Abstract
Ziehl-Neelsen (ZN) staining for the diagnosis of tuberculosis (TB) is time-consuming and operator dependent and lacks sensitivity. A new method is urgently needed. We investigated the potential of an electronic nose (EN) (gas sensor array) comprising 14 conducting polymers to detect different Mycobacterium spp. and Pseudomonas aeruginosa in the headspaces of cultures, spiked sputa, and sputum samples from 330 culture-proven and human immunodeficiency virus-tested TB and non-TB patients. The data were analyzed using principal-component analysis, discriminant function analysis, and artificial neural networks. The EN differentiated between different Mycobacterium spp. and between mycobacteria and other lung pathogens both in culture and in spiked sputum samples. The detection limit in culture and spiked sputa was found to be 1 × 104 mycobacteria ml−1. After training of the neural network with 196 sputum samples, 134 samples (55 M. tuberculosis culture-positive samples and 79 culture-negative samples) were used to challenge the model. The EN correctly predicted 89% of culture-positive patients; the six false negatives were the four ZN-negative and two ZN-positive patients. The specificity and sensitivity of the described method were 91% and 89%, respectively, compared to culture. At present, the reasons for the false negatives and false positives are unknown, but they could well be due to the nonoptimized system used here. This study has shown the ability of an electronic nose to detect M. tuberculosis in clinical specimens and opens the way to making this method a rapid and automated system for the early diagnosis of respiratory infections.
The World Health Organization (WHO) has declared tuberculosis (TB) a global emergency. It is estimated that one-third of the world's population is infected with Mycobacterium tuberculosis. An estimated 8 to 9 million new cases occur each year, with 2 to 3 million deaths (4). The majority of these new infections and deaths occur in developing countries. The human immunodeficiency virus (HIV) epidemic has massively contributed to the worldwide tuberculosis problem.
The usual method of diagnosing TB in low-income countries is by detection of acid-fast bacteria in sputum by direct microscopy. When done properly, 60 to 70% of all adults with pulmonary TB can be identified using the Ziehl-Neelsen (ZN) staining procedure, followed by microscopic examination (11, 19). However, in areas of endemicity, laboratories are often overloaded with samples for smear examination. Therefore, a new simple and rapid diagnostic test should directly replace microscopy with similar specificity and sensitivity (19). In the past, research was mainly focused on the development of either antibody/antigen detection assays or the development of nucleic acid amplification reactions.
Against this background, we have investigated the potential of a gas sensor array (“electronic nose” [EN]) to detect M. tuberculosis in culture and sputum. It is well known that smell can be used to diagnose diseases, and it has been used by both the Greeks and the Chinese since 2,000 BC (12). Electronic nose is the colloquial name for an instrument made up of chemical sensors combined with a pattern recognition system (5). The reversible adsorption of volatile organic compounds (VOCs) to the sensor surface leads to a change of physical properties (conductivity, resistance, and frequency) of the sensor, which is measured. The key function of an EN is to mimic the human olfactory system by combining nonspecific gas sensors with a pattern recognition system to analyze and characterize complex odors without separation of the mixture into individual components. In the EN, the human olfactory receptors have their analogues in chemical sensors that produce an electrical signal (similar to nerve cells). Each sensor within the array is characterized by partial and overlapping specificities to VOCs. Due to the partial and overlapping specificities, a unique response curve is recorded during the measurement by each sensor containing the vital information to allow discrimination of the different samples. To describe this information, the response curve is described by mathematical terms expressed as maximum absorption rate, desorption rate, maximum response (or divergence), and area under the response curve. These mathematical terms are subsequently analyzed by pattern recognition software. The pattern recognition software corresponds to the cerebral cortex of the brain and is able to classify and memorize odors (1, 18).
Electronic noses have been applied mainly in the food industry to characterize the odors of beverages (2, 17) or olive oil (7). More recently, researchers discovered the potential of electronic noses as a diagnostic tool for the detection of Mycobacterium bovis in badgers and cattle (3). A review of medical applications is given elsewhere (15, 23).
