these 51 genotypes are highly similar to previously characterized GAS genotypes. The remaining 11 genotypes had emm type associations with multilocus and/or sof sequence types that have not been documented elsewhere. Although st1731 was first observed among the two isolates described here (Table 1), multilocus sequence types and/or sof sequence types are not yet available for a group of subsequent isolates with new st1731 subtypes in the CDC emm database that were recently discovered in the United States, Belgium, the Czech Republic, and Ethiopia (4).
Potential selection against strains arising from emm and sof exchanges between sof-negative and sof-positive strains.
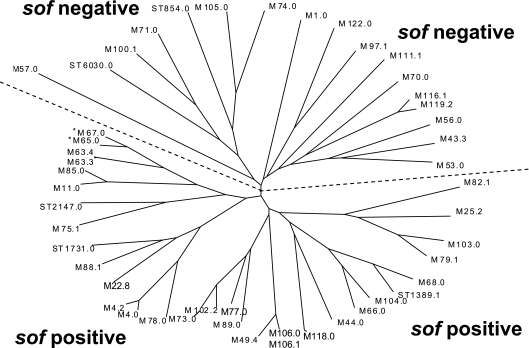
Extensive analysis has demonstrated that the presence or absence of sof within a GAS strain can be accurately determined based upon emm sequence relationships with known sof-positive and sof-negative strains (1, 9, 23, 24). This observation held true for the types discovered in this study, with the exception of types emm65 and emm67, which were sof PCR negative and still clustered fairly close to emm63 subtypes from sof-positive strains (Fig. 1). Interestingly, we have observed that the CDC emm65 reference strain yields a truncated sof gene upon sof-specific PCR (unpublished data).
FIG. 1.
Sequence relationships between deduced N-terminal M protein sequences from study set isolates. M protein sequences consisting of 22 signal sequence residues and 66 N-terminal residues were aligned using the Wisconsin Package 10.3 PileUp program. A phylogram was generated as described in Materials and Methods, using the Wisconsin Package version 10.3 Distances and GrowTree programs followed by entering the data into the TreeView program, selecting the unrooted tree option. The asterisks displayed for M65 and M67 indicate the sole exceptions in this study of M sequences from sof-negative and sof-positive strains clustering on the same half of the dendrogram. It is also interesting that one CDC emm65 reference strain apparently contains an aberrant (deletion) sof gene derivative (www.cdc.gov/ncidod/biotech/strep/emmdata.htm#emm65).
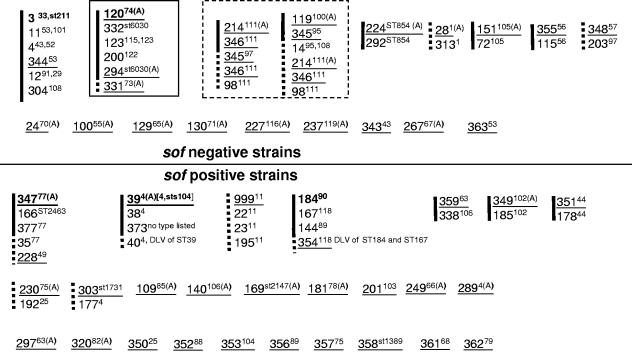
In light of previous published data that indicate the free exchange of MLST alleles between sof-positive and sof-negative strains (11, 16), relationships between strains from this study with other known strains would indicate that sof and emm gene exchange between sof-negative and sof-positive GAS is not generally followed by the successful expansion of resultant recombinants (Fig. 1 and 2). In addition to segregation of sof-positive from sof-negative strains based upon emm sequence relationships (Fig. 1), with only one exception STs from this study and related STs from the website www.mlst.net form distinct segregated sets corresponding to emm types exclusively associated with sof-positive or sof-negative strains (Fig. 2). The single exception involved an ST set that included a single clonal type from this study determined to be sof gene positive (emm73; ST331) and several clonal types previously determined to be sof gene negative (Fig. 2). This was the only example of a clonal set that included STs in this study that displayed genetic relatedness between sof-positive and sof-negative strains.
FIG. 2.
