Abstract
The Roche PGMY primer-based research prototype line blot assay (PGMY-LB) is a convenient tool in epidemiological studies for the detection and typing of human papillomavirus (HPV) DNA. This assay has been optimized and is being commercialized as the Linear Array HPV genotyping test (LA-HPV). We assessed the agreement between LA-HPV and PGMY-LB for detection and typing of 37 HPV genotypes in 528 anogenital samples (236 anal, 146 physician-collected cervical, and 146 self-collected cervicovaginal swabs) obtained from human immunodeficiency virus-seropositive individuals (236 men and 146 women). HPV DNA was detected in 433 (82.0%) and 458 (86.7%) samples with PGMY-LB and LA-HPV (P = 0.047), respectively, for an excellent agreement of 93.8% (kappa = 0.76). Of the 17,094 HPV typing results, 16,562 (1,743 positive and 14,819 negative results) were concordant between tests (agreement = 96.9%; kappa = 0.76). The mean agreement between tests for each type was 96.4% ± 2.4% (95% confidence interval [CI], 95.6% to 97.2%; range, 86% to 100%), for an excellent mean kappa value of 0.85 ± 0.10 (95% CI, 0.82 to 0.87). However, detection rates for most HPV types were greater with LA-HPV. The mean number of types per sample detected by LA-HPV (4.2 ± 3.4; 95% CI, 3.9 to 4.5; median, 3.0) was greater than that for PGMY-LB (3.4 ± 3.0; 95% CI, 3.1 to 3.6; median, 2.0) (P < 0.001). The number of types detected in excess by LA-HPV in anal samples correlated with the number of types per sample (r = 0.49 ± 0.06; P = 0.001) but not with patient age (r = 0.03 ± 0.06; P = 0.57), CD4 cell counts (r = 0.06 ± 0.06; P = 0.13), or the grade of anal disease (r = −0.11 ± 0.06; P = 0.07). LA-HPV compared favorably with PGMY-LB but yielded higher detection rates for newer and well-known HPV types.
Infection by human papillomavirus (HPV) causes squamous intraepithelial lesions and invasive cancer of the uterine cervix and anus (3). HPV testing relies on the detection and analysis of viral DNA. Epidemiological studies and vaccine clinical trials require reliable and reproducible identification and genotyping of genital HPV infections. Since only a fraction of the 40 HPV genotypes infecting the anogenital tract are associated with malignant lesions, the detection method has to identify types individually. Specific genotyping also provides information on mixed HPV infections (26). Type-specific PCR assays are impractical for epidemiological studies because of the multiplicity of relevant genotypes infecting the anogenital tract. Consensus PCR assays that target conserved regions of the HPV genome have been devised to amplify all relevant genital types in one reaction, with analysis of amplicons by direct sequencing, restriction fragment length polymorphism analysis, or type-specific hybridization.
The most common PCR methods use the consensus primer set MY09/MY11/HMB01 (20, 25), GP5+/GP6+ (9, 21), PGMY09/PGMY11 (13, 34), or SPF10 (30, 34). Convenient assays for detection and typing of HPV have been developed for all of these primer sets. HPV amplicons generated by PGMY or MY primers can easily be detected and typed by a nonisotopic reverse hybridization assay, the line blot assay (14). In previous studies, the reverse line blot assay combined with PCR using the MY09-MY11 or PGMY09-PGMY11 primers compared favorably to a dot blot assay using type-specific radiolabeled oligonucleotide probes for HPV typing of PCR-amplified products (6, 7, 23).
Consensus L1 PGMY09/11 primers were reported to improve the sensitivity for HPV detection over that of the MY09/MY11/HMB01 primers (6, 12, 13, 23). The research prototype version of the assay, the PGMY line blot assay (PGMY-LB), has been further developed and is now commercially available from Roche Molecular Systems under the designation Linear Array HPV genotyping test (LA-HPV). The reagents used for LA-HPV are now standardized and produced under quality-controlled conditions. Amplification profiles and reagents have been optimized to increase the sensitivity and reproducibility, mainly by avoiding competition during coamplification of β-globin and HPV DNA (J. Kornegay, personal communication). The use of standardized protocols for HPV detection and typing increases the reproducibility of results and should facilitate the comparison of results between studies (22).
The aim of the present study was to compare results obtained with PGMY-LB and LA-HPV for 528 anogenital specimens collected from 236 men and 146 women for HPV DNA detection and typing. The potential to separately identify types in multiple-type infections, a frequent occurrence in human immunodeficiency virus (HIV)-seropositive hosts, was also explored. The rates of detection of the 37 genotypes were compared between assays. This information is useful in view of the wider use of LA-HPV by numerous research groups as well as its potential application in diagnostic laboratories for HPV typing.
MATERIALS AND METHODS
Cell lines and clinical specimens.
