Abstract
A lack of anti-HBc antibodies during chronic hepatitis B virus infection is a serological pattern that is rarely observed. In our series of 39 patients with such a confirmed profile, mutations within the precore/core gene were rarely found and the lack of antibody detection was mostly explained by concurrent immunosuppression and the low sensitivities of the serological assays.
Chronic carriage of hepatitis B virus (HBV) is sometimes associated with an atypical serological pattern characterized by the presence of hepatitis B surface antigen (HBsAg) without antibodies to the HB core antigen (HBcAb). The aim of our work was to study the reasons for the lack of HBcAb in HBsAg-positive patients. We selected 39 patients who had this peculiar serological profile on at least two visits. These 39 patients were among 2,169 chronically infected patients confirmed to be positive for HBsAg in La Pitié-Salpêtrière Hospital (Paris, France) between January 1994 and September 2005. The stringent selection of the patients with at least two samples without HBcAb permitted exclusion of any technical artifacts.
Serological profiles.
Detection of HBV serological markers was routinely performed by using specific Axsym tests (HBsAg v2.0, Core, HBe; Abbott Diagnostics, Les Ulis, France), and viral load (VL) was estimated by the HBV Hybrid Capture (Digene-Abbott, Les Ulis, France) or the HBV Monitor (Roche, Meylan, France) assay.
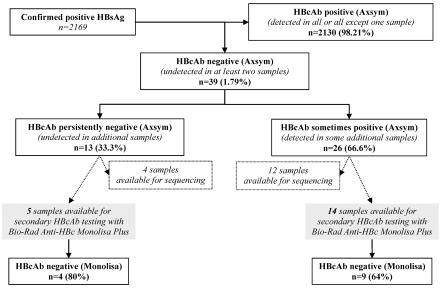
Among 2,169 confirmed HBsAg-positive chronically HBV-infected patients, 39 (1.79%) who lacked HBcAb were identified. For 26 of the 39 patients (66.6%), at least one other additional sample proved to be HBcAb positive, while for the other 13 patients, HBcAb was never detected in any of the available samples (Fig. 1). We have studied 14 and 5 samples from these two groups of patients with and without HBcAb detection in any additional samples, respectively (Fig. 1). The selection of these 19 samples was solely based on the availability of a sufficient volume that had been stored under proper conditions to perform all complementary analyses. The main characteristics of these 19 patients are summarized in Table 1. All samples were positive for HBeAg, and most samples had high HBV viral loads.
FIG. 1.
HBcAb detection during hepatitis B virus infection. Among 39 chronic hepatitis B patients without HBcAb on at least two occasions, additional samples from 13 were persistently negative for HBcAb, while at least one additional HBcAb sample from 26 of them was positive. Nineteen samples were further studied by a second HBcAb assay (Monolisa; Bio-Rad), and for 16 of them the precore and core gene sequences were analyzed.
TABLE 1.