Nepal GAS STs in the context of clonal sets defined by eBURST of all known STs and inclusion of double-locus variants of Nepal STs. All STs found during this study are underlined. Clonal sets were determined by eBURST for all STs listed at www.mlst.net and by inclusion of all known double-locus variants of STs found during this study. eBURST forms groups in which each member shares six of seven MLST loci with at least one other member of the set. The solid black line indicates members of each eBURST group. Added double-locus variants of STs obtained in the study are indicated by dashed lines or dashed extensions of the eBURST groups. emm types associated with each ST at the global database (21) are indicated in superscript. Instances where there are multiple emm types for a given ST documented at www.mlst.net are indicated by brackets. Bold STs indicate predicted founder STs as defined by eBURST. These STs are defined as the ST within an eBURST group that has the greatest number of single-locus variants. The superscript A indicates that the ST-emm association is shared by another GAS strain(s) recovered in other geographic locations. ST999 is a six-allele profile that shares four of six identical loci with the other indicated STs. We were unable to amplify the yqiL allele from the emm11 isolates in this study. The single clonal set within a solid square (ST120 founder) includes a single clonal type determined to be sof gene positive (ST331) and several clonal types previously determined to be sof gene negative. This is the only example of a clonal set in this study associated with both sof-positive and sof-negative strains (ST331 was the sole sof-positive strain in the set). The two clonal sets within the dashed square each include the same four study STs depicted in relation to two different eBURST sets.
Association of projected mga locus patterns with skin and throat recovery sites.
The distribution of GAS in this study did not conform to previous observations, where pattern A to C strains were usually recovered from the throat, pattern D strains were usually recovered from skin, and pattern E strains displayed no site preference (Table 3). Ten of the 13 pattern A to C isolates were recovered from skin sites (Table 3), with 9 of these 10 skin isolates corresponding to the globally prevalent ST28 emm1 clone (18, 21) (Table 1). The 34 projected pattern D isolates from this study, representing 17 emm types, were nearly evenly distributed between skin (16 isolates) and throat (18 isolates) recovery sites. Finally, projected pattern E isolates, representing 25 different emm types, accounted for 46 throat isolates and only 27 skin isolates.
Erythromycin resistance was rare and tetracycline resistance was common among GAS recovered from children in Nepal.
Only 1 of 120 isolates (a single emm11, ST999 throat isolate) was erythromycin resistant (data not shown), even after two annual treatments with azithromycin (12, 13). This contrasts to a frequency of 3.8 to 4.3% macrolide resistance among U.S. pharyngitis isolates from children (22). Forty-five of 65 (69%) isolates tested were tetracycline resistant (data not shown), and tetracycline resistance was common within both mga pattern D and pattern E isolates. It was interesting that all 13 mga pattern A to C isolates were tetracycline sensitive.
DISCUSSION
Each of the 45 distinct emm sequence types found in this study of throat and skin lesion isolates recovered from children in a remote region of Nepal was highly conserved with emm types obtained from GAS isolates recovered elsewhere. Our current ongoing invasive and noninvasive GAS surveillance in the United States has uncovered very few new emm sequence types during the past 5 years (17, 20). In addition, even though we have documented markedly increased usage of the CDC emm database during the past few years and deposition of many new emm subtypes from many different developed and developing countries, we have concurrently observed a greatly decreased rate of deposition of sequences meeting the criteria for new emm types. We feel that these observations indicate that most emm types of significant frequency within the population have been discovered throughout the world. Similarly, most of the genotypes (40 of 51) included in this study had emm type-ST and/or emm type-sof gene associations that were shared by strains characterized previously from other locations (1, 2, 4, 18, 21). These data are consistent with the possibility that much of the existing genetic diversity within group A streptococci has already been documented.
The vast majority of GAS emm types that we typically encounter during U.S. surveillance are a small subset within 117 types recognized by an international panel (within the sequence of emm1 through emm124) (9). During recent U.S. surveillance, we found only 3 of 1,061 invasive isolates and 4 of 1,975 pharyngitis isolates were of emm types not within the sequence of emm1 to emm124 (17, 20). In the present study, 40 of the 45 types were of these recognized emm types. Although at the current time there are more than 70 additional emm sequence types (designated as “st” instead of “emm”) in the CDC database that have been documented from group A streptococci, many have been recorded only from single isolates and may not represent sizeable GAS populations. All five emm sequence types outside of the sequence emm1 to emm124 (st854, st6030, st2147, st1731, and st1389) were of sequence types that we have encountered in one or more countries outside of Nepal (4) and of the same subtypes as the original reference strains (st854.0, st6030.0, st1731.0, and st1389.0).