The aim of this study was to investigate the potential of an electronic nose to detect Mycobacterium tuberculosis and other pathogens in both culture and patients' sputa as a first step toward simple breath analysis for the specific, rapid, and noninvasive diagnosis of diverse lung infections.
MATERIALS AND METHODS
Liquid cultures.
All bacteria (Mycobacterium tuberculosis, RIVM myc 4514; Mycobacterium avium, RIVM myc 3875; Mycobacterium scrofulaceum, RIVM myc 3442; and Pseudomonas aeruginosa, AMC 23123) were cultured in Middlebrook 7H9 medium with oleic acid-albumin-dextrose-catalase enrichment. The bacteria were incubated at 37°C until an optical density (420 nm) of 0.30 (≈2 × 108 bacteria ml−1) was reached.
Sputum samples.
The study was approved by the ethics committee of the Academic Medical Center, Amsterdam, The Netherlands, and The Saint Francis Hospital in Katete, Zambia. All subjects gave written permission for sputum sampling after oral and written information was provided.
Sputum samples were collected from 280 patients with suspected TB either from The Saint Francis Hospital in Katete, Zambia (n = 80), or from the WHO Sputum Bank (n = 200) (through the WHO Specimen Bank). In addition, sputum samples were collected at the Academic Medical Center, Amsterdam, The Netherlands, from 7 patients with proven pneumonia (caused by Streptococcus pneumoniae) and from 50 patients with proven non-TB (serving as negative controls). All patients were examined by chest X ray, and their sputum samples were examined by ZN staining and liquid culture (either by BacT/ALERT from bioMérieux, France, or by Bactec MGIT 960 from Becton Dickenson). Culture was used as the “gold standard” in this study as both inclusion and exclusion criteria for TB. Furthermore, the HIV status and the smoking habits of the suspected TB patients were investigated. The collected sputum samples were stored at −70°C until the analysis was performed.
Spiked sputum samples.
A pool of 25 sputum samples (each 1 ml) was made from the above-mentioned non-TB patients (n = 50) for spiking purposes. The sputum pool was spiked with various numbers of different bacterial isolates, including M. tuberculosis, M. avium, and P. aeruginosa, as well as a mixture (50:50) of M. tuberculosis and P. aeruginosa, which served as a mixed-infection sample.
Sample preparation and headspace analysis.
The liquid cultures were cooled to 4°C and allowed to equilibrate for at least 20 min to minimize the loss of volatiles during the transfer into smaller-headspace vials. Two milliliters of the “cold” culture was transferred into a 5-ml-headspace vial (Macherey and Nagel, United Kingdom) and immediately sealed with a silicon/Teflon crimp cap (Jaytee Bioscience Ltd., United Kingdom). The headspace was allowed to equilibrate for 45 min at 37°C.
The frozen sputum samples were defrosted on ice to minimize the loss of volatiles; 0.5 ml of “well-mixed” sputum was transferred into a sterile 5-ml-headspace vial, mixed with 0.5 ml of a 1 M NaCl solution (4°C), and subsequently sealed.
Spiked sputum samples were prepared by mixing 0.5 ml of non-TB sputa (individual samples or a pool) with 0.5 ml of “cold and equilibrated” bacterial suspension containing 1 M NaCl. Negative control samples were prepared by mixing 0.5 ml of individual non-TB sputum samples with 0.5 ml of a 1 M NaCl solution (4°C). All control samples were prepared in 5-ml-headspace vials as described above. All sputum samples were incubated at 37°C for 330 min prior to the headspace analysis. Two cycles of freezing and thawing had no influence on the results (not shown).
Gas-sensing system and headspace sampling.
For this study, an electronic nose (Bloodhound BH-114; Bloodhound Sensors, Leeds, United Kingdom) that employed 14 conducting polymers based on polyaniline was used. The sensor unit automatically set two calibration points. The first was the baseline, which was obtained when activated-carbon-filtered (Carbon Cap 150; Whatman) air was passed over the sensor at a flow rate of 4 ml min−1. The second calibration point was a reference point obtained from the headspace of a control sample vial containing 9 ml of distilled water.
