Abstract
The 5′ noncoding region (5′ NCR) of the hepatitis C virus (HCV) has become the standard for genotyping even though several reports show that its use can result in classification errors. The purpose of this study was to perform genotyping based on sequence analysis of the NS5b region in a set of 357 HCV strains isolated from blood donors in France in 2002 and 2003. Results were compared with those previously obtained using 5′ NCR analysis, and HCV subtype distribution was reevaluated. Twenty-six of 120 strains (∼22%) initially identified as genotype 1b by 5′ NCR region sequence analysis were reclassified as genotype 1a by NS5b region sequence analysis. Similarly, 14 of 23 strains (∼61%) initially identified as 2a/2c were reclassified as non-2a and non-2c subtypes, and 12 of 22 strains (∼45%) initially identified as 4c/4d subtypes were reclassified as non-4c and non-4d subtypes. Sequence analysis of the NS5b region also revealed 5 putative new subtype 2 variants and 2 putative new subtype 4 variants. Although these findings demonstrated full agreement between 5′ NCR and NS5b sequence analysis with regard to type classification, genotyping based on phylogenetic analysis of the NS5b region is more accurate for subtype determination than genotyping based on analysis of the 5′ NCR. Sequence analysis of the NS5b region is mandatory for epidemiologic studies.
Hepatitis C virus (HCV) is a common human pathogen considered as the major cause of parenterally transmitted hepatitis (6). It is an enveloped virus with a positive-sense RNA genome containing a single large open reading frame composed of over 9,000 nucleotides (6, 11). Sequencing of HCV isolates has identified 6 genotypes and more than 70 subtypes (19, 25, 28, 32). Accurate HCV genotyping is important for predicting response to antiviral therapy, since genotypes 1 and 4 are less likely to respond to interferon than genotypes 2 and 3 (14, 21). Genotyping is also an essential tool for epidemiological studies, since HCV genotypes vary according to epidemic history in different geographical regions (23, 24, 30, 40). Epidemiologic studies of HCV strains from blood donors (7, 18), drug addicts (1, 12, 13), and hospital patients (22, 35) have demonstrated a correlation between some subtypes and risk factors.
Since the 5′ noncoding region (5′ NCR) is one of the most highly conserved regions of HCV, it has historically been used for virus detection and is now one of the best-characterized regions. For practical reasons, the 5′ NCR was also chosen as the target for various genotyping methods, including the InnoLipa HCV II test (33, 34), sequencing (8, 9, 10, 26), and the duplex mobility assay (39). However, recent studies involving NS5b region analysis (3, 29, 35) show that 5′ NCR sequence analysis can lead to genotyping errors (5, 35). In this study, we used NS5b region sequence analysis to determine the HCV subtype distribution of 357 isolates collected from blood donors in France between 2002 and 2003. All samples had already been genotyped based on 5′ NCR analysis. Results of the two sequence analysis methods were compared to identify genotyping errors associated with 5′ NCR analysis.
MATERIALS AND METHODS
Study population.
A total of 357 HCV-positive blood samples collected from French blood donors between 2002 and 2003 were analyzed. These strains (named Frbd for French blood donor), collected for the national surveillance of HCV-positive blood donors, came from the overall collection of confirmed HCV-positive blood donations in France, i.e., seropositive for anti-HCV and HCV viremic by nucleic acid testing. Plasmas were aliquoted and stored at −80°C under RNase-free conditions until analysis.
The distribution of HCV types determined using the InnoLipa HCV II genotyping kit (Innogenetics, Zwijnaarde, Belgium) (33, 34) and sequencing of a 280-bp amplicon in the 5′ NCR using InnoLipa kit primers (8, 34) was as follows: subtype 1b, 120 strains (33.6%); subtype 1a, 91 strains (25.5%); subtype 3a, 71 strains (19.9%); type 2, 33 strains (9.2%); type 4, 33 strains (9.2%); subtype 5a, 7 strains (2%) (Table 1).
TABLE 1.