On the basis of the data presented here, it is possible that there is much more emm type diversity among western Nepal GAS carriage isolates than among GAS associated with pharyngitis and invasive disease within the United States (45 types among 120 isolates compared to 35 types among 1,975 pharyngitis isolates or 41 types among 1,061 invasive isolates) (17, 20); however, an accurate comparison of emm type diversity would require a much larger sampling of GAS in western Nepal.
The emm type diversity among current clinical isolates in the United States would be well covered by a current 26-valent M type-specific vaccine formulation (14), which would theoretically target approximately 86% of pharyngitis-causing isolates (20) and 77% of invasive isolates (17). In contrast, of the 45 emm types remarkably found among a collection of just 120 throat and skin lesion isolates recovered from Nepalese children, only 7 were of types included in the 26-valent vaccine. These seven types (emm types 1, 11, 22, 43, 75, 77, and 89) were found in only 23 of the 120 isolates (19.2%) described in this study. Determining the feasibility of a multivalent, type-specific M protein-based vaccine in a region such as this one would require knowledge of the emm type distribution. In addition, fluctuations in emm type distributions would need to be monitored over extended periods. For example, pharyngitis and invasive disease surveillance within the United States reveals marked emm type turnover within specific regions from year to year; however, the replacement types are generally types that are predominant within the country and are also targeted by the 26-valent vaccine (20; CDC, unpublished data).
Remarkably, within this Nepal study, 32 of the 49 subtypes obtained (those designated with subtype 0, e.g., emm1.0) shared the exact same deduced 50-residue N-terminal M protein sequence with CDC reference strains collected 6 to 60 years ago (Table 1). The 17 subtypes displaying variation from CDC reference strains generally showed modest changes of one to three amino acid substitutions within the 50-residue M protein N terminus. Within U.S. surveillance, we have also found very modest subtype diversity (17, 20). Recent studies showed that 26-valent vaccine-induced antibodies were bactericidal against all subtypes tested within targeted types (6).
Our data were in agreement with previous work indicating that in tropical areas mga pattern D is much more common and patterns A to C are much less common among GAS strains than in temperate regions (2). For example, in tropical Australia, 43% of isolates in a community with a high occurrence of streptococcal impetigo had mga locus pattern D (2), compared with 28.3% (34/120; 17 emm types) of the isolates in this Nepal study that were projected to have these patterns (Table 3). In contrast, in a recent collection of 1,061 invasive U.S. isolates (15), only 73 isolates (6.9%) were predicted to be mga pattern D on the basis of past pattern associations with emm types (17, 20). Pattern A to C isolates were relatively uncommon in this Nepal study (13/120; 10.8%) and were uncommon in tropical Australia (16%) (2). In U.S. invasive surveillance, mga patterns A to C were projected to account for 42.5% of invasive isolates (451 of 1,061 isolates; 11 different emm types) (17).
In earlier studies, marked associations of patterns A to C with throat carriage, pattern D with impetigo lesions, and pattern E with either GAS reservoir were observed (2, 3, 7). In contrast, this Nepal study showed a marked association of pattern A to C isolates with skin lesions; pattern D isolates showed no site preference, and pattern E isolates were more commonly recovered from the throat (Table 3). In both a tropical Australia study (2) and this Nepal study, there were only three emm types highly associated with patterns A to C (among 141 and 120 isolates, respectively). In contrast to the Australia study, where positive throat cultures for GAS were rare from asymptomatic individuals (2), we recovered throat GAS isolates from 17% of oropharyngeal swabs taken. However, in this and the previous study (2), GAS were recovered from more than 50% of impetigo lesions, and there was an obviously high frequency of streptococcal impetigo apparent in both locations (2; A. Fry, personal observations). In contrast to previous studies, the mga locus patterns would not be likely to serve as a genotypic marker for skin or throat reservoirs for these isolates recovered in Nepal (Table 3). At the present time we lack sufficient information to speculate upon the basis of the profoundly different observations drawn from this study compared to other independent studies (2, 3, 7), and we also lack information concerning asymptomatic GAS throat isolates recovered in the United States. It is interesting that while the percentage of predicted mga pattern D isolates is low among invasive isolates collected in the United States, they are still in a strikingly higher proportion than those from throat cultures collected from pharyngitis patients (6.9% [73/1,061 invasive isolates] versus 0.2% [6/1,975 pharyngitis isolates]) (16, 19).
