Abstract
Serum hepatitis B virus (HBV) DNA was extracted from a chronically infected patient with cocirculation of hepatitis B surface antigen (HBsAg) and anti-HBs antibodies. Direct PCR and clone-derived sequences of the S and overlapped P genes were obtained. DNA sequences and phylogenetic analysis ascribed this isolate to genotype A (serotype adw2). Five of six HBV DNA clones exhibited point mutations inside and outside the major hydrophilic region, while the sixth clone exhibited a genotype A “wild-type” amino acid sequence. Observed replacements included both humoral and/or cellular (major histocompatibility complex class I [MHC-I] and MHC-II) HBV mutated epitopes, such as S45A, P46H, L49H, C107R, T125A, M133K, I152F, P153T, T161S, G185E, A194T, G202R, and I213L. None of these mutants were individually present within a given clone. The I213L replacement was the only one observed in the five clones carrying nonsynonymous mutations in the S gene. Some of the amino acid substitutions are reportedly known to be responsible for the emergence of immune escape mutants. C107R replacement prevents disulfide bonding, thus disrupting the first loop of the HBsAg. Circulation of some of these mutants may represent a potential risk for the community, since neither current hepatitis B vaccines nor hyperimmune hepatitis B immune globulin are effectively prevent the liver disease thereto associated. Moreover, some of the recorded HBsAg variants may influence the accuracy of the results obtained with currently used diagnostic tests.
In addition to acute infections, hepatitis B virus (HBV) may be associated with chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Both host- and virus-related factors have been involved in viral persistence. Among the latter, viral variability is regarded as a key strategy in the attempt to avoid both humoral and cellular immune responses (reviewed in reference 33). Several reports have documented the appearance of escape mutants from protective anti-HBs antibodies as well as from cytotoxic T lymphocyte-specific clones. Spontaneous mutations at T-cell receptor contact sites within individual viral epitopes can abrogate or antagonize the recognition of the corresponding wild-type epitope. Such mutations may contribute to viral persistence (antagonism for T cell receptor), though its clinical relevance appears to be limited (31, 41; reviewed in references 10, 32, and 33).
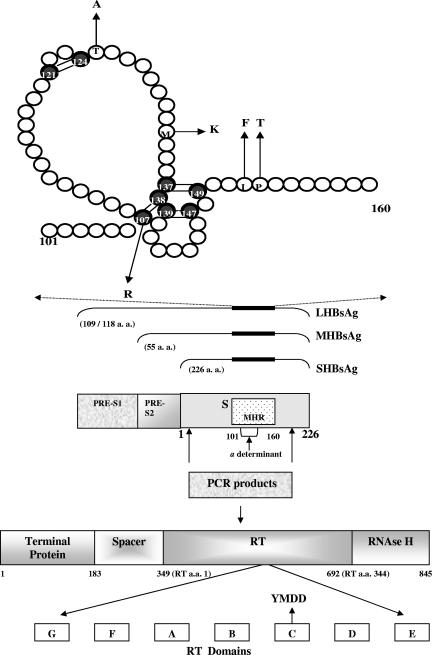
The HBV envelope is composed of host-derived lipids and three related proteins: the large (LHBsAg), middle (MHBsAg), and small (SHBsAg) proteins (24).The S protein is the major one; it is coded for by the S gene and made up of 226 amino acids (aa) (41). As observed in Fig. 1, all three envelope proteins contain the SHBsAg antigenic sites. Antibodies directed against HBsAg confer protective immunity and are crucial for HBV clearance in patients as well as being markers associated with both active and passive prophylactic protection (31).
FIG. 1.
Schematic representation of the S protein (positions 101 to 160), including the first and second loops, according to the second model proposed by Carman et al. (6). Shadowed circles represent Cys residues. Disulfide bonds are shown by double bars. Mutated residues as detected in clone sequences are indicated by arrows pointing to the respective amino acid replacement. A representation of the overlapped S and P genes and the genomic region amplified by PCR is shown. S gene-encoded envelope proteins (large, middle, and small HBsAg) are depicted at the bottom. The MHR is shown between aa 101 and 160 and is also indicated as a thicker zone on lines corresponding to S, pre-S2, and pre-S1 proteins. The a determinant is placed inside the MHR. The catalytic site of the HBV RT is indicated with an arrow pointing to the YMDD amino acidic region of the RT C domain.