Comparison of genotyping results obtained with 5′ NCR analysis versus the NS5b and E1 region analyses
| NS5b subtype | No. of strains with 5′ NCR genotyping result
|
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1a | 1b | 2a/2c | 2 | 2b | 3a | 4 | 4c/4d | 4e | 4f | 5a | ||
| 1 | 1 | 1 | |||||||||||
| 1a | 91 | 26 | 117 | ||||||||||
| 1b | 2 | 93 | 95 | ||||||||||
| 2a | 8 | 8 | |||||||||||
| 2b | 6 | 6 | |||||||||||
| 2c | 1 | 1 | 2 | ||||||||||
| 2i | 6 | 6 | |||||||||||
| 2k | 4 | 4 | |||||||||||
| 2l | 1 | 1 | |||||||||||
| 2(1) | 1 | 1 | |||||||||||
| 2(2) | 1 | 1 | |||||||||||
| 2(3) | 1 | 1 | |||||||||||
| 2(4) | 1 | 1 | 2 | ||||||||||
| 2(5) | 1 | 1 | |||||||||||
| 3a | 71 | 71 | |||||||||||
| 4aa | 4 | 8 | 12 | ||||||||||
| 4d | 2 | 12 | 14 | ||||||||||
| 4f | 2 | 2 | |||||||||||
| 4h | 1 | 1 | 2 | ||||||||||
| 4r | 1 | 1 | |||||||||||
| 4(1) | 1 | 1 | |||||||||||
| 4(2) | 1 | 1 | |||||||||||
| 5a | 7 | 7 | |||||||||||
| Total | 2 | 91 | 120 | 23 | 4 | 6 | 71 | 8 | 22 | 1 | 2 | 7 | 357 |
Six strains were not identified in the NS5b region but were amplified in the E1 region.
Amplification and sequencing.
A 339-nucleotide segment from position 8370 to 8708 in the NS5b region was amplified and sequenced (3). If amplification in the NS5b region failed, a 357-nucleotide segment from position 1029 to 1385 in the E1 region was analyzed (2).
RNA extraction from 200 μl of plasma, reverse transcription using random hexaprimers, and PCR were performed as previously reported (2). Amplicons in the NS5b and E1 regions were sequenced directly with the amplification primers, the d-rhodamine DNA sequencing kit, and an ABI Prism 377 sequencer (Perkin-Elmer).
Phylogenetic analysis.
All nucleotide sequences from HCV strains were aligned using the ClustalW 1.8 software program (36) with a reference panel of reported sequences available in the HCV sequence database (http://hcv.lanl.gov/content/hcv-db/index) provided by the Los Alamos National Laboratory (15). Pairwise evolutionary distance matrices for the E1 and NS5b nucleotide sequences were computed using the p-distance algorithm of the MEGA software package (version 3.0, 2005; Pennsylvania State University, University Park, PA) (16). Sequence distance matrices were analyzed with conventional statistical software (SYSTAT, v10.2; Systat Software, Inc., Point Richmond, CA) using files derived from Mega distance tables. For NS5b sequences, a p-distance of 0.12 was found to delineate strains of the same subtype and strains of a different subtype.
Phylogenetic analysis was performed with the MEGA software package using the p-distance algorithm for distance determination and the neighbor-joining method for tree drawing. The reliability of phylogenetic classification was evaluated by a 1,000-replication bootstrap test using the same software package.
Genotyping was performed using the nomenclature defined by Simmonds et al. (31).
Statistical analysis.
The distribution of HCV genotypes was compared by using the chi-square test and Fisher test. Differences were considered to be statistically significant when the P value was <0.05.
RESULTS
Analysis of E1 and NS5b regions: HCV genotype distribution.
Genotyping based on NS5b region sequence analysis was successful in 351 of the 357 strains previously typed with InnoLipa HCV II test. Amplification of the NS5b region failed in 6 type 4 strains which, however, were successfully subtyped as 4a isolates by analyzing the E1 region.