The cervical carcinoma cell line HeLa (which contains 40 copies of HPV type 18 [HPV-18] DNA per cell) was obtained from the American Type Culture Collection (Rockville, Md.). Overall, 528 genital specimens were obtained from 382 participants (146 women and 236 men) in two different cohort studies on the natural history of anogenital HPV infection during the course of HIV infection. Each participant in both studies gave written informed consent for HPV testing and completed a standardized questionnaire. Both studies were approved by the local research ethics committees of participating institutions. Male subjects were participants in a cohort study of the natural history of anal intraepithelial neoplasia (AIN). Anal swabs obtained at the inclusion visit for the cohort were collected from men and agitated in 1.5 ml of Preservcyt (Cytyc Corporation, Boxborough, MA) (33). After centrifugation at 13,000 × g for 15 min at 22°C, the supernatant was discarded, and the cell pellet was left to dry and resuspended in 300 μl of 20 mM Tris buffer, pH 8.3 (31, 33). Female subjects were from The Canadian Women's HIV Study, conducted across Canada to evaluate relationships between HPV and HIV infection and cervical disease (18, 28). Self-collected cervicovaginal (n = 146) and physician-collected cervical (n = 146) swabs were obtained from the same women to evaluate self-sampling for HPV detection, as described previously (28). After being sampled, each swab was agitated in a tube containing 1 ml of 10 mM Tris, pH 8.3, with 0.1 mM EDTA and then discarded. Purification of DNA was done with a Master Pure extraction kit (Epicenter, Madison, WI) for all samples (17). Five microliters of processed sample diluted in 45 μl of sterile water was tested in each PCR assay, since 50 μl of processed sample collected in Preservcyt is tested in the commercial LA-HPV protocol. All samples yielded a β-globin amplicon when amplified individually with 10 pmol each of the PC04 and GH20 primers, as previously described, and migrated in an ethidium bromide-stained agarose gel, confirming the presence of amplifiable DNA (2, 8).
LA-HPV.
PCR was performed in a final reaction volume of 100 μl with 50 μl of kit working master mix containing MgCl2, KCl, AmpliTaq Gold DNA polymerase, uracil-N-glycosylase, dATP, dCTP, dGTP, dUTP, dTTP, and biotinylated PGMY primers and β-globin primers GH20 and PC04. The mixture was incubated in a TC 9700 thermal cycler set at maximum ramp speed for 2 min at 50°C and 9 min at 95°C, followed by 40 cycles of 30 s at 95°C, 1 min at 55°C, and 1 min at 72°C, with a final extension at 72°C (ramp rate set at 50%) for 5 min. Biotinylated amplicons were denatured in 0.4 N NaOH and hybridized to an immobilized probe array containing probes for 37 HPV genotypes according to the protocol provided by Roche Molecular Systems. Positive hybridization reactions were detected by streptavidin-horseradish peroxidase-mediated color precipitation on the membrane at the probe line. The probe for detection of HPV-52 amplicons on the array was a cross-reactive probe that also hybridized with types 33, 35, and 58. Samples positive with the HPV-52 cross-reactive probe and containing at least one of these types were also tested with a real-time PCR assay specific for type 52 validated in the laboratory of F. Coutlée (unpublished data; see below). Only samples reactive in the HPV-52 real-time PCR assay were considered HPV-52 positive.
PGMY-LB.
HPV DNA was amplified with the L1 consensus HPV PGMY09/PGMY11 primer set, as previously described (13). The amplification mixture contained 4 mM MgCl2, 50 mM KCl, 7.5 units of AmpliTaq Gold DNA polymerase (Perkin-Elmer, Foster City, CA), a 200 μM concentration (each) of dATP, dCTP, and dGTP, 600 μM of dUTP, 100 pmol of each biotinylated PGMY primer, and 2.5 pmol each of the 5′-biotinylated β-globin primers GH20 and PC04. HPV was amplified in a TC9600 thermal cycler at 95°C for 9 min, followed by denaturation for 1 min at 95°C, annealing for 1 min at 55°C, and extension at 72°C for 1 min for a total of 40 cycles. Amplification was followed by a 5-minute terminal extension step at 72°C. HPV genotyping was performed with a reverse line blot detection system as previously described (14). PCR products were denatured in 0.4 N NaOH and hybridized to an immobilized probe array containing probes for 37 mucosotropic HPV genotypes. The array was similar to that used with LA-HPV but contained an HPV-52-specific probe. Positive hybridization was detected by streptavidin-horseradish peroxidase-mediated color precipitation on the membrane at the probe line.
Real-time PCR assay for HPV-16.
Each 20-μl reaction mixture contained 10 mM Tris-HCl, pH 8.0, 50 mM KCl, a 200 μM concentration (each) of dATP, dGTP, and dCTP, 400 μM dUTP, 0.05 μM of TaqMan probe HPV 16-TM, 0.3 pmol each of primers HPV 16-U and HPV 16-L (targeting the E6 gene), 3.0 mM MgCl2, and 5 units of AmpliTaq Gold DNA polymerase (15, 16). Capillaries were placed in a Light Cycler system (Roche Molecular Systems) and amplified at 95°C for 10 min, followed by 50 cycles at 95°C for 15 s and 55°C for 30 s. Ten copies of an HPV-16-expressing plasmid in 500 ng of cellular DNA served as a weak positive control.
Real-time PCR assay for HPV-52.
Each 20-μl reaction mixture contained 10 mM Tris-HCl, pH 8.0, 50 mM KCl, a 200 μM concentration (each) of dATP, dGTP, and dCTP, 400 μM dUTP, 0.05 μM of TaqMan probe 52-TM (CGTGCAGGGTCCGGGGTC), 0.3 pmol each of primers 52JA-3 (GAACACAGTGTAGCTAACGCACG) and 52JA-4 (GCATGACGTTACACTTGGGTCA) (targeting the E6 gene), 2.0 mM MgCl2, and 5 units of AmpliTaq Gold DNA polymerase. The primers had been used for PCR-sequencing experiments in previous work (1). Capillaries were placed in a Light Cycler system and amplified at 95°C for 10 min, followed by 50 cycles at 95°C for 15 s and 60°C for 60 s. Ten copies of an HPV-52-expressing plasmid in 500 ng of cellular DNA served as a weak positive control. This assay was shown to be sensitive and specific for HPV-52 DNA detection (F. Coutlée, unpublished data).