Main patient characteristics
| HBcAb result for additional samples and patient no. | Age (yr) | Sexa | HBeAg result | HbeAb result | Anti-HBc Monolisa PLUS resultb | HBV DNA load (log copy no./ml) | No. of samples negative or positive for HBcAb among all available samplesc
|
Genotype | Mutationd | Concomitant pathology | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | |||||||||||
| Negative | ||||||||||||
| 1 | 47 | M | + | − | − | 4.41 | 4 | 0 | A | HIV | ||
| 2 | 40 | M | + | − | − | >6 | 5 | 0 | D | HIV | ||
| 3 | 35 | M | + | − | − | >6 | 2 | 0 | E | PC (S11T) | HCV | |
| 4 | 49 | M | + | − | − | 4.21 | 6 | 0 | NAe | Kidney transplant | ||
| 5 | 42 | M | + | − | + | >6 | 2 | 0 | D | Kidney transplant | ||
| Positive | ||||||||||||
| 6 | 35 | M | + | − | − | >6 | 6 | 2 | A | Immunosuppressive treatment | ||
| 7 | 31 | M | + | − | − | >6 | 2 | 1f | A | HIV | ||
| 8 | 38 | M | + | − | − | >6 | 7 | 1 | A | HIV | ||
| 9 | 39 | M | + | − | − | >6 | 4 | 4 | A | HIV | ||
| 10 | 38 | F | + | − | − | >6 | 2 | 2 | C | Bone marrow transplant | ||
| 11 | 53 | M | + | − | − | >6 | 17 | 5 | D | Heart transplant | ||
| 12 | 44 | M | + | − | − | >6 | 3 | 3 | G | PC (W28stop) | HIV | |
| 13 | 35 | M | + | − | − | >6 | 4 | 1 | A | HIV | ||
| 14 | 43 | M | + | − | − | >6 | 6 | 1f | NA | HIV | ||
| 15 | 29 | M | + | − | + | >6 | 2 | 4 | A | HIV | ||
| 16 | 39 | M | + | − | + | 2.67 | 3 | 2 | D | HIV | ||
| 17 | 40 | M | + | − | + | >6 | 6 | 5 | NA | HIV | ||
| 18 | 49 | M | + | − | + | >6 | 2 | 3 | A | HIV | ||
| 19 | 29 | M | + | − | + | 4.33 | 2 | 2 | A | PC (W28stop), C (E64stop) | HIV | |
M, male; F, female.
In samples with HBsAg without HBcAb as detected by Axsym assay.
Result were obtained with the Axsym assay with all available additional samples.
Only mutations not described among reference and classical PC mutations.
NA, not available.
Weakly positive.
The absence of HBcAb, as identified by our routine screening test, based on a competitive immunoassay (Axsym Core), was confirmed by a second technique based on a direct sandwich enzyme-linked immunosorbent assay (anti-HBc Monolisa PLUS; Bio-Rad, Marnes la Coquette, France) for 13 of the 19 available samples (68.4%). Although we may have introduced a bias since all samples were selected on the basis of an absence of HBcAb, as measured by the Axsym assay, the data collected and presented in Table 1 seem to indicate that the Abbott assay has a slightly lower sensitivity than the Bio-Rad assay. It is noteworthy that this finding was also observed for two patients who presented with acute hepatitis; that is, the Bio-Rad assay became positive for the samples earlier than the Abbott assay did (data not shown).
Genotypes and mutations.
Purification of the HBV genome from 16 samples was done with a QIAamp MinElute Virus Spin kit (QIAGEN, Les Ulis, France). These available samples were from 4 patients in whom HBcAb was never detected and 12 patients in whom HBcAb was detected in some additional samples. The fragment encoding the basic core promoter, precore (PC), and most of the core regions (positions 1609 to 2401) was amplified by PCR (sense primer, GCATGGAGACCACCGTGAACG; antisense primer, TCTGCGAGGCGAGGGAGTTCT), and both strands were sequenced with the same primers. The sequences were then compared to a selection of different genotype reference sequences in GenBank, as reported by Stuyver et al. (11).
The genotype distribution obtained by phylogenetic analysis for the 16 patients was heterogeneous (genotypes A, C, D, E, and G) (Table 1) and representative of those detected in the patients monitored in our hospital units. Particular genotypic-specific sequences could therefore not account for the lack of HBcAb detection in the samples that were studied. The classical PC mutation at codon 28 was found in two samples (from patients 12 and 19) (Table 1). Comparison of the core sequences from the samples to those in the reference database (11) did not highlight any particular mutations that could explain a lack of reactivity of the assays, and most of the few mutations that were found were silent or already described in reference sequences. We therefore ruled out the hypothesis that a modified core epitope would have precluded the recognition of HBcAb by these commercial assays. Moreover, even though information had been difficult to obtain from the two companies, the two manufacturers seem to use different recombinant core antigens in their assays. Thus, it would be difficult to explain the absence of reactivity of these two assays solely on the basis of a lack of specificity.