The exact three-dimensional structure of the S protein is unknown (3). However, it is widely accepted that HBs antigenicity is mainly dependent on the conformation of the a determinant, placed within the major hydrophilic region (MHR) of the HBsAg (Fig. 1). The MHR encompasses amino acids 101 to 160 of the HBsAg and is exposed on the surface of both virions and subviral particles (Fig. 1, top). This region is highly immunogenic and is potentially under selective pressure of the immune system. The MHR includes a complex conformational region named the a determinant (Fig. 1) which is dependent on disulfide bonding among highly conserved Cys residues. It is thought that the a determinant consists of two loops maintained by disulfide bridges between Cys 107 and 138 and Cys 139 and 147 (Fig. 1, top). A large proportion of serum anti-HBs is directed against this major determinant, the main neutralization epitope. Amino acid substitutions within the a determinant can lead to conformational changes, which in turn, can affect the binding of the neutralizing antibodies (6, 27). However, the clinical significance of most of these mutants is still uncertain (37). The epidemiological importance of such HBs mutants is supported by reports from Taiwan, where the HBV vaccination program was associated with an increased prevalence of HBsAg mutants, concurrent with a 10-fold decrease in the HBs carrier rate in children (18). These data would imply that the selective pressure induced by vaccination might promote the emergence of vaccine-resistant strains (40). However, a recent study carried out in Pacific Island countries has suggested that vaccine escape variants are not an important cause for failing to prevent HBV transmission in this geographical area (1).
Some S mutants can affect the HBV polymerase protein sequence due to the overlapping nature of both open reading frames (ORFs) (Fig. 1). As a result, mutations in the S gene may or may not affect the catalytic domain of the polymerase gene and vice versa. Mutations within the a determinant and the corresponding fragment of the viral polymerase (A and B regions within the reverse transcriptase [RT] domain) (Fig. 1) are more frequently observed among chronic carriers with anti-HBc antibodies as the only serological marker for HBV compared with HBsAg-positive patients (39).
Mutations outside the MHR (Fig. 1), around codons 44 to 49 and 152 to 213 of the S protein, were also described, thus affecting several B-cell and major histocompatibility complex class I (MHC-I) and MHC-II T-cell epitopes that might be associated with viral persistence (2, 8, 14, 24, 30, 36).
In this study, we report further evidence of chronic hepatitis B infection despite the cocirculation of usually protective anti-HBs antibodies. Moreover, the simultaneous detection of mutated S- and P-derived MHC-I and MHC-II epitopes in a genotype A HBV-infected patient is described.
MATERIALS AND METHODS
Patient.
A 43-year-old Argentine male (C) with chronic hepatitis B was studied. He had not been vaccinated against HBV and had no known risk factors for contracting viral hepatitis, such as intravenous drug abuse, transfusions, transplants, sexual preferences, surgeries, and/or prolonged stay in areas of HBV endemicity. However, his sexual partner (unvaccinated for HBV) proved positive for anti-HBs, anti-HBe, and total anti-HBc antibodies and negative for HBsAg. She received a blood transfusion in 1986, the putative source of a persistent hepatitis C virus (HCV) infection. Symptoms related to viral hepatitis were absent to date. None of her three descendants (at present 20, 16, and 14 years old, respectively) exhibit serologic markers of HBV infection. The infection source of any member of the couple remains unknown.
In June 2002, the patient received medical care at Argerich Hospital, in the city of Buenos Aires, exhibiting reactive arthritis, myalgia, elevated serum transaminases (aspartate aminotransferase, 98 IU/ml; alanine aminotransferase, 242 IU/ml) and seropositivity for HBsAg, HBeAg, total anti-HBc, and anti-HBs antibodies (33.6 mIU/ml) and negative for anti-HBe antibodies. Serological markers for HCV and human immunodeficiency virus were negative.
The simultaneous seropositivity for HBsAg and anti-HBs antibodies was evaluated three times and independently performed in two laboratories.
Clinical symptoms disappeared after 3 weeks, and the patient remained asymptomatic thereafter. A liver biopsy was performed in June 2003, and histology showed chronic hepatitis with moderate necroinflammatory activity, fibrosis, and steatosis. In February 2004, the viral load was 1.87 × 105 genomes per milliliter of serum (Amplicor HBV monitor; Roche Diagnostic Systems, Branchburg, NJ). Serum transaminases remained still high (aspartate aminotransferase, 181 IU/ml; alanine aminotransferase, 406 IU/ml). The patient had not received treatment during the follow-up up to October 2004. Taking into account the simultaneous detection of both HBsAg and anti-HBs antibodies, the raised transaminase levels, and the circulating HBV DNA, the patient was then referred to the Hepatitis Laboratory, Department of Microbiology, Faculty of Medicine, University of Buenos Aires, to perform molecular biology studies. After that, the patient started treatment with pegylated alpha 2b interferon (120 μg of polyethylene glycol-INTRON weekly for 6 months). Initial falls in both viral load and serum transaminases were observed. After 1 year, the patient is still under clinical and virological evaluation. Serum transaminase levels remain still elevated.