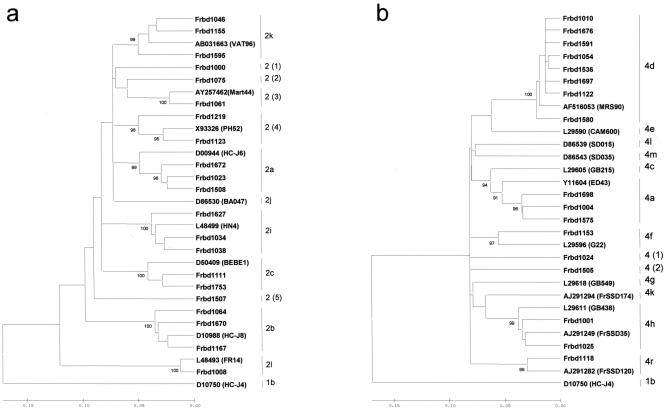
The distribution of HCV subtype can be summarized as follows: subtype 1a, 117 strains (32.8%); subtype 1b, 95 strains (26.6%); type 2, 33 strains (9.2%); subtype 3a, 71 strains (19.9%); type 4, 33 strains (9.2%), subtype 5a, 7 strains (2%). The 33 type 2 strains were classified into 11 subtypes (Fig. 1a). Twenty-seven strains clustered within 6 previously described subtypes: 2a, 8 strains (24.2%); 2b, 6 strains (18.2%); 2c, 2 strains (6.1%); 2i, 6 strains (18.2%); 2k, 4 strains (12.1%); 2l, 1 strain (3.0%). The remaining six strains, including the Frbd1000, Frbd1075, Frbd1061, Frbd1219/Frbd1123, and Frbd1507 strains, could not be assigned to any previously described subtype and may constitute five new subtype variants tentatively designated 2 (1), 2 (2), 2 (3), 2 (4), and 2 (5), respectively. The 33 type 4 strains were classified into 7 subtypes (Fig. 1b), including subtype 4a (12 strains, 36.4%) and subtype 4d (14 strains, 42.4%). Subtypes 4 h and 4f were represented by 2 strains each (Table 1). The Frbd1118 strain showing similarity with the FrSSD120 strain (20) was classified as 4r (15). The Frbd1024 and Frbd1505 strains could not be assigned to any existing type 4 subtype and may constitute new subtypes variants tentatively designated 4 (1) and 4 (2), respectively.
FIG. 1.
Phylogenetic tree showing partial NS5b nucleotide sequences in representative HCV subtype 2 (a) and 4 (b) strains isolated from blood donors together with panels of reference strains identified by their GenBank reference numbers. Numbers on the right correspond to prototype strain subtypes as published by Simmonds et al. (31). Numbers in parentheses indicate new subtypes. Numbers at the nodes are percentages of 1,000 bootstrap replicates greater than 90%. Scale bars indicate p-distances.
Comparison of HCV genotyping methods.
This study demonstrated full agreement between 5′ NCR and NS5b sequence analysis with regard to type classification.
Analysis of type 3 and 5 strains.
Comparison of HCV genotyping based on NS5b sequence analysis showed no difference in the classification of the 71 subtype 3a strains and 7 subtype 5a strains initially identified by 5′ NCR analysis between 2002 and 2003.
Analysis of type 1 strains.
A total of 120 strains were initially identified as subtype 1b based on 5′ NCR analysis. Sequence analysis of the NS5b region was concordant for 93 strains (77.5%). The remaining strains were reclassified as subtype 1a in 26 cases (21.7%) and subtype 1d in 1 case. Sequencing of the 5′ NCR showed that all discordant isolates had a G in position −99 (data not shown).
A total of 91 strains were initially identified as subtype 1a based on 5′ NCR analysis. Sequence analysis of the NS5b was concordant in all cases.
In the two type 1 strains that could not be subtyped using 5′ NCR analysis, NS5b region sequence analysis demonstrated subtype 1b. In the Frbd1531 strain, failure to achieve genotyping was due to the presence of an adenine in position −92 that prevented fixation of the amplicon to probes 5, 6, 7, and 8 of the InnoLipa HCV II kit (Fig. 2). In the Frbd1735 strain, failure of genotyping was due to fixation of the amplicon to probe 9 that is specific for type 2.