The interaction of the VOCs with the conducting polymer surface produced a change in resistance over time, which was measured and subsequently displayed on a computer screen for each sensor. The curve was similar to the classical Langmuir adsorption curve. Two sensor parameters were selected to study the sensor response: divergence (maximum response) and area under the response curve. The sampling profile was set at 6 seconds of absorption and 14 seconds of desorption for the analysis of liquid samples; for the analysis of spiked sputum, we used 7 seconds of absorption and 21 seconds of desorption.
For the analysis of the unknown headspace, the sample vials were connected to the electronic nose by inserting a needle into the headspaces of the sample vials. The unknown headspace was passed over the sensor surface at a flow rate of 20 ml min−1, which was automatically set by the sensor unit. Between each pair of measurements, a time delay of 2 min was set. The individual samples in each experiment were tested in a randomized, blinded fashion.
Data analysis.
Principal-component analysis (PCA), discriminant function analysis (DFA), and an artificial neural network (ANN) were applied to analyze the multivariate data. The sensor response was normalized prior to the multivariate data analysis (except for the determination of the detection limit). To perform these analyses Excel add-in software (XLstat version 3.4) was used. Two sensor parameters, namely, the maximum response and the area under the response curve (Langmuir adsorption spectrum), were used to perform the analysis.
Principal-component analysis is a method aimed at reducing the amount of data when there is a correlation present. The idea is to find principal components (PC), which are linear combinations of the original variables (sensor responses) describing each specimen. In other words, PCA projects the original data matrix from a high-dimensional space into a lower-dimensional space (a three-dimensional space or plane) without losing essential information (variance). The relationship between samples can be visualized by plotting individual principal components against each other (13).
Discriminant function analysis is a supervised classification method aimed at finding a formal decision boundary between classes. The idea is to find linear discriminant functions (S1, S2, … , Sn), which are linear combinations of the original variables. The classification model (DFA) was built on the first four PC, which normally account for over 90% of the variance (information) of the original data matrix. In each case, the DFA model was cross-validated as follows. Data from individual samples (culture or sputum) were withheld, and a DFA model was built on the remaining data set (training set). The data from the withheld samples (testing set) were then inserted into the discriminant functions and subsequently assigned to the class for which the centroid had the smallest Euclidean distance to the unknown sample. The result could be visualized by plotting the individual discriminant functions against each other (13).
Artificial neural networks are attempts to mimic the neurons in the human brain and were used here for classification of sputum samples into TB and non-TB. Such networks have a number of linked layers of artificial neurons, including input, hidden, and output layers. The ANN is trained using a large training set of sputum samples from suspected TB patients. In this study, a back propagation network with a sigmoid transfer function was applied. During the training period, the weights connecting individual neurons were adjusted so that the error between output signal and target signal was minimized (13). The performance of the ANN was evaluated using the test set of sputum samples. Only the sputum samples from the suspected TB patients (n = 280) and from the proven non-TB cases (n = 50) were included in the ANN analysis.
RESULTS
Specificity of the EN.
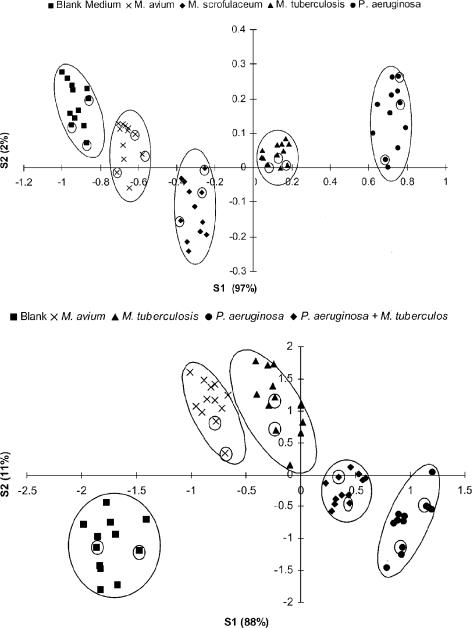
The “smells” of three Mycobacterium sp. cultures (M. tuberculosis, M. avium, and M. scrofulaceum) and one Pseudomonas aeruginosa culture were analyzed and compared to the “odor” of blank medium. The raw EN data were analyzed by PCA (data not shown), followed by DFA. The DFA model was built on the first four principal components. Figure 1 shows the results of the DFA analysis. It was possible to distinguish between the different bacterial classes using the first two discriminant functions (S1 and S2). The three different Mycobacterium spp. were grouped closely together but still allowed discrimination (Fig. 1, top). The DFA model was validated by the analysis of 15 “unknown” samples. All unknown samples were correctly classified as either one of the three Mycobacterium spp., P. aeruginosa, or blank medium.