Written informed consent was provided by the patient to carry out all the studies herein described. Results shown in this study correspond to a blood sample obtained before interferon treatment.
Serology.
Serological tests for HBsAg, anti-HBs, HBeAg, anti-HBe, and total anti-HBc antibodies were carried out by using commercially available standard microparticle enzyme immune assay procedures (AxSYM; Abbott Laboratories, Ill.). Serological tests for human immunodeficiency virus and HCV were also performed by following the manufacturer's instructions (Abbott).
PCR amplification.
Viral DNA was extracted from 100 μl serum by using a DNA extraction kit (Macherey-Nagel, Germany) according to the manufacturer's instructions. PCR amplification of the S gene (541 bp spanning nucleotide positions 256 to 796) was performed by following a previously described protocol (22, 25) using Taq DNA polymerase and primers P7 (sense, 5′-GTG GTG GAC TTC TCT CAA TTT TC-3′) and P8 (antisense, 5′-CGG TA [A/T] AAA GGG ACT CA [A/C] GAT-3′). PCR products encompass an overlapping region of both S and P genes (Fig. 1). Kwok and Higuchi rules (21) were strictly followed. Their effectiveness was routinely assessed by testing known negative serum samples and reagent controls.
Cloning.
The PCR products corresponding to the HBV S gene from the patient serum were cloned into the pGEM-T Easy vector system (Promega, Madison, WI) according to the manufacturer's instructions. Transformed Escherichia coli colonies were initially screened for the HBV S gene DNA by PCR, and the plasmid DNA from PCR-positive colonies was purified by using the modified alkaline lysis-polyethylene glycol precipitation procedure.
DNA sequencing.
DNA from each clone and from the PCR product obtained from serum was bidirectionally sequenced by using Big-Dye termination chemistry (Applied Biosystems) with P7 and P8 primers, and the sequencing products were analyzed with an ABI 3100-Avant (Applied Biosystems) capillary-based automatic sequencer. The PCR-derived sequence was obtained from three independent sequencing reactions.
The potential Taq-dependent DNA misincorporation rate was investigated by bidirectionally sequencing PCR products derived from a GB virus C/hepatitis G virus clone as previously described (25). Briefly, a cDNA fragment of 325 bp was amplified in triplicate and sequenced with specific forward and reverse primers.
HBV phylogenetic analysis.
Initially, an alignment of the DNA sequences obtained from clones as well as from the PCR product and from selected isolates deposited in the GenBank database ascribed to each of the eight HBV genotypes was carried out. The phylogenetic analysis was subsequently performed by using several programs included within the Phylip package (version 3.5.c).
Nucleotide sequence accession number.
Nucleotide sequences have been deposited in GenBank under the following accession numbers: clones 1 to 6, DQ350602 to DQ350607, respectively; PCR-derived sequence, DQ350608.
RESULTS
HBV cloning, sequencing, and phylogenetic analysis.
Phylogenetic analysis of the DNA sequences corresponding to the HBV S gene obtained from six clones, as well as from PCR products, assigned genotype A to this isolate (data not shown). All 16 amino acids that determine the adw2 subtype of the HBsAg (4) proved conserved when the DNA sequence was translated.
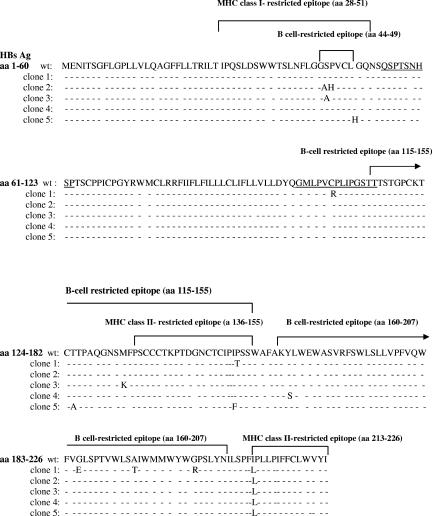
Hepatitis B surface variants were detected in 5 of the 6 analyzed clones, whereas the remaining one exhibited a wild-type virus. Neither insertions nor deletions were recorded within the S gene. Nonsynonymous nucleotide point mutations were associated with replacements at codons 45, 46, 49, 107, 125, 133, 152, 153, 161, 185, 194, 202, and 213, some of them also affecting the overlapping polymerase ORF (Fig. 1; Table 1). A summary of the involvement of such substitutions in B-, MHC-I-, and MHC-II-restricted epitopes is depicted in Fig. 3.