FIG. 2.
Alignment of 37 sequences in the 5′ noncoding region of type 1, 2, and 4 HCV strains (adapted from reference 34 with permission). Boxes indicate the regions covered by InnoLipa HCV II probes. The first position is indicated in the bottom left hand corner of each box. The reactivity of each isolate is indicated in the alignment of the sequence by the number of the probe (34). The results of testing using the InnoLipa HCV II kit followed by the genotype obtained by sequencing of the NS5b region are shown in parentheses.
Using NS5b region analysis as the reference technique, the estimated sensitivity and specificity of 5′ NCR analysis were 97.8% and 90%, respectively, for detection of subtype 1b and 77.7% and 100%, respectively, for detection of subtype 1a. The prevalence of subtypes 1a and 1b were 25.5% versus 33.6%, respectively, based on 5′ NCR analysis, compared to 32.7% versus 26.6%, respectively, based on NS5b region sequence analysis. The differences in prevalence obtained using the two techniques were statistically significant (χ2 = 6.15; P = 0.013).
Analysis of type 2 strains.
A total of 33 type 2 strains were studied. Based on 5′ NCR analysis, these strains were classified as subtype 2a/2c in 23 cases and subtype 2b in 6 cases. The remaining 4 type 2 strains could not be subtyped. For all 6 subtype 2b strains, sequence analysis based on the NS5b region was concordant with 5′ NCR analysis.
For the 23 strains identified as subtypes 2a/2c, NS5b region sequence analysis confirmed only 8 strains as subtype 2a and 1 strain as subtype 2c. The other strains were identified as subtype 2i in 6 cases, subtype 2k in 4 cases, and as subtype 2 (1), 2 (3), 2 (4), and 2 (5) in one case each. (Table 1). These findings indicate that the 2a/2c profile in the 5′ NCR corresponds to 8 subtypes in the NS5b region.
The 4 type 2 strains that could not be subtyped based on 5′ NCR analysis were classified by NS5b region sequence analysis as subtype 2 (2), 2 (4), 2c, and 2l in one case each (Table 1). These strains were identified as type 2 due to the presence of three different sequences in the 5′ NCR (Fig. 2). The first sequence was found in the Frbd1753 and Frbd1219 strains. The second and third sequences were found in the Frbd1075 and Frbd1008 strains, respectively.
Analysis of type 4 strains.
Thirty-three type 4 strains were studied. Analysis of the 5′ NCR identified these strains as subtype 4 in 8 cases, subtype 4c/4d in 22 cases, subtype 4e in 1 case, and subtype 4f in 2 cases.
Sequence analysis of the NS5b and E1 regions in the 22 subtype 4c/4d strains demonstrated subtype 4a in 8 cases, subtype 4d in 12 cases, subtype 4 h in 1 case, and subtype 4 (1) in 1 case. The 8 type 4 strains were classified as type 4 (2) in 1 case, subtype 4a in 4 cases, subtype 4d in 2 cases, and subtype 4 h in 1 case. The Frbd1118 strain initially classified as subtype 4e by 5′ NCR analysis was reclassified as subtype 4r by NS5b region sequence analysis (Table 1).
Subtyping of type 4 strains was prevented by the presence of five different sequences in the 5′ NCR (Fig. 2). The first sequence was found in the Frbd1003, Frbd1058, Frbd1505, and Frbd1676 strains. The second, third, fourth, and fifth sequences were found in the Frbd1001, Frbd1063, Frbd1152, and Frbd1575 strains, respectively.
DISCUSSION
Genotyping of HCV is routinely performed in many laboratories for three indications. The first indication involves clinical decision making, i.e., to make recommendations and provide counseling regarding treatment. For this indication, type determination is sufficient. The second indication involves classification and epidemiology, i.e., to monitor the distribution of virus strains and identify risk factors for transmission. The third indication involves legal issues, i.e., to investigate contamination between individuals. For the second and third indications, accurate subtype determination is necessary.