FIG. 1.
(Top) DFA analysis of liquid cultures of M. tuberculosis (12 samples), M. avium (12 samples), M. scrofulaceum (12 samples), and P. aeruginosa (12 samples) and blank medium (12 samples). Cross-validation: 15 samples (3 from each group) were withheld from building the DFA model but were subsequently assigned correctly once the model was built (encircled symbols). S1 and S2, discriminant functions 1 and 2. The numbers in parentheses indicate the percentages of the data matrix described by the relevant functions. The circles were added by the authors. (Bottom) DFA analysis of sputum samples spiked with M. tuberculosis (12 samples), M. avium (12 samples), P. aeruginosa (12 samples), mixed infection (12 samples), and blank sputum (12 samples). Cross-validation: 10 samples (2 from each group) were withheld from building the DFA model but were subsequently assigned correctly once the model was built (encircled symbols).
Similarly, when the negative pooled sputum samples were spiked with M. tuberculosis, M. avium, P. aeruginosa, and a mixture of M. tuberculosis and P. aeruginosa (at a final concentration of 1 × 108 bacteria ml−1) and analyzed, it was possible to distinguish between “unspiked” sputum and “spiked” sputum samples (Fig. 1, bottom). Within the spiked sputum samples, a difference in smell was observable for the different bacterial classes. The DFA model was validated by the analysis of 10 unknown samples. All unknown samples were correctly identified as unspiked sputum or spiked sputum. Within the spiked sputum samples, all unknown samples were correctly assigned to one of the four “subclusters” representing the different bacterial classes (Fig. 1, bottom).
Analytical sensitivity of the EN.
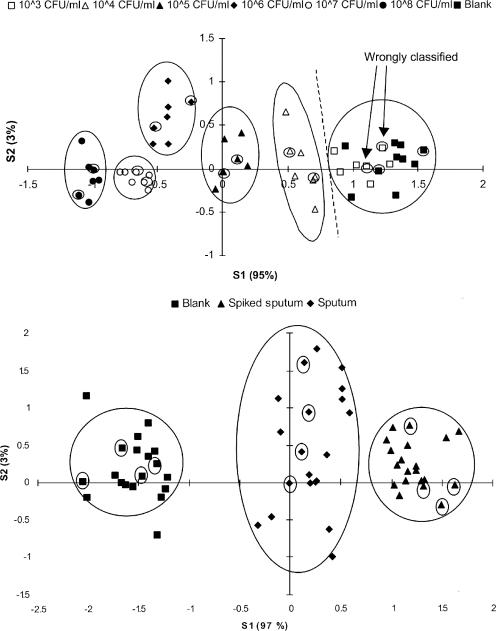
Six different concentrated M. tuberculosis suspensions (1 × 103 mycobacteria ml−1 to 1 × 108 mycobacteria ml−1) were analyzed and compared to blank medium. The raw EN data were analyzed by PCA (data not shown), followed by DFA. The results of the DFA analysis are shown in Fig. 2, top. The DFA model was validated by the analysis of 14 “unknown” samples. Two out of 14 unknown samples were incorrectly classified as blank medium (Fig. 2, top). Both incorrectly classified samples belonged to the group containing 1 × 103 mycobacteria ml−1. All other unknown samples were correctly identified. Therefore, the detection limit was determined to be as low as 1 × 104 mycobacteria ml−1.