TABLE 1.
Amino acid changes in the HBsAg and the overlapping polymerase open reading frame of the 5 clones showing variations within the S genea
| Clone no. | HBsAg
|
HBV polymerase
|
||
|---|---|---|---|---|
| Amino acid position | Amino acid change | Amino acid position | Amino acid change | |
| 1 | 107 | C→R | 463 | L→S |
| 153 | P→T | 509 | P→H | |
| 185 | G→E | 541 | Wild type | |
| 194 | A→T | 550 | S→N | |
| 202 | G→R | 558 | G→E | |
| 213 | I→L | 569 | Y→F | |
| 2 | 45 | S→A | 401 | I→S |
| 46 | P→H | 402 | Wild type | |
| 213 | I→L | 569 | Y→F | |
| 3 | 45 | S→A | 401 | I→S |
| 133 | M→K | 489 | Y→STOP | |
| 213 | I→L | 569 | I→F | |
| 4 | 161 | Y→S | 517 | Wild type |
| 213 | I→L | 569 | I→F | |
| 5 | 49 | L→H | 405 | Wild type |
| 125 | T→A | 481 | H→R | |
| 140 | Wild type | 496 | K→E | |
| 152 | I→F | 508 | H→L | |
| 213 | I→L | 569 | Y→F | |
Amino acid positions of both HBsAg and polymerase reflect the corresponding number for each residue at the same location of both out-of-frame overlapping ORFs, as represented in Fig. 1A; i.e., position 107 of HBsAg corresponds to position 463 of polymerase and likewise for the rest of the corresponding positions. Note that most substitutions in HBsAg affect the polymerase amino acid sequence and vice versa.
FIG. 3.
Mutated B- and T-cell-restricted epitopes derived from the S protein are shown. Pol-mutated epitopes (the overlapped region within the S protein is underlined) were observed within amino acids 455 to 463 (L463S in clone 1) and 504 to 512 (P509H in clone 1 and H508L in clone 5) for MHC-I peptides and within amino acids 503 to 517 (P509H in clone 1 and H508L in clone 5) for the overlapped MHC-II peptide.
Analysis of the HBV S ORF in the 5 clones with surface variants.
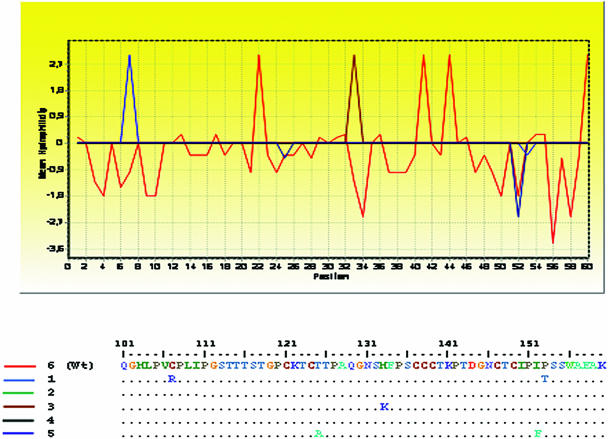
The amino acid sequences of the S antigen as deduced from the DNA sequence analysis of these variants proved diverse. As shown in Table 1, all mutated clones exhibited at least one substitution within the HBsAg, either inside and/or outside the MHR, in addition to the amino acid change I213L. Mutations within the S gene were more frequent outside than inside this region. Most amino acid replacements within the a determinant occurred in the first loop (Fig. 1, top). Some of these substitutions profoundly affected the hydrophilicity profile of the S protein, as observed in Fig. 2.
FIG. 2.
Hydrophilicity patterns obtained for HBV clones. A partial analysis of the S protein (amino acid positions 101 to 160) encompassing the MHR is shown (BioEdit program, 1999). The amino acid sequence corresponding to the wild-type (wt) sequence (as observed in clone 6) is drawn in red. Profiles depicting mutated clones are shown in blue, green, brown, black, and dark blue and correspond to clones 1, 2, 3, 4, and 5, respectively.