As shown in previous studies (2, 9, 23, 24), we noted clustering of sof-positive strains (all projected to be mga pattern E) and sof-negative strains (all projected to be patterns A to C or pattern D) into clusters based upon 5′ emm sequence comparisons (Fig. 2). Additionally, there were 15 different examples of emm types from sof-positive isolates in this study that had been documented previously with different sof types, suggesting horizontal genetic transfer events between different sof-positive strains (Table 1). We have never encountered emm types associated with both sof-positive and sof-negative strains. It is also interesting that, with the exceptions of emm65.0 and emm67.0 (Fig. 1 legend), the strains from this study were also segregated into either sof-positive or sof-negative groups on the basis of relationships between multilocus sequence types (Fig. 1). The simplest explanation for this apparent genetic segregation between sof-negative and sof-positive strains would be the existence of biologic barriers preventing horizontal transfer events between these two populations; however, past analysis of MLST data has indicated high recombination rates among housekeeping genes and demonstrated a complete absence of congruence among the gene tree topologies for the seven MLST loci encompassing GAS genotypes representing all mga pattern groups (11, 16). The alternative possibility is that there are not biologic barriers preventing sof and emm gene transfers between the sof-negative and sof-positive populations, but there could be strong selection against expansion of mga pattern A to C and D strains in which sof or emm is introduced from sof-positive (mga pattern E) donors. Similarly, it is possible that there is strong selection against the expansion of sof-positive strains that are recipients of emm genes from mga pattern A to C and D strains. While there is evidence of past transfer of MLST loci between sof-negative and sof-positive GAS populations, there still could be relatively inefficient horizontal transfer between sof-negative and sof-positive GAS populations. Laboratory experimentation assessing the efficiency of selective marker exchange between different GAS genotypes could shed some light upon this issue.
Acknowledgments
We thank H. C. Jha, J. S. P. Chaudhary, and R. C. Bhatta of the Geta Eye Hospital in far-western Nepal for their assistance in the field. We thank Karen McGregor and Adrian Whatmore for their unpublished information concerning eight of the STs encountered during this work. We thank Delois Jackson for her assistance in serotyping and antibiotic susceptibility testing and Lingling Chen for assistance with genotyping.
REFERENCES
- 1.Beall, B., G. Gherardi, M. Lovgren, R. R. Facklam, B. A. Forwick, and G. J. Tyrrell. 2000. emm and sof gene sequence variation in relation to serological typing of opacity factor positive group A streptococci. Microbiology 146:1195-1209. [DOI] [PubMed] [Google Scholar]
- 2.Bessen, D. E., J. R. Carapetis, B. Beall, R. Katz, M. Hibble, B. J. Currie, T. Collingridge, M. W. Izzo, D. A. Scaramuzzino, and K. S. Sriprakash. 2000. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J. Infect. Dis. 182:1109-1116. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, D. E., C. M. Sotir, T. L. Readdy, and S. K. Hollingshead. 1996. Genetic correlates of throat and skin isolates of group A streptococci. J. Infect. Dis. 173:896-900. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2006. CDC emm sequence database. [Online.] http://www.cdc.gov/ncidod/biotech/strep/emmtypes.htm.