FIG. 2.
(Top) Determination of the detection limit of the electronic nose for M. tuberculosis in liquid culture. Six different concentrations ranging from 1 × 103 to 1 × 108 mycobacteria ml−1 were analyzed (seven samples for each concentration and seven blanks). Cross-validation: 14 samples (2 samples from each group) were withheld from building the DFA model. Samples containing more than 1 × 104 mycobacteria ml−1 were correctly assigned (encircled symbols). In contrast, blank medium and samples containing 1 × 103 mycobacteria ml−1 could not be distinguished from each other. (Bottom) Determination of the detection limit of the electronic nose for M. tuberculosis in spiked sputum. The sputum was spiked with 1 × 104 mycobacteria ml−1. Twenty replicates were analyzed for each group. Cross-validation: 12 samples (4 samples from each group) were withheld from building the DFA model but were subsequently assigned correctly once the model was built (encircled symbols).
The detection limit for M. tuberculosis in spiked pooled sputum samples was also determined. For this purpose, a sputum pool was divided into two parts. One part was spiked with an M. tuberculosis suspension containing 1 × 104 mycobacteria ml−1. The raw EN data were analyzed by PCA (data not shown), followed by DFA. The bottom panel of Fig. 2 shows that it was possible to distinguish between spiked and unspiked samples. The DFA model was validated by the analysis of 12 “unknown” samples. All “unknown” samples were correctly classified (Fig. 2, bottom). The variability within the unspiked samples was higher than within the spiked samples.
Performance of the EN using clinical samples.
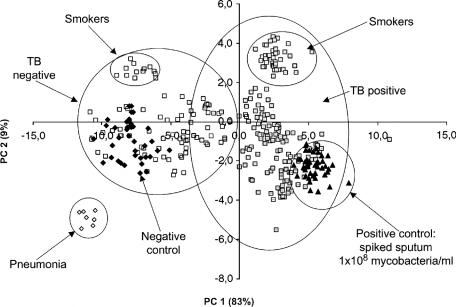
The PCA analysis of 50 positive control samples (i.e., the individual non-TB sputum samples spiked with 108 M. tuberculosis cells ml−1), 50 negative non-TB sputum samples from The Netherlands, and 280 clinical samples from Africa (Table 1) is shown in Fig. 3. It was possible to obtain good discrimination between TB and non-TB samples. As shown in Fig. 1, TB-negative samples were found on the left-hand side, whereas TB positive samples were on the right-hand side. Nevertheless, no complete separation could be obtained, as indicated by the overlapping circles in Fig. 3. In both groups (TB positive and TB negative), a subcluster could be identified. These subclusters contained the samples of smoking patients. However, not all smokers were present in this subcluster (89% were present). Patients suffering from pneumonia formed a separate cluster.
TABLE 1.
Details of patients that provided clinical samples, showing TB status (culture), sex, HIV status, and smoking habits
| Status | Total no. | % Males (no.) | % HIV+ (no.) | % Smokers |
|---|---|---|---|---|
| TB+ | 188 | 67.0 (126) | 53.7 (101) | 31.4 (59) |
| TB− | 142 | 59.2 (84) | 29.6 (42) | 9.2 (13) |
| Total | 330 | 63.6 (210) | 43.3 (143) | 21.8 (72) |
FIG. 3.
PCA plot showing the analysis of negative (Neg.) control samples (50 samples), positive (Pos.) control samples (50 samples), confirmed pneumonia (7 samples), and clinical samples (92 samples, TB negative; 188 samples, TB positive). PC 1 and PC 2 are the first two principal components; the numbers in parentheses represent the percentages of information described by each principal component (circles were added by the authors).
After training the neural network with 196 of the original samples (133 TB and 63 non-TB samples), the remaining 134 samples were used to validate the model. Among the 134 samples were 55 culture-confirmed TB samples, of which 51 were ZN positive and 4 were ZN negative. The results from the ANN are summarized in Table 2. The neural network was able to predict 49 TB-positive patients out of 55 correctly. Six suspected TB culture-positive samples gave a false-negative result. Among these six false negatives were four ZN-negative and two ZN-positive patients. Three of the four ZN-negative false negatives were HIV positive, and one was a smoker. Among the ZN-positive false negatives was one HIV-positive patient, who was also a smoker.