Statistical methods.
The crude percent agreement between both detection methods was the percentage of samples with identical results in both methods. Percentages of agreement for overall positivity (HPV DNA-positive samples), for positivity for high-risk HPV types as a group (types 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, and 82), and for positivity for each HPV type were calculated. Low-risk types were types 6, 11, 40, 42, 54, 61, 70, 72, 81, and CP6108, and types with unknown risk were types 55, 62, 64, 67, 69, 71, 73, 83, 84, and IS39 (HPV-82 subtype). The unweighted kappa statistic was calculated to adjust for chance agreement between HPV detection methods (11). In general, kappa values above 0.75 indicate excellent agreement, values between 0.40 and 0.75 indicate fair to good agreement, and values below 0.40 represent poor agreement beyond chance. The mean number of types detected per sample by each PCR test was compared using the sign test, since the frequencies of types per sample were not normally distributed. Two-sided McNemar's chi-square analysis for matched pair data was performed to analyze contingency tables comparing both PCR tests. Proportions were compared with the Z statistic.
RESULTS
A total of 528 anogenital samples from 378 HIV-seropositive participants were tested in one laboratory using the PGMY-LB and LA-HPV systems. The characteristics of these 378 participants are shown in Table 1. The human β-globin gene was detected with the high-concentration probe in 528 (100%) and 527 (99.8%) samples and with the low-concentration probe in 497 (94.1%) and 466 (88.3%) samples by LA-HPV and PGMY-LB, respectively (P = 0.001). All 528 samples had tested positive for β-globin after amplification with 10 pmol of GH20 and PC04 and gel electrophoresis of the amplicons. Of the 31 β-globin-negative samples with the low probe concentration in LA-HPV, 22 (71.0%) were HPV positive. Of the 62 β-globin-negative samples with the low probe concentration in PGMY-LB, 44 (71.0%) were HPV positive. A total of 12 samples tested negative with the low probe concentration for β-globin in both assays. The number of types detected per specimen was greater for β-globin-positive than β-globin-negative samples in LA-HPV (median and mean, 4.0 and 4.3 ± 3.4 versus 2.0 and 2.0 ± 2.3; P = 0.001) or PGMY-LB (median and mean, 3.0 and 3.5 ± 3.0 versus 2.0 and 2.2 ± 2.2; P = 0.001).
TABLE 1.
Clinical characteristics of 378 HIV-seropositive participants
| Characteristica | Value (no. of subjects [%], unless indicated otherwise)b
|
|
|---|---|---|
| CWHIS cohort | HIPVIRG cohort | |
| Age (median yrs [range]) | 32.5 (18.0-68.3) | 43.4 (26.8-66.5) |
| Age categories (yrs) | ||
| 10-19 | 3 (2.1) | 0 (0) |
| 20-29 | 48 (32.9) | 7 (3.0) |
| 30-39 | 70 (47.9) | 62 (26.3) |
| 40-49 | 17 (11.6) | 109 (46.2) |
| 50-59 | 5 (3.4) | 50 (21.2) |
| 60-69 | 1 (0.7) | 5 (2.1) |
| Unknown | 2 (1.4) | 3 (1.2) |
| Gender | ||
| Female | 46 (100) | 0 (0) |
| Male | 0 (0) | 236 (100) |
| Mean CD4 cell count ± SD | 507 ± 318 | 443 ± 261 |
| CD4 count categories | ||
| <200 cells/μl | 22 (15.1) | 37 (15.7) |
| 200-400 cells/μl | 40 (27.4) | 107 (45.3) |
| >400 cells/μl | 82 (56.2) | 87 (36.9) |
| Not tested | 2 (1.4) | 5 (2.1) |
| Ethnicity | ||
| Caucasian | 94 (64.4) | 218 (92.4) |
| African descent | 36 (24.7) | 3 (1.3) |
| Asian | 2 (1.4) | 3 (1.2) |
| First Nation | 9 (6.2) | 1 (0.4) |
| Unknown | 5 (3.4) | 11 (4.7) |
| Smoking status | ||
| Current smoker | 40 (47.6) | 88 (37.3) |
| Not currently smoking | 44 (52.4) | 148 (62.7) |
| Median no. of lifetime sexual partners | 7 | NAc |
| Cytology smear results | ||
| Normal | 124 (84.9) | 80 (33.9) |
| ASCUS | 2 (1.4) | 76 (32.3) |
| LSIL | 15 (10.3) | 65 (27.5) |
| HSIL | 0 (0.0) | 14 (5.9) |
| Not done | 5 (3.4) | 1 (0.4) |
| Histology results | ||
| Normal | NA | 57 (24.2) |
| AIN grade 1 | NA | 108 (45.8) |
| AIN grades 2 and 3 | NA | 70 (29.6) |
| Not done | NA | 1 (0.4) |
ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions; AIN, anal intraepithelial neoplasia.
NA, not available.
Median of two to five male sexual partners per year.