- 5.Courtney, H. S., D. L. Hasty, Y. Li, H. C. Chiang, J. L. Thacker, and J. B. Dale. 1999. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol. Microbiol. 32:89-98. [DOI] [PubMed] [Google Scholar]
- 6.Dale, J. B., T. Penfound, E. Y. Chiang, V. Long, S. T. Shulman, and B. Beall. 2005. Multivalent group A streptococcal vaccine elicits bactericidal antibodies against variant M subtypes. Clin. Diagn. Lab. Immunol. 12:833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dicuonzo, G., E. Fiscarelli, G. Gherardi, G. Lorino, F. Battistoni, S. Landi, M. De Cesaris, T. Petitti, and B. Beall. 2001. Erythromycin-resistant pharyngeal isolates of Streptococcus pyogenes recovered in Italy. Antimicrob. Agents Chemother. 46:3987-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facklam, R. R., A. Efstratiou, P. Kriz, G. Tyrrell, M. Lovgren, T. Thompson, S. Gowan, D. Martin, E. Kaplan, and B. Beall. 2002. Continued extension of the Lancefield classification scheme for group A streptococci by addition of 22 new M protein gene sequence types: emm103 to emm124. Clin. Infect. Dis. 34:28-38. [DOI] [PubMed] [Google Scholar]
- 10.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 86:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry, A. M., H. C. Jha, T. M. Lietman, J. S. P. Chaudhary, R. C. Bhatta, J. Elliott, T. Hyde, A. Schuchat, B. Gaynor, and S. F. Dowell. 2002. Adverse and beneficial secondary effects of mass treatment with azithromycin to eliminate blindness due to trachoma in Nepal. Clin. Infect. Dis. 35:395-402. [DOI] [PubMed] [Google Scholar]
- 13.Holm, S. O., H. C. Jha, R. C. Bhatta, J. S. Chaudhary, B. B. Thapa, D. Davis, R. P. Pokhrel, M. Yinghui, M. Zegans, J. Schachter, K. D. Frick, L. Tapert, T., and M. Lietman. 2001. Comparison of two azithromycin distribution strategies for controlling trachoma in Nepal. Bull. W. H. O. 79:194-200. [PMC free article] [PubMed] [Google Scholar]
- 14.Hu, M. C., M. A. Walls, S. D. Stroop, M. A. Reddish, B. Beall, and J. B. Dale. 2002. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect. Immun. 70:2171-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeng, A., V. Sakota, Z. Li, V. Datta, B. Beall, and V. Nizet. 2003. Molecular genetic analysis of a group A streptococcal operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185:1208-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalia, A., B. G. Spratt, M. C. Enright, and D. E. Bessen. 2002. Influence of recombination and niche separation on the population genetic structure of the pathogen Streptococcus pyogenes. Infect. Immun. 70:1971-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Z., V. Sakota, D. Jackson, A. R. Franklin, B. Beall, et al. 2003. Array of M protein gene subtypes in 1064 recent invasive group A streptococcus isolates recovered from the active bacterial core surveillance. J. Infect. Dis. 188:1587-1592. [DOI] [PubMed] [Google Scholar]
- 18.McGregor, K. F., B. G. Spratt, A. Kalia, A. Bennett, N. Bilek, B. Beall, and D. E. Bessen. 2004. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J. Bacteriol. 186:4285-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakonjac, J. V., J. C. Robbins, and V. A. Fischetti. 1995. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shulman, S. T., R. R. Tanz, W. Kabat, K. Kabat, E. Cederlund, D. Patel, Z. Li, V. Sakota, J. B. Dale, B. Beall, et al. 2004. Group A streptococcal pharyngitis serotype surveillance in North America, 2000-2002. Clin. Infect. Dis. 39:325-332. [DOI] [PubMed] [Google Scholar]
- 21.Streptococcus pyogenes Database. 2005. Streptococcus pyogenes multilocus sequence typing. [Online.] http://spneumoniae.mlst.net/#.
- 22.Tanz, R. R., S. T. Shulman, V. D. Shortridge, W. Kabat, K. Kabat, E. Cederlund, J. Rippe, J. Beyer, S. Doktor, B. W. Beall, et al. 2004. Community-based surveillance in the united states of macrolide-resistant pediatric pharyngeal group A streptococci during 3 respiratory disease seasons. Clin. Infect. Dis. 39:1794-1801. [DOI] [PubMed] [Google Scholar]
- 23.Teixera, L. M., R. R. Barros, A. C. D. Castro, J. M. Peralta, M. D. G. S. Carvalho, D. F. Talkington, A. M. Vivoni, R. R. Facklam, and B. Beall. 2001. Genetic and phenotypic features of Streptococcus pyogenes strains isolated in Brazil that harbor new emm sequences. J. Clin. Microbiol. 39:3290-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whatmore, A. M., V. Kapur, D. J. Sullivan, J. M. Musser, and M. A. Kehoe. 1994. Non-congruent relationships between variation in emm gene sequences and the population genetic structure of group A streptococci. Mol. Microbiol. 14:619-631. [DOI] [PubMed] [Google Scholar]