TABLE 2.
Performance of the electronic-nose-neural-network system in comparison to culture
| Status | No. (%) with culture-confirmed TB | No. (%) culture TB negative | Total |
|---|---|---|---|
| EN positive | 49 (89.9) | 7 (8.9) | 56 |
| EN negative | 6 (10.9) | 72 (91.1) | 78 |
| Total | 55 (100) | 79 (100) | 134 |
The neural network was also able to predict 72 suspected TB-negative patients correctly. Seven TB-negative patients gave a false-positive result. One false-positive patient was HIV positive and a smoker.
The sensitivity for the detection of culture-proven TB was 89% (95% confidence interval [CI], 80 to 97%), the specificity was 91% (95% CI, 85 to 97%), and the positive and negative predictive values were 88% (95% CI, 78 to 96%) and 92% (95% CI, 86 to 98%), respectively.
DISCUSSION
Current global TB control depends on the diagnosis of cases, followed by adequate treatment. The available laboratory methods for the detection of M. tuberculosis do not fully meet the need in environments with high TB and HIV prevalences (19).
This study showed that volatile detection through electronic-nose technology is able to identify M. tuberculosis in both cultures and sputum samples. It has long been established that smell can be used to diagnose diseases, such as diabetes and uremia (10, 20). Pavlou and Turner (15) and Pavlou et al. (14, 16) were among the first to apply electronic noses in medical diagnostics. They showed that different bacteria, such as Helicobacter pylori, E. coli, and M. tuberculosis, generate a unique “smell” and can therefore be differentiated from each other, allowing a diagnosis. Recently, the same EN used in this study was shown to be able to diagnose M. bovis infection in badgers and cattle (3).
Electronic-nose technology offers certain advantages, such as a low detection limit (5 to 0.1 ppm) (21), cost- and time effectiveness, robustness, simplicity, and operator independence, in contrast to molecular or immunologically based assays. Therefore, we investigated the ability of an electronic nose to detect Mycobacterium spp. and other lung pathogens in culture and sputum. In this study, we showed that Mycobacterium spp. and Pseudomonas aeruginosa emit characteristic volatiles, allowing discrimination between the different bacterial classes in both culture and sputum (Fig. 1). The intragroup (class) variability is greater in sputum than in culture. The reasons for this observation are not clear. The volatility of molecules is influenced by parameters such as sample viscosity, equilibrium temperature, and concentration (22). Since the incubation parameters for liquid and sputum samples were similar, we assume that the higher viscosity, the heterogeneity, and/or a stronger background “smell” of sputum might be responsible for the variability. As shown in Fig. 1, bottom, and 2, bottom, by adding M. tuberculosis to sputum, an additional “odor” was introduced into the sample headspace, leading to a reduced intragroup variability. This indicates that mycobacteria release enough volatiles into the headspace, even in a complex matrix (sputum), to allow a diagnosis at low concentrations (1 × 104 mycobacteria ml−1). This result is of extreme importance for clinical diagnosis, where sputum is the usual source for TB detection.
Electronic noses show a linear relationship between sensor response and concentration (8). This concentration dependency was exploited here to determine the detection limit (Fig. 2). This relationship might also reveal relevant clinical information. For the treatment itself, it is not important how many bacteria are present in sputum, but the number of bacteria present greatly influences the infectiousness of patients. This opens the possibility to predict not only the presence of TB, but also the risk for patients to transmit the disease.