Interestingly, the absence of a mutation or a deletion in the same genomic region has already been shown in the sera of blood donors or HBV carriers presenting with the same serological pattern (4-6, 12). However, in our study, two previously unreported mutations were detected in two samples. In one case, it was an S11T change in the precore-core open reading frame (patient 3); in the second case, it was an E64stop in the core region (patient 19) (Table 1). The latter change should be considered with caution because it is the consequence of a single G deletion after a set of G repeats, and it may be due to a Taq polymerase error during amplification rather than a true deletion. It is hardly likely that the C (E64stop) nonsense mutation influences the core protein or HBcAb synthesis because HBcAb was detected in the sample by the Bio-Rad assay and in additional samples from the same patient by the Abbott test. Therefore, in this human immunodeficiency virus (HIV)-infected patient, we can rule out the absence of detection of HBcAb because of truncated core synthesis. Mutation S11T, which was observed in the sample from patient 3, is uncommon and deserves further study to determine whether it may interfere with the core protein conformation or may modify a T- or B-cell-particular epitope. Indeed, we may speculate that changes in protein antigenicity may lead to reduced reactivity in analytical assays. Moreover, as described previously, mutations within B- or T-cell epitopes might lead to more virulent HBV strains (13).
Concomitant pathologies.
It is striking that a common feature to all the patients was a state of relative immunosuppression. For most of them (24/39), coinfection with HIV was the cause for depressed immunity, while for the remaining patients, treatment for kidney (4/39), heart (7/39), or bone marrow (2/39) transplantation or systemic inflammatory disease (1/39) could potentially explain the decreased antibody production. One patient was coinfected with hepatitis C virus (HCV). Several follow-up samples were available from some of these patients, and the absence of HBcAb was concomitant with a more severe immunosuppression, as revealed by a CD4-cell count less than 50/mm3 in association with a rather high HIV plasma VL above 100,000 copies per ml, while HBcAb was detectable when the level of immunosuppression was less significant.
Failure to produce HBcAb during HBV infection has been observed in three different circumstances. First, HBV variants, especially those with deletions in the core gene, which leads to the synthesis of a truncated core protein, could cause infections with a low level of production or a lack of detection of HBcAb (3, 13). Second, a lack of HBcAb production has been reported in infants born to HBeAg-positive carrier mothers and has been linked to helper T-cell tolerance to HBcAg and HBeAg induced by transplacental maternal HBeAg (7, 10). Third, a lack of responsiveness to HBcAg has been encountered in immunocompromised patients (1, 8, 9). The last hypothesis was confirmed by our observations for most patients. Finally, the diagnostic tests have continuously improved over the last few decades; however, sensitivity and specificity differences exist between assays and may sometimes explain the discrepant results for some patients. In immunosuppressed patients, the choice of the most sensitive assay will yield the most reliable diagnostic approach. It is noteworthy that the absence of HBcAb has also been attributed to a new virus, termed HBV2 (2). However, discrepant data have been produced since this first report without further characterization of this virus. Our data are in agreement with those recently described by Terada et al. (12), who failed to identify any particular mutation in patients presenting a HBV2-like serological profile and who also came to the conclusion that this feature is likely due to an immune response abnormality of the host.
We conclude that in addition to the early phase following HBV infection, the detection of HBsAg without HBcAb may occur in highly immunocompromised patients, as seen in the context of transplantation or an uncontrolled HIV infection. Our study strongly suggests that this rare atypical serological pattern is not linked to a specific HBV genotype or to mutations that would translate into a peculiar core protein and subsequently into antibody production with atypical specificity. Finally, in some cases, it might be connected to a sensitivity default of the analytical assay. We may expect that these patients will eventually develop antibodies against the core antigen when the immunosuppression is less pronounced. Although an absence of HBcAb during chronic HBV infection is usually not linked to a more severe disease course, we should be cautious because of the risk of sometimes severe flare-ups associated with the development of an efficient immune response to the HBV antigen (8). Monitoring of the HBcAb level may be a useful tool for the identification of such clinically relevant events.
Acknowledgments
This work was supported by grants from Assistance Publique-Hôpitaux de Paris and Université Pierre et Marie Curie, Paris 6.