Results from both assays were first compared for HPV DNA detection, irrespective of type(s). As shown in Table 2, HPV DNA was detected in 433 (82.0%) and 458 (86.7%) of 528 samples by PGMY- LB and LA-HPV, respectively (P = 0.047). There was high agreement for HPV DNA by both methods for 495 (93.8%) samples (kappa = 0.76) (Table 2). Similar results were obtained when only the 447 β-globin-positive samples in both assays were considered in the comparison: 373 were positive and 47 were negative for HPV in both assays (concordance, 94.0%; kappa, 0.76; 95% confidence interval [CI], 0.69 to 0.84). Ninety-nine samples reacting with the HPV-52 cross-reactive probe and containing HPV-33, HPV-35, or HPV-58 DNA were retested with the HPV-52-specific real-time PCR assay. Sixty-three were shown to contain HPV-52 DNA.
TABLE 2.
Comparison of PGMY-LB and LA-HPV assays for detection of HPV DNAs from 37 genotypes in 528 anogenital samples
| Test result | No. (%) of samples with indicated resulta
|
Total | |
|---|---|---|---|
| PGMY-LB positive | PGMY-LB negative | ||
| LA-HPV positive | 429 (81.2) | 29 (5.5) | 458 (86.7) |
| LA-HPV negative | 4 (0.8) | 66 (12.5) | 70 (13.3) |
| Total | 433 (82.0) | 95 (18.0) | 528 (100) |
Absolute agreement, 93.8%; kappa = 0.76 (95% CI, 0.68 to 0.85).
We then compared typing results for 37 genotypes for the 462 specimens found to be HPV positive with at least one of the PCR tests, for a total of 17,094 type-specific results. Overall, 1,743 positive results for the same type and 14,819 negative results were obtained in both PCR tests, while 492 positive typing results were obtained only with LA-HPV and 40 were obtained only with PGMY-LB. The overall agreement was 96.9% (16,562 of 17,094 results), with a kappa value of 0.76 (95% CI, 0.68 to 0.85).
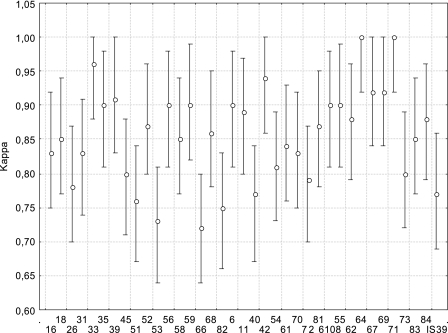
Since these agreement calculations are influenced by the high rates of negative typing results in both tests, we compared the specific identification of individual genotypes by each of the two methods for all samples (Fig. 1). The mean agreement reached 97% ± 2.5% (95% CI, 95.8% to 98.5%) and ranged from 93.2% to 100%. The mean kappa value reached 0.86 ± 0.07 (95% CI, 0.83 to 0.88). Kappa values below 0.75 were obtained only for types 66 and 53. Since histological diagnosis was available only for men, we then compared agreements between tests, considering the AIN grades for the eight most frequent types (types 16, 18, 53, 58, 6, 42, 61, and 84). The AIN grade did not modify agreement between tests, as the mean agreement for men without disease was 94.6% ± 5.4% (kappa, 0.83 ± 0.13), that for men with AIN grade 1 was 94.3% ± 2.7% (kappa, 0.84 ± 0.07), and that for men with AIN grades 2 and 3 was 94.6% ± 3.4% (kappa, 0.86 ± 0.11). These differences were not significant (P > 0.10 for each comparison). HPV-67 was the only type for which PGMY-LB detected a greater number of samples than LA-HPV, with 25 samples being positive in both tests, 1 testing positive in LA-HPV only, and 3 testing positive in PGMY-LB only.
FIG. 1.
Agreement between results obtained for 528 anogenital samples with LA-HPV and PGMY-LB. Kappa coefficients are presented, with 95% CI.
For each type but HPV-64 and HPV-71, at least one specimen was positive only by LA-HPV. HPV-16 was the type for which we obtained the lowest specificity with respect to the results obtained with PGMY-LB, measured at 92.1% (95% CI, 89.0% to 94.3%). The mean specificity obtained for all types reached 95.5% ± 6.3% (95% CI, 93.5% to 97.4%). For HPV-16, 113 samples were positive in both tests, 33 were positive by LA-HPV only, none were positive by PGMY-LB only, and 382 were negative in both tests. All samples had been tested initially before this evaluation, using PGMY-LB, but without coamplification of β-globin (6): 10 (30.3%) of the 33 LA-HPV-positive, PGMY-LB-negative samples for HPV-16 had tested positive with PGMY-LB without coamplification. Additional testing was performed in duplicate on these 33 samples which were positive only with LA-HPV by a real-time PCR assay for HPV-16. Thirty (90.9%) of these samples tested positive for HPV-16.