The sensitivity of the ZN stain compared to culture under field conditions is at most 50 to 60% (19). With the method presented, we achieved a specificity of 91% and a sensitivity of 89% compared to culture (Table 2). The electronic nose was unable to detect four ZN-negative but culture-positive specimens (false negatives). The bacterial loads in these four specimens were most likely below the detection limit of the electronic nose under the current setup, but larger numbers of ZN-negative but culture-positive specimens need to be tested. The detection limit of the electronic nose for M. tuberculosis in spiked sputum was 104 mycobacteria ml−1 (Fig. 2, top). However, the two remaining false-negative specimens had a positive ZN stain (1+) and should therefore contain enough mycobacteria to cause a sufficient sensor response. At present, we do not know the reasons for either the false negatives or false positives. They could well be due to the nonoptimized system used here or to sample degradation during storage.
Clinical specimens are more diverse than spiked samples. It is assumed that the viscosity, the background smell, and especially, the heterogeneity of sputa influence the outcome of the analysis. Interestingly, smoking itself did not affect the analysis in terms of diagnosing TB. However, not all smokers were grouped in the subclusters shown in Fig. 3. The individual smoking habits (number of cigarettes per day and last cigarette before sample taking) could not be established. We assume that certain smoke ingredients give rise to a slightly different sensor response, allowing separation.
Seven cases of pneumonia were among the clinical specimens. As shown in Fig. 3, they formed a separate cluster. This indicates that the causative agent (in this case, S. pneumoniae) for pneumonia generates a different volatile profile (smell) than mycobacteria. This is of clinical importance, showing the potential to differentiate between TB cases and cases of other respiratory diseases.
To date, it is unknown which volatile compounds are responsible for the sensor response. We assume that the response is caused by the combined effects of (i) microbial metabolites and (ii) volatile cellular compounds. In the past, many research groups tried to identify volatile substances emitted from microorganism using gas chromatography or gas chromatography-mass spectroscopy. Each Mycobacterium species synthesizes a unique set of mycolic acids, among other substances, which might allow discrimination between different Mycobacterium species (Fig. 1) (9). In contrast, Pseudomonas aeruginosa emits sulfur compounds and esters (6).
The described method is not yet fully optimized for “field” application. Nevertheless, it potentially fulfils all requirements for a new diagnostic tool for TB (19), including robustness, simplicity, sensitivity, and cost-effectiveness. Among many advantages are the simple sample preparation and its amenability to automation. Together with an appropriate classification model, this method has the potential to become a rapid and automated system for the early diagnosis of respiratory diseases through sputum or even breath analysis. It might also be possible to improve or modify currently available sensors toward specific M. tuberculosis markers, which would simplify the optimization of such a system.
Acknowledgments
This work was financially supported by a TDR grant (A 20565) from the World Health Organization.
We thank Sjoukje Kuijper for her help during the experimental phase of this work.
REFERENCES
- 1.Bartlett, P. N., J. M. Elliot, and J. W. Gardner. 1997. Application of, and developments in, machine olfaction. Ann. Chim. 87:33-44. [Google Scholar]
- 2.Di Natale, C., F. A. M. Davide, A. D'Amico, P. Nelli, S. Groppelli, and G. Sberveglieri. 1996. An electronic nose for the recognition of the vineyard of a red wine. Sens. Actuators B 33:83-88. [Google Scholar]