The highly conserved 5′ NCR of the HCV has been used for development of effective diagnostic tests and for genotyping based either on hybridization using subtype-specific probes (34) or sequence analysis (8). Commercially available test kits have been instrumental in determining the relative prevalence of viral types and subtypes in various geographical regions. Findings have shown that subtype 1a accounts for most infections in North America, while subtype 1b is primarily found in Japan and Europe, including France. Genotyping has also shown that subtype 1a is associated with drug addiction and subtype 1b with transfusion (40).
However, several studies have shown that testing based on the 5′ NCR is a source of subtyping errors (5, 35). The purpose of this study using HCV isolates from 357 blood donors was to compare results previously obtained using 5′ NCR sequence analysis with those obtained by NS5b region sequence analysis.
The choice of the NS5b region in phylogenetic analysis relies on its highly informative character (32, 38). Indeed, the degree of sequence variation of NS5b correlates well with HCV subtype definition (37, 38), by contrast with the 5′ NCR, which is highly conserved and, hence, does not provide a tool for differentiation between subtypes. Conserved primers in the NS5b have been described previously that allow the amplification of all genotypes of HCV (3, 17, 20). Moreover, the existence of a previously defined reference panel in HCV databases for the NS5b regions considerably facilitates subtype identification (15, 25, 31).
Findings showed that the NS5b region achieved amplification in all but a few subtype 4a strains. This study demonstrated full agreement between 5′ NCR and NS5b sequence analysis with regard to type classification. However, subtyping was discordant for 63 of the 357 strains (17.6%). Subtyping differences involved types 1, 2, and 4. Classification by the two techniques was concordant for 99% of subtype 1b strains but discordant for a nonnegligible proportion (22%) of subtype 1a strains classified as subtype 1b by 5′ NCR analysis. Similarly, 61% of strains initially identified as 2a/2c were reclassified as non-2a and non-2c subtypes, and 45% of strains initially identified as 4c/4d subtypes were reclassified as non-4c and non-4d subtypes. Analysis based on the 5′ NCR achieved accurate genotyping only for subtypes 1b, 2b, 3a, 4f, and 5a.
A notable finding of this study is that the InnoLipa HCV II kit did not identify any of the 12 type 4a strains accurately. Four strains were classified as unsubtyped type 4, and 8 strains were classified as 4c/4d. The apparent explanation is that the sequence defining subtype 4a (strain Z4) in the InnoLipa HCV II test is different from the fully sequenced subtype 4a reference strain (ED43) (4). Strain Z4 was recently attributed to subtype 4r (15, 20, 31). In this regard it is noteworthy that another strain classified as subtype 4r in our study (Fig. 1b), i.e., Frbd1118, was classified as 4e using the InnoLipa HCV II test.
This study also showed that 5′ NCR analysis was unable to detect the great diversity of types 2 and 4 in French blood donors. The 23 strains classified as 2a/2c were broken down into 7 subtypes, i.e., 2a, 2c, 2i, 2k, and 3 putative new subtypes. All strains identified as 2a, 2i, and 2k by NS5b region sequence analysis had been identified as 2a/2c by 5′ NCR analysis using the InnoLipa HCV II kit. Similarly, NS5b and E1 region sequence analysis broke down the 22 strains classified as subtype 4c/4d by 5′ NCR analysis into 4 subtypes, i.e., 4a, 4d, 4h, and one putative new subtype. Analysis of the NS5b region could not assign six type 2 strains and two type 4 strains to any previously described subtype (Fig. 1). The Frbd1123 and Frbd1219 strains belonging to subtype 2 (4) were phylogenetically close to the PH52 strain isolated in Guinea Conakry (27). The Frbd1061 strain classified as subtype 2 (3) belonged to the same subtype as the Mart44 strain from the Caribbean island of Martinique (17). These findings are in agreement with a previous study showing a high variety of subtype 2 in a population of blood donors from southeastern France (3).