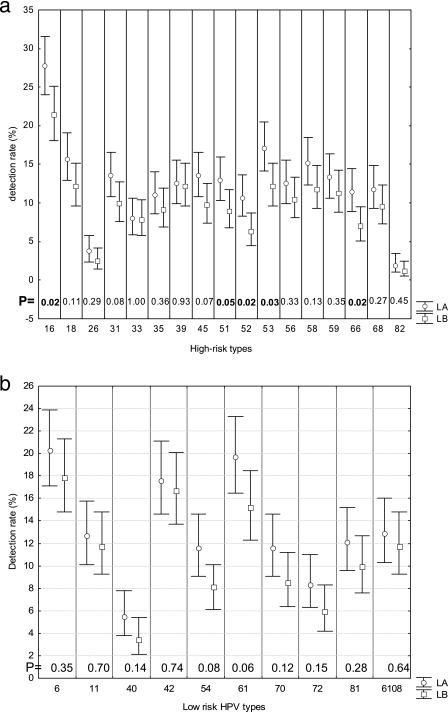
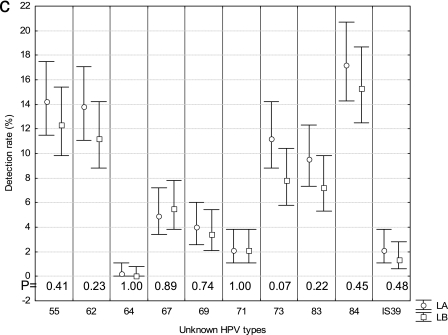
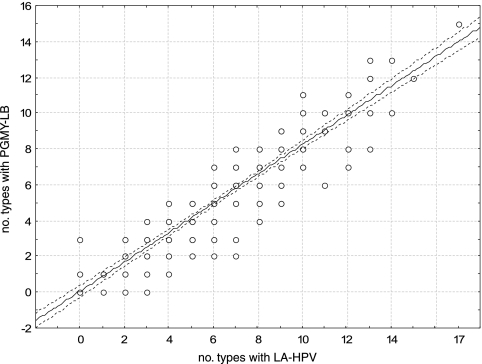
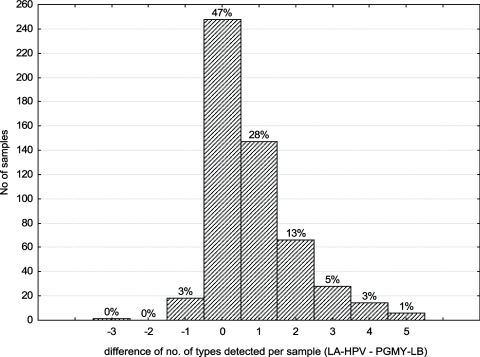
We then determined if the use of LA-HPV over PGMY-LB could improve the type-specific detection rate for HPV infections (Fig. 2a, b, and c). The most frequent HPV type was HPV-16. For all but two types (67 and 71), the rate of detection was higher with LA-HPV. This difference reached statistical significance for types 16, 51, 52, 53, 66, and 61 as well as the high-risk types as a group (P < 0.05). More than one HPV type per sample was detected in 223 (94.5%) of 236 anal swab samples and 167 (57.2%) of 292 cervical or cervicovaginal swab samples (P < 0.001). The median numbers of HPV types per specimen detected in anal swab samples and cervical or cervicovaginal swabs with PGMY-LB were 5 (range, 0 to 18) and 1 (range, 0 to 12), respectively. Figure 3 demonstrates a strong correlation between the numbers of types per sample detected with LA-HPV and PGMY-LB. However, a greater number of types per sample was identified with LA-HPV (mean, 4.2 ± 3.4; 95% CI, 3.9 to 4.5; median, 3.0; range, 0 to 17) than with PGMY-LB (mean, 3.4 ± 3.0; 95% CI, 3.1 to 3.6; median, 3.0; range, 0 to 15) (P < 0.001). Figure 4 shows the distribution of the numbers of types identified per sample in excess with LA-HPV. Although nearly half of the samples contained the same number of types with both assays, the distribution of differences shows a shift towards a greater number of types per sample detected by LA-HPV.
FIG.2.
Detection rates for HPV types by LA-HPV and PGMY-LB. (a) High-risk types; (b) low-risk types; (c) types with unknown risk. Rates of detection were compared, and P values are shown at the bottom of each graph near the axis. Bars represent 95% CI.
FIG. 3.
Correlation between numbers of types detected in each sample by PGMY-LB and LA-HPV. The dashed lines represent the 95% CI of the regression line.
FIG. 4.
Differences in numbers of types detected per sample by LA-HPV and PGMY-LB. The number of types detected with PGMY-LB was subtracted from the number of types detected with LA-HPV for each sample. The percentage of the total number of samples is shown above each column.
We then investigated the determinants of detecting a greater number of types per sample with LA-HPV for the anal cohort study (n = 236), since histology data had been obtained only for these individuals. The number of types per sample found in excess with LA-HPV correlated with the total number of types per sample (r = 0.49 ± 0.06; P = 0.001) but not with age (r = 0.03 ± 0.06; P = 0.57), blood T-lymphocyte CD4 counts (r = 0.06 ± 0.06; P = 0.13), or AIN grade (r = −0.11 ± 0.06; P = 0.07). LA-HPV detected 0.66 ± 1.04 (95% CI, 0.46 to 0.87) type in excess compared to PGMY-LB in samples containing five types of fewer and 1.41 ± 1.42 (95% CI, 1.17 to 1.66) types in excess for samples containing more than five types (P = 0.001). A greater number of types per sample was also associated with having more than two types detected by LA-HPV over PGMY-LB (odds ratio, 1.4; 95% CI, 1.2 to 1.6), controlling for age, AIN grade, and blood T-lymphocyte CD4 count by logistic regression.
DISCUSSION
The precise typing of HPV isolates is essential for the performance of epidemiological studies. It is an essential tool for measuring the impact of HPV vaccination on the risk of acquisition of individual HPV types. The oncogenic potential of some rare or new types also needs to be assessed. Persistent infections by high-risk HPV types are relevant to identifying women at greater risk of progressing to cervical cancer. The transmission of HPV between sexual partners as well as determinants of disease progression is still under study in large-scale cohort studies.