- 3.Fend, R., R. Geddes, S. Lesellier, H.-M. Vordermeier, L. A. L. Corner, E. Gormley, E. Costello, R. G. Hewinson, D. J. Marlin, A. C. Woodman, and M. A. Chambers. 2005. Use of an electronic nose to diagnose Mycobacterium bovis infection in badgers and cattle. J. Clin. Microbiol. 43:1745-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frieden, T. R., T. R. Sterling, S. S. Munsiff, C. J. Watt, and C. Dye. 2003. Tuberculosis. Lancet 362:887-899. [DOI] [PubMed] [Google Scholar]
- 5.Gardner, J. W., and P. N. Bartlett. 1994. A brief history of electronic noses. Sens. Actuators B 18:210-220. [Google Scholar]
- 6.Gibson, T. D., O. Prosser, J. N. Hulbert, R. W. Marshall, P. Corcoran, P. Lowery, E. A. Ruck-Keene, and S. Heron. 1997. Detection and simultaneous identification of microorganism from headspace samples using an electronic nose. Sens. Actuators B 44:413-422. [Google Scholar]
- 7.Guadarrama, A., M. L. Rodríguez-Méndez, J. A. de Saja, J. L. Ríos, and J. M. Olías. 2000. Array of sensors based on conducting polymers for the quality control of the aroma of the virgin olive oil. Sens. Actuators B 69:276-282. [Google Scholar]
- 8.Hudon, G., C. Guy, and J. Hermia. 2000. Measurement of odor intensity by an electronic nose. Air Waste Manag. Assoc. 50:1750-1758. [DOI] [PubMed] [Google Scholar]
- 9.Jantzen, E., T. Tangen, and J. Eng. 1989. Gas chromatography of mycobacterial fatty acids and alcohols: diagnostic applications. APMIS 97:1037-1045. [DOI] [PubMed] [Google Scholar]
- 10.Lin, Y. J., H. Guo, and Y. Chang. 2001. Application of the electronic nose for uraemia diagnosis. Sens. Actuators B 76:177-180. [Google Scholar]
- 11.Lipsky, B. A., J. A. Gates, F. C. Tenover, and J. J. Plorde. 1984. Factors affecting the clinical value of microscopy for acid-fast bacilli. Rev. Infect. Dis. 6:214-222. [DOI] [PubMed] [Google Scholar]
- 12.Mitruka, B. M. 1975. Presumptive diagnosis of infectious diseases, p 349-374. In B. M. Mitruka (ed.), Gas chromatographic applications in microbiology and medicine. John Wiley and Sons, New York, N.Y.
- 13.Otto, M. 1999. Chemometrics: statistics and computer application in analytical chemistry. John Wiley and Sons, New York, N.Y.
- 14.Pavlou, A., N. Magan, C. McNulty, J. Meecham Jones, D. Sharp, J. Brown, and A. P. F. Turner. 2002. Use of an electronic nose system for diagnosis of urinary tract infections. Biosens. Bioelectron. 17:893-899. [DOI] [PubMed] [Google Scholar]
- 15.Pavlou, A. K., and A. P. Turner. 2000. Sniffing out the truth: clinical diagnosis using the electronic nose. Clin. Chem. Lab. Med. 38:99-112. [DOI] [PubMed] [Google Scholar]
- 16.Pavlou, A. K., N. Magan, J. Meecham Jones, J. Brown, P. R. Klatser, and A. P. F. Turner. 2004. Use of an electronic nose system for diagnoses of urinary tract infections. Biosens. Bioelectron. 20:538-544. [DOI] [PubMed] [Google Scholar]
- 17.Pearce, T. C., J. W. Gardner, and S. Friel. 1993. Electronic nose for monitoring the flavours of beers. Analyst 118:371-377. [Google Scholar]
- 18.Pearce, T. C. 1997. Computational parallels between the biological olfactory pathway and its analogue “the electronic nose.” Part II. Sensor based machine olfaction. Biosystems 41:69-90. [DOI] [PubMed] [Google Scholar]
- 19.Perkins, M. D. 2000. New diagnostic tools for tuberculosis. Int. J. Tuberc. Lung. Dis. 4:182-188. [PubMed] [Google Scholar]
- 20.Ping, W., H. Yin, X. Haibao, and S. Farong. 1997. A novel method for diabetes diagnosis based on electronic noses. Biosens. Bioelectron. 12:1031-1036. [DOI] [PubMed] [Google Scholar]
- 21.Schiffman, S., B. Kermani, and H. Nagle. 1997. Analysis of medication off-odors using an electronic nose. Chem. Senses 22:119-128. [DOI] [PubMed] [Google Scholar]
- 22.Seto, Y. 1994. Determination of volatile substances in biological samples by headspace gas chromatography. J. Chromatogr. A 674:25-62. [Google Scholar]
- 23.Woodman, A. C., and R. Fend. 2004. Electronic-nose technology: potential applications in point-of-care clinical diagnosis and management, p. 61-70. In C. P. Price, A. St. John, and J. M. Hicks (ed.), Point of care testing, 2nd ed. AACC Press, Washington, D.C.