In contradiction with the generally accepted notion that subtype 1b was predominant in French blood donors, this study shows that subtype 1a was the most common. This finding is in agreement with a recent study (3) showing not only that subtype 1a strains were predominant but also that their distribution has been stable in blood donors in southeastern France since 1995. These findings suggest that it may be necessary to reevaluate the validity of the previous studies based on 5′ NCR analysis which established that subtype 1b was more prevalent than subtype 1a (range of 27 to 34% versus range of 16% to 20%, respectively) (7, 18, 22). Revision of previously reported prevalence rates assuming that 5′ NCR analysis overestimates subtype 1b by 20% at the expense of subtype 1a led to results similar to those reported in the present study with a predominance of subtype 1a.
This study confirms that genotyping based on phylogenetic analysis of NS5b region sequences is still the most reliable technique for subtyping. This finding does not reduce the value of commercially available genotyping test kits based on 5′ NCR, since subtyping is not important for routine clinical purposes. The main implication of this study is that genotyping based on NS5b region sequence analysis could be a major improvement for epidemiologic studies and medicolegal investigations by eliminating the 17.6% of classification errors that occur using 5′ NCR analysis. Most classification errors using the 5′ NCR region are related to poor discriminating power and to the absence of target motifs specific for some subtypes. Sequence analysis of the NS5b region must be used instead of 5′ NCR analysis for epidemiologic studies.
Acknowledgments
This study was supported by grant no. 2003-09 from the “Conseil Scientifique de l'Etablissement Français du Sang.”
We thank all colleagues from blood banks for their active participation in transmission of blood samples.
REFERENCES
- 1.Bourliere, M., J. M. Barberin, M. Rotily, V. Guagliardo, I. Portal, L. Lecomte, S. Benali, C. Boustiere, H. Perrier, M. Jullien, G. Lambot, R. Loyer, O. LeBars, R. Daniel, H. Khiri, and P. Halfon. 2002. Epidemiological changes in hepatitis C virus genotypes in France: evidence in intravenous drug users. J. Viral. Hepat. 9:62-70. [DOI] [PubMed] [Google Scholar]
- 2.Cantaloube, J.-F., H. Venault, J.-P. Zappitelli, P. Gallian, M. Touinssi, H. Attoui, P. Biagini, X. de Lamballerie, and P. de Micco. 2000. Molecular analysis of HCV type 1 to 5 envelope gene: application to investigations of posttransfusion transmission of HCV. Transfusion 40:712-717. [DOI] [PubMed] [Google Scholar]
- 3.Cantaloube, J.-F., P. Gallian, H. Attoui, P. Biagini, P. De Micco, and X. de Lamballerie. 2005. Genotype distribution and molecular epidemiology of hepatitis C virus in blood donors from Southeast France. J. Clin. Microbiol. 43:3624-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamberlain, R. W., N. Adams, A. A. Saeed, P. Simmonds, and R. M. Elliott. 1997. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J. Gen. Virol. 78:1341-1347. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, A. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elghouzzi, M. H., F. Bouchardeau, J. Pillonel, E. Boiret, C. Tirtaine, V. Barlet, P. Montcharmont, P. Maisonneuve, M. C. du Puy-Montbrun, D. Lyon-Caen, and A. M. Couroucé. 2000. Hepatitis C virus: routes of infection and genotypes in a cohort of anti-HCV-positive French blood donors. Vox Sang. 79:138-144. [DOI] [PubMed] [Google Scholar]