In this evaluation, results obtained with LA-HPV were compared with those generated with PGMY-LB, a prototype assay that uses the same primers. In previous studies, PGMY-LB was shown to be a convenient, sensitive, and reproducible alternative to PCR assays based on radiolabeled probes (6, 7). Cycle sequencing has been reported to detect some HPV types better than PGMY-LB but did not perform as well on samples containing several HPV types (35). Moreover, PGMY-LB generally detected more HPV-positive samples than did cycle sequencing (35). One study reported that PGMY-LB performed similarly to the SPF10 Line Probe assay, a PCR-based system that also uses reverse hybridization for genotyping (34). An international proficiency study had also shown the reliability of PGMY-LB (22). We carried out the present study to assess the relative gain in diagnostic yield of using the commercial LA-HPV assay instead of PGMY-LB for the detection and typing of HPV DNA in 528 anogenital samples from HIV-seropositive individuals.
Based on these analyses, we found substantial agreement between the assays for the detection of HPV DNA. Our evaluation also demonstrated very good agreement between assays for consideration of each of the 37 genotypes individually. Our results indicate that HPV detection and typing can be significantly improved to various degrees for most genital types with the use of LA-HPV, including the most frequent genotype, HPV-16. Most samples positive for HPV-16 only with LA-HPV also tested positive with an HPV-16-specific real-time PCR assay, demonstrating a greater sensitivity of LA-HPV than of PGMY-LB. More HPV types per specimen were also identified with LA-HPV, especially with samples that contained over five genotypes.
The 93% to 100% agreement between the LA-HPV and PGMY-LB systems, whether considering all results or those for each of the 37 genotypes, is greater than the levels of agreement reported in the first evaluations of PGMY primers and in other studies comparing two consensus L1 PCR systems (4, 12, 13, 19, 27, 29, 32, 35). Since both PGMY systems use the same primer sets, a greater level of agreement was expected. Moreover, type-specific probes were identical between the two assays, except those for type 52. The high prevalence of HPV infection and the presence of multiple-type infections in our population allowed us to derive levels of agreement with relatively narrow 95% confidence intervals for most types studied. However, four types (HPV-82, -64, and -71 and IS39) were detected in <10 samples each. Since all individuals recruited for the anal cohort study had high-resolution anoscopies, we could assess if the correlation between assays was dependent on histological diagnosis. For this limited subset of samples, correlation was similar, irrespective of the grade of disease.
Several reasons could explain the increased ability of LA-HPV to detect HPVs compared with PGMY-LB. The oligonucleotide probes and consensus PGMY primers were identical for both systems, making it less likely that the excess detection rate for LA-HPV was due to nonspecific hybridization or amplification. Moreover, when samples positive for HPV-16 with LA-HPV only were further investigated with a real-time PCR assay, most were confirmed as positive for HPV-16 DNA. In contrast to PGMY-LB, the LA-HPV protocol uses standardized reagents that are quality controlled and cycling parameters that are optimized, along with an optimal concentration of β-globin primers to minimize competition due to coamplification of β-globin and HPV DNAs. Interestingly, previous runs using PGMY-LB without coamplification of β-globin on the same samples confirmed the results obtained with LA-HPV for HPV-16 DNA in a substantial proportion of samples.
Coamplification of HPV with β-globin could reduce the level of sensitivity of consensus L1 PCR assays for HPV detection, especially for multiple-type infections (7, 35). Moreover, all samples had tested positive initially for β-globin in a PCR assay amplifying β-globin separately from HPV DNA sequences (6, 7). Samples that tested negative for β-globin in at least one PGMY assay contained HPV DNA. More samples were positive for β-globin with LA-HPV. Since β-globin is coamplified with HPV in PGMY assays, these assays could also be more sensitive to the effects of inhibitors than an assay testing only for β-globin sequences with a higher primer concentration (5). This hypothesis could be investigated further with internal controls for β-globin. Sample degradation could also have occurred since the initial testing for β-globin. Nevertheless, more samples were considered adequate by testing positive for β-globin with LA-HPV. Optimized amplification parameters could also have facilitated HPV typing for multiple-type infections, a frequent occurrence in samples from HIV-seropositive subjects.
Detection and genotyping of HPVs become more complex in samples containing multiple genotypes because of competition for reagents during amplification and the discrimination of types amplified by PCR. The correlation between the number of types contained in a sample and the number of additional types detected with LA-HPV suggests that less competition during amplification was encountered with LA-HPV than with PGMY-LB.
One limitation of the current study is the use of specimens from HIV-seropositive individuals. HIV-infected individuals have a high prevalence of HPV infection and are also infected more frequently with more than one HPV type. The test panel thus cannot be representative of the prevalence of HPV in a random population. On the other hand, our decision to use only samples from HIV-infected individuals afforded the opportunity to scrutinize assay performance under the more extreme conditions seen with specimens containing several HPV types. HIV-seropositive individuals are not only prone to acquiring multiple-type infections but also tend to harbor higher HPV viral loads (24). Another consideration may be that the specimens analyzed were stored for several months, and there could be some specimen degradation over time. The same sample preparations were evaluated in each assay concurrently, and degradation was thus not an issue here.
In conclusion, LA-HPV will be of great value for epidemiological studies and clinical trials to monitor HPV infection at the genotype level in genital samples. Since high-risk HPV persistence is the most important predictor of cervical cancer, genotyping assays could prove useful for assessing viral persistence, but further clinical studies are required before precise recommendations can be made. Eventually, women with persistent infections caused by the same types could be monitored more closely (10, 26). There was good agreement between these two assays for HPV DNA detection and HPV typing. Still, LA-HPV had a superior ability to detect HPV DNA and individual types compared with PGMY-LB. In light of the present results, studies currently using PGMY-LB will have to assess the opportunity to use LA-HPV because of the greater sensitivity of LA-HPV. Nevertheless, although there are some detection gains from using the LA-HPV commercial assay, the agreement with the prototype PGMY-LB assay is excellent, and therefore previous epidemiological studies using the prototype reagents are unlikely to suffer from severe misclassification bias. Further evaluations should be performed with HIV-seronegative subjects.