The authors have no conflict of interest in this publication.
REFERENCES
- 1.Bhat, R. A., P. P. Ulrich, and G. N. Vyas. 1990. Molecular characterization of a new variant of hepatitis B virus in a persistently infected homosexual man. Hepatology 11:271-276. [DOI] [PubMed] [Google Scholar]
- 2.Coursaget, P., B. Yvonnet, C. Bourdil, Y. Buisson, J. Chotard, R. N'Doye, C. Molinie, I. Diop-Mar, and J. P. Chiron. 1990. Hepatitis B surface antigen reactivity in man due to a new variant of hepatitis B virus. Vaccine 8(Suppl.):S15-S17. [DOI] [PubMed] [Google Scholar]
- 3.Fiordalisi, G., D. Primi, E. Tanzi, E. Magni, C. Incarbone, A. R. Zanetti, and E. Cariani. 1994. Hepatitis B virus C gene heterogeneity in a familial cluster of anti-HBc negative chronic carriers. J. Med. Virol. 42:109-114. [DOI] [PubMed] [Google Scholar]
- 4.Gotoh, K., S. Mima, T. Uchida, T. Shikata, K. Yoshizawa, M. Irie, and M. Mizui. 1995. Nucleotide sequence of hepatitis B virus isolated from subjects without serum anti-hepatitis B core antibody. J. Med. Virol. 46:201-206. [DOI] [PubMed] [Google Scholar]
- 5.Laperche, S., C. Guitton, W. Smilovici, and A. M. Courouce. 2001. Blood donors infected with the hepatitis B virus but persistently lacking antibodies to the hepatitis B core antigen. Vox Sang. 80:90-94. [DOI] [PubMed] [Google Scholar]
- 6.Lee, J. H., T. G. Paglieroni, P. V. Holland, and J. B. Zeldis. 1992. Chronic hepatitis B virus infection in an anti-HBc-nonreactive blood donor: variant virus or defective immune response? Hepatology 16:24-30. [DOI] [PubMed] [Google Scholar]
- 7.Lee, S. D., K. J. Lo, Y. T. Tsai, J. C. Wu, and T. C. Wu. 1989. HBsAg carrier infants with serum anti-HBc negativity. Hepatology 9:102-104. [DOI] [PubMed] [Google Scholar]
- 8.Melegari, M., M. C. Jung, R. Schneider, T. Santantonio, S. Bagnulo, N. Luchena, G. Pastore, G. Pape, P. P. Scaglioni, E. Villa, and H. Will. 1991. Conserved core protein sequences in hepatitis B virus infected patients without anti-HBc. J. Hepatol. 13:187-191. [DOI] [PubMed] [Google Scholar]
- 9.Möller, B., U. Hopf, R. Stemerowicz, G. Henze, and H. Gelderblom. 1989. HBcAg expressed on the surface of circulating Dane particles in patients with hepatitis B virus infection without evidence of anti-HBc formation. Hepatology 10:179-185. [DOI] [PubMed] [Google Scholar]
- 10.Ni, Y. H., H. Y. Hsu, M. H. Chang, D. S. Chen, and C. Y. Lee. 1993. Absence or delayed appearance of hepatitis B core antibody in chronic hepatitis B surface antigen carrier children. J. Hepatol. 17:150-154. [DOI] [PubMed] [Google Scholar]
- 11.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 12.Terada, T., M. Moriyama, T. Uchida, and Y. Arakawa. 2001. Nucleotide sequence of the precore/core gene and X gene of hepatitis B virus DNA in asymptomatic hepatitis B virus carriers who are negative for serum hepatitis B core antibody. Intervirology 44:243-249. [DOI] [PubMed] [Google Scholar]
- 13.Zoulim, F., X. Zhang, C. Pichoud, and C. Trepo. 1996. Heterogeneity of hepatitis B virus (HBV) core gene in a patient with HBV-associated cirrhosis and serum negativity for anti-HBc. J. Hepatol. 24:155-160. [DOI] [PubMed] [Google Scholar]