- 8.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germer, J. J., D. W. Majewski, M. Rosser, A. Thompson, P. S. Mitchell, T. F. Smith, S. Elagin, and J. D. Yao. 2003. Evaluation of the TRUGENE HCV 5′NC genotyping kit with the new GeneLibrarian module 3.1.2 for genotyping of hepatitis C virus from clinical specimens. J. Clin. Microbiol. 41:4855-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halfon, P., P. Trimoulet, M. Bourliere, H. Khiri, V. de Ledinghen, P. Couzigou, J. M. Feryn, P. Alcaraz, C. Renou, H. J. Fleury, and D. Ouzan. 2001. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J. Clin. Microbiol. 39:1771-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inchauspe, G., S. Zebedee, D. H. Lee, M. Sugitani, M. Nasoff, and A. M. Prince. 1991. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc. Natl. Acad. Sci. USA 88:10292-20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalinina, O., H. Norder, T. Vetrov, Z. Zhdanov, M. Barzunova, V. Plotnikova, S. Mukomolov, and L. O. Magnius. 2001. Shift in predominating subtype of HCV from 1b to 3a in St. Petersburg mediated by increase in injecting drug use. J. Med. Virol. 65:517-524. [PubMed] [Google Scholar]
- 13.Kalinina, O., H. Norder, S. Mukomolov, and L. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshy, A., J. P. Madda, P. Marcellin, and M. Martinot. 2002. Treatment of hepatitis C virus genotype 4-related cirrhosis: ribavirin and interferon combination compared with interferon alone. J. Clin. Gastroenterol. 35:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos HCV sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 17.Martial, J., Y. Morice, S. Abel, A. Cabie, C. Rat, F. Lombard, A. Edouard, S. Pierre-Louis, P. Garsaud, O. Bera, R. Chout, E. Gordien, P. Deny, and R. Cesaire. 2004. Hepatitis C virus (HCV) genotypes in the Caribbean island of Martinique: evidence for a large radiation of HCV-2 and for a recent introduction from Europe of HCV-4. J. Clin. Microbiol. 42:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinot-Peignoux, M., F. Roudot-Thoraval, I. Mendel, J. Coste, J. Izopet, G. Duverlie, C. Payan, J. M. Pawlotsly, C. Defer, M. Bogard, V. Gerolami, P. Halfon, Y. Buisson, B. Fouqueray, P. Loiseau, J. Lamoril, J. J. Lefrere, and P. Marcellin. 1999. Hepatitis C virus genotypes in France: relationship with epidemiology, pathogenicity and response to interferon therapy. J. Viral. Hepat. 6:435-443. [DOI] [PubMed] [Google Scholar]
- 19.Mellor, J., E. C. Holmes, L. M. Jarvis, P. L. Yap, and P. Simmonds. 1995. Investigation of the pattern of hepatitis C sequence diversity in different geographical regions: implications for virus classification. J. Gen. Virol. 76:2493-2507. [DOI] [PubMed] [Google Scholar]
- 20.Morice, Y., D. Roulot, V. Grando, J. Stirnemann, E. Gault, V. Jeantils, M. Bentata, B. Jarrousse, O. Lortholary, C. Pallier, and P. Deny. 2001. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J. Gen. Virol. 82:1001-1012. [DOI] [PubMed] [Google Scholar]
- 21.Pawlotsky, J. M. 2003. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antivir. Res. 59:1-11. [DOI] [PubMed] [Google Scholar]
- 22.Payan, C., F. Roudot-Thoraval, P. Marcellin, N. Bled, G. Duverlie, I. Fouchard-Hubert, P. Trimoulet, P. Couzigou, D. Cointe, C. Chaput, C. Henquell, A. Abergel, J. M. Pawlotsky, C. Hezode, M. Coude, A. Blanchi, S. Alain, V. Loustaud-Ratti, P. Chevallier, C. Trepo, V. Gerolami, I. Portal, P. Halfon, M. Bourliere, M. Bogard, E. Plouvier, C. Laffont, G. Agius, C. Silvain, V. Brodard, G. Thiefin, C. Buffet-Janvresse, G. Riachi, F. Grattard, T. Bourlet, F. Stoll-Keller, M. Doffoel, J. Izopet, K. Barange, M. Martinot-Peignoux, M. Branger, A. Rosenberg, P. Sogni, M. L. Chaix, S. Pol, V. Thibault, P. Opolon, A. Charrois, L. Serfaty, B. Fouqueray, J. D. Grange, J. J. Lefrere, and F. Lunel-Fabiani. 2005. Changing of hepatitis C virus genotype patterns in France at the beginning of the third millennium: The GEMHEP GenoCII Study. J. Viral. Hepat. 12:405-413. [DOI] [PubMed] [Google Scholar]