Acknowledgments
We thank Jean-Marc Trépanier and Serge Coté for maintenance of the database for the HIPVIRG study and for sampling of men and Diane Gaudreault and Diane Bronsard for processing genital samples.
This work was supported by Roche Molecular Systems, which supplied reagents for PGMY assays, and by the Réseau FRSQ-SIDA Maladies Infectieuses. The National Cancer Institute of Canada supports the HIPVIRG cohort. The Canadian Institutes for Health Research and Health and Welfare Canada supported the Canadian Women's HIV Study. F.C. is a clinical research scholar supported by the Fonds de Recherche en Santé du Québec. E.F. holds a Distinguished Scientist Award from the Canadian Institutes for Health Research. S.W. holds career scientist salary support from the Ontario HIV Treatment Network.
REFERENCES
- 1.Aho, J., C. Hankins, C. Tremblay, F. Lang, P. Forest, K. Pourreaux, F. Rouah, The Canadian Women's HIV Study Group, and F. Coutlée. 2003. Molecular analysis of human papillomavirus type 52 isolates detected in the genital tract of HIV-seropositive and seronegative women. J. Infect. Dis. 188:1517-1527. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, H. M., Y. Ting, C. E. Greer, J. C. Chambers, C. J. Tashiro, J. Chimera, A. Reingold, and M. M. Manos. 1991. Genital human papillomavirus infection in female university students as determined by a PCR-based method. JAMA 265:472-477. [PubMed] [Google Scholar]
- 3.Bosch, F. X., A. Lorincz, N. Munoz, C. J. L. M. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castle, P. E., M. Schiffman, P. E. Gravitt, H. Kendall, S. Fishman, H. Dong, A. Hildesheim, R. Herrero, M. C. Bratti, M. E. Sherman, A. Lorincz, J. E. Schussler, and R. D. Burk. 2002. Comparisons of HPV DNA detection by MY09/11 PCR methods. J. Med. Virol. 68:417-423. [DOI] [PubMed] [Google Scholar]
- 5.Coutlée, F., M. de Ladurantaye, C. Tremblay, J. Vincelette, L. Labrecque, and M. Roger. 2000. An important proportion of genital samples submitted for Chlamydia trachomatis detection by PCR contain small amounts of cellular DNA as measured by β-globin gene amplification. J. Clin. Microbiol. 38:2512-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutlée, F., P. Gravitt, J. Kornegay, C. Hankins, H. Richardson, N. Lapointe, H. Voyer, and E. Franco. 2002. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J. Clin. Microbiol. 40:902-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutlée, F., P. Gravitt, H. Richardson, C. Hankins, E. Franco, N. Lapointe, H. Voyer, and The Canadian Women's HIV Study Group. 1999. Nonisotopic detection and typing of human papillomavirus DNA in genital samples by the line blot assay. J. Clin. Microbiol. 37:1852-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutlée, F., C. Hankins, N. Lapointe, J. Gill, B. Romanowski, S. Shafran, R. Grimshaw, D. Haase, W. Schlech, J. Sellors, F. Smaill, M. Boucher, M. Chateauvert, J. Falutz, R. Lalonde, J. Macleod, G. Noel, J. P. Routy, E. Toma, G. Garber, G. Victor, S. Trottier, P. Berger, L. Friedland, and D. Keystone. 1997. Comparison between vaginal tampon and cervicovaginal lavage specimens collection for detection of human papillomavirus DNA by the polymerase chain reaction. J. Med. Virol. 51:42-47. [DOI] [PubMed] [Google Scholar]
- 9.de Roda, H., J. M. Walboomers, A. J. van den Brule, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 10.Ferenczy, A., and E. L. Franco. 2002. Persistent human papillomavirus infection and cervical neoplasia. Lancet Oncol. 3:11-16. [DOI] [PubMed] [Google Scholar]
- 11.Fleiss, J. L. 1981. Statistical methods for rates and proportions. John Wiley and Sons Inc., New York, N.Y.