- 23.Pistello, M., F. Maggi, C. Fornai, A. Leonildi, A. Morrica, M. L. Vatteroni, and M. Bendinelli. 1999. Classification of hepatitis C virus type 2 isolates by phylogenetic analysis of core and NS5 regions. J. Clin. Microbiol. 37:2116-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu, D., O. Hantz, M. Gouy, L. Vitvitski, J.-S. Li, F. Berby, S.-P. Tong, and C. Trépo. 1994. Heterogeneity of hepatitis C virus genotypes in France. J. Gen. Virol. 75:1063-1070. [DOI] [PubMed] [Google Scholar]
- 25.Robertson, N., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gashen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-i, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 26.Ross, R. S., S. O. Viazov, C. D. Holtzer, A. Beyou, A. Monnet, C. Mazure, and M. Roggendorf. 2000. Genotyping of hepatitis C virus isolates using CLIP sequencing. J. Clin. Microbiol. 38:3581-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruggieri, A., C. Argentini, K. Kourouma, P. Chionne, E. D'Ugo, E. Spada, S. Dettori, S. Sabbatini, and M. Rapicetta. 1996. Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry). J. Gen. Virol. 77:2073-2076. [DOI] [PubMed] [Google Scholar]
- 28.Samokhvalov, E. I., M. Hijikata, R. I. Gylka, D. K. Lvov, and S. Mishiro. 2000. Full-genome nucleotide sequence of a hepatitis C variant (isolate VAT-96) representing a new subtype within the genotype 2 (arbitrarily 2k). Virus Genes 20:183-187. [DOI] [PubMed] [Google Scholar]
- 29.Sandres-Saune, K., P. Deny, C. Pasquier, V. Thibaut, G. Duverlie, and J. Izopet. 2003. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J. Virol. Methods 109:187-193. [DOI] [PubMed] [Google Scholar]
- 30.Simmonds, P. 2004. Genetic diversity and evolution of hepatitis C virus-15 years on. J. Gen. Virol. 85:3173-3188. [DOI] [PubMed] [Google Scholar]
- 31.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 32.Stuyver, L., W. van Arnhem, A. Wyseur, F. Hernandez, E. Delaporte, and G. Maertens. 1994. Classification of hepatitis C virus based on phylogenetic analysis of envelope 1 and nonstructural 5b regions and identification of five additional subtypes. Proc. Natl. Acad. Sci. USA 91:10134-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuyver, L., A. Wyseur, W. van Arnhem, F. Lunel, P. Laurent-Puig, J. M. Pawlotsky, B. Kleter, L. Bassit, J. Nkengasong, L. J. Van Doorn, and G. Maertens. 1995. Hepatitis C genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 38:137-157. [DOI] [PubMed] [Google Scholar]
- 34.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second generation line probe assay for hepatitis C virus genotyping. 1996. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamalet, C., P. Colson, H. Tissot-Dupont, M. Henry, C. Tourres, N. Tivoli, D. Botta, I. Ravaux, I. Poizot-Martin, and N. Yahi. 2003. Genomic and phylogenetic analysis of hepatitis C virus isolates: a survey of 535 strains circulating in southern France. J. Med. Virol. 7:391-398. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokita, H., H. Okamoto, H. Izuka, J. Kishimoto, F. Tsuda, L. A. Lesmana, Y. Miyakawa, and M. Mayumi. 1996. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J. Gen. Virol. 77:293-301. [DOI] [PubMed] [Google Scholar]
- 38.Tokita, H., H. Okamoto, H. Iizuka, J. Kishimoto, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1998. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7-9 and comparison with those in the other eight genetic groups. J. Gen. Virol. 79:1847-1857. [DOI] [PubMed] [Google Scholar]
- 39.White, P. A., X. Zhai, I. Carter, Y. Zhao, and W. D. Rawlinson. 2000. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J. Clin. Microbiol. 38:477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]