- 12.Giovannelli, L., A. Lama, G. Capra, V. Giordano, P. Arico, and P. Ammatuna. 2004. Detection of human papillomavirus DNA in cervical samples: analysis of the new PGMY-PCR compared to the hybrid capture II and MY-PCR assays and a two-step nested PCR assay. J. Clin. Microbiol. 42:3861-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravitt, P., C. L. Peyton, T. Q. Alessi, C. Wheeler, F. Coutlée, A. Hildesheim, M. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravitt, P., C. L. Peyton, R. J. Apple, and C. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravitt, P. E., R. D. Burk, A. Lorincz, R. Herrero, A. Hildesheim, M. E. Sherman, M. C. Bratti, A. C. Rodriguez, K. J. Helzlsouer, and M. Schiffman. 2003. A comparison between real-time polymerase chain reaction and hybrid capture 2 for human papillomavirus DNA quantitation. Cancer Epidemiol. Biomarkers Prev. 12:477-484. [PubMed] [Google Scholar]
- 16.Gravitt, P. E., C. Peyton, C. Wheeler, R. Apple, R. Higuchi, and K. V. Shah. 2003. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J. Virol. Methods 112:23-33. [DOI] [PubMed] [Google Scholar]
- 17.Habis, A. H., S. D. Vernon, D. R. Lee, M. Verma, and E. R. Unger. 2004. Molecular quality of exfoliated cervical cells: implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol. Biomarkers Prev. 13:492-496. [PubMed] [Google Scholar]
- 18.Hankins, C., F. Coutlée, N. Lapointe, P. Simard, T. Tran, J. Samson, L. Hum, and The Canadian Women's HIV Study Group. 1999. Prevalence of risk factors associated with human papillomavirus infection in women living with HIV. Can. Med. Assoc. J. 160:185-191. [PMC free article] [PubMed] [Google Scholar]
- 19.Harnish, D. G., L. M. Belland, E. E. Scheid, and T. E. Rohan. 1900. Evaluation of human papillomavirus consensus primers for HPV detection by the polymerase chain reaction. Mol. Cell. Probes 13:9-21. [DOI] [PubMed] [Google Scholar]
- 20.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, R. J. Kurman, and M. M. Manos. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, M. V., P. J. F. Snidjers, A. J. C. van den Brule, T. J. M. Helmerhorst, C. Meijer, and J. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornegay, J., M. Roger, P. O. Davies, A. P. Shepard, N. A. Guerrero, B. Lloveras, D. Evens, and F. Coutlée. 2003. International proficiency study of a consensus L1 PCR assay for the detection and typing of HPV DNA: accuracy and intralaboratory and interlaboratory agreement evaluation. J. Clin. Microbiol. 41:1080-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornegay, J. R., A. P. Shepard, C. Hankins, E. Franco, N. Lapointe, H. Richardson, The Canadian Women's HIV Study Group, and F. Coutlée. 2001. Nonisotopic detection of human papillomavirus DNA in clinical specimens using a consensus PCR and a generic probe mix in an enzyme-linked immunoassay format. J. Clin. Microbiol. 39:3530-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefevre, J., C. Hankins, K. Pourreaux, The Canadian Women's HIV Study Group, and F. Coutlée. 2004. Human papillomavirus type 16 viral load is increased in HIV-seropositive women with high-grade squamous intraepithelial lesions compared to those with normal cytology smears. J. Clin. Microbiol. 42:2212-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. Broker, and S. M. Wolinski. 1989. Use of the polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells 7:209-215. [Google Scholar]
- 26.Perrons, C., R. Jelley, B. Kleter, W. Quint, and N. Brink. 2005. Detection of persistent high risk human papillomavirus infections with hybrid capture II and SPF10/LiPA. J. Clin. Virol. 32:278-285. [DOI] [PubMed] [Google Scholar]
- 27.Perrons, C., B. Kleter, R. Jelley, H. Jalal, W. Quint, and R. Tedder. 2002. Detection and genotyping of human papillomavirus DNA by SPF10 and MY09/11 primers in cervical cells taken from women attending a colposcopy clinic. J. Med. Virol. 67:246-252. [DOI] [PubMed] [Google Scholar]
- 28.Petignat, P., C. Hankins, S. Walmsley, D. Money, D. Provencher, K. Pourreaux, J. Kornegay, F. Rouah, and F. Coutlee. 2005. Self-sampling is associated with increased detection of human papillomavirus DNA in the genital tract of HIV-seropositive women. Clin. Infect. Dis. 41:527-534. [DOI] [PubMed] [Google Scholar]
- 29.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quint, W. G. V., G. Scholte, L. J. van Doorn, B. Kleter, P. H. M. Smits, and J. Lindeman. 2002. Comparative analysis of human papillomavirus infections in cervical scrapes and biopsy specimens by general SPF10 PCR and HPV genotyping. J. Pathol. 194:51-58. [DOI] [PubMed] [Google Scholar]
- 31.Sherman, M. E., M. H. Schiffman, A. T. Lorincz, R. Herrero, M. L. Hutchinson, C. Bratti, D. Zahniser, J. Morales, A. Hildesheim, K. Helgesen, D. Kelly, M. Alfaro, F. Mena, I. Balmaceda, L. Mango, and M. Greenberg. 1997. Cervical specimens collected in liquid buffer are suitable for both cytologic screening and ancillary human papillomavirus testing. Cancer 81:89-97. [PubMed] [Google Scholar]
- 32.Tate, J. E., Y. C. Yang, J. Shen, C. M. McLachlin, E. E. Sheets, and C. P. Crum. 1996. A comparison of early (E7) and late (L1) primer-mediated amplification of papillomaviral DNA in cervical neoplasia. Mol. Cell. Probes 10:347-351. [DOI] [PubMed] [Google Scholar]
- 33.The ALTS Group. 2000. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J. Natl. Cancer Inst. 92:397-402. [DOI] [PubMed] [Google Scholar]
- 34.van Doorn, L.-J., W. Quint, B. Kleter, A. Molijn, B. Colau, M.- T. Martin, Kravang-In, N. Torrez-Martinez, C. L. Peyton, and C. M. Wheeler. 2002. Genotyping of human papillomavirus in liquid cytology cervical specimens by the PGMY line blot assay and the SPF10 line probe assay. J. Clin. Microbiol. 40:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernon, S. D., E. R. Unger, and D. Williams. 2000. Comparison of human papillomavirus detection and typing by cycle sequencing, line blotting, and hybrid capture. J. Clin. Microbiol. 38:651-655. [DOI] [PMC free article] [PubMed] [Google Scholar]