As shown in Table 1 and Fig. 3, three of six clones exhibited amino acid substitutions within a reportedly known region (positions 28 to 51) encompassing a class I T-cell epitope of the HBsAg (10, 23, 36), as were S45A (two clones), P46H, and L49H. Moreover, as observed in Fig. 3, two clones exhibited amino acid exchanges in class II T helper epitopes (positions 136 to 155) (2), as were I152F and P153T, while 5 clones showed the I213L replacement in another class II T-cell epitope (positions 213 to 226) (2).
Five clones (Table 1 and Fig. 3) showed variations within B-cell epitopes (HBsAg 44 to 49, 115 to 155, 124 to 147, and 160 to 207) (7, 14, 30).
One clone exhibited Arg instead of Cys at position 107 (C107R) (Fig. 1, top), which avoids first loop formation according to the last model of Carman et al. (6).
Analysis of the overlapping HBV polymerase ORF in the 5 clones with surface variants.
As the P gene is in the +1 reading frame with respect to the overlapping S gene, the deduced amino acid sequence of the corresponding fragment of the viral polymerase was also examined. This fragment represents an important part of the RT domain, including regions B and C, which may be associated with resistance to nucleoside analogues such as lamivudine. The YMDD motif, which is essential for the enzymatic activity of many RTs, was conserved in the 5 clones showing variations in the surface antigen. Several residues of the HBV DNA polymerase fragment that overlaps with the ORF encoding HBsAg were affected by surface variants in this study. Replacement at position 569 (I569F) was detected in the 5 clones showing variations in the S antigen but was involved in neither B- nor T-cell epitopes. This position corresponds to the amino acid 213 of HBsAg. A single clone exhibited a stop codon at position 489 (i.e., position 133 of the S antigen).
One clone showed an amino acid exchange in the 455 to 463 MHC-I-restricted epitope (L463S) (Fig. 3). The two clones mentioned above exhibiting amino acid substitutions in the MHC-II-restricted 136 to 155 S epitope (I152F and P153T) also showed the corresponding substitutions (H508L and P509H) in both the MHC-II 503 to 517 Pol epitope (26) and the nested MHC-I 504 to 512 Pol epitope (Fig. 3) (26). To date, no B-cell Pol epitopes have been reported so far within the PCR-amplified region herein described (43).
Direct sequencing of PCR-derived products: analysis of HBV S and polymerase ORF.
As expected after sequence analysis of the clones, mutations were also detected within the PCR products at positions 45 (S45A) and 213 (I213L) of the S gene. The observed nonsynonymous mutations corresponded to changes at the first base of both codons: a TCA→GCA transversion for the former replacement, and an ATA→TTA transversion for the latter. Coincidentally, such replacements were represented within the P ORF (I401S and I569F) as well. In contrast, none of the remaining substitutions observed as mutations within the genome of a given clone (of six studied) proved detectable within the sequence of PCR products. No evidence of mixed HBV populations at a given nucleotide position was recorded throughout the analyzed genomic region. Twelve nucleotides could not be faithfully assigned, although all of them were uniformly the same base in the corresponding position in all 6 analyzed clones. Nevertheless, none of these unassigned nucleotides in the PCR-derived sequence were located at mutated positions as observed in the studied clones. Thus, PCR sequence ambiguities were undoubtedly solved by gene cloning.
DISCUSSION
In this study, a total of 13 S gene nonsynonymous amino acid exchanges (S45A, P46H, L49H, C107R, T125A, M133K, I152F, P153T, Y161S, G185E, A194T, G202R, and I213L) (Table 1) were detected in five of six analyzed clones in a patient who exhibited chronic active hepatitis despite the presence of (usually neutralizing) anti-HBs antibodies with cocirculating HBsAg. The PCR-derived sequence confirmed both S45A and I213L substitutions. Interestingly, when the consensus sequence was obtained (data not shown), the S45A replacement could not be detected, reflecting the fact that only 2 of 6 clones exhibited the mutated base, therefore emphasizing the intrinsic value of sequencing PCR products. It seems likely that if more clones had been sequenced, the consensus sequence and the PCR-derived sequence could have been identical. Since S45A exchange is placed at both a T- and a B-cell epitope (positions 44 to 49), this replacement might be crucial for immune escape, as the case might be for P46H and L49H as well.
The remaining substitutions mentioned above do not seem to be PCR artifacts, as we have considered the following items: (i) the reportedly known misincorporation rate of Taq DNA polymerase per polymerized nucleotide is estimated within the range of 12.1 × 10−5 (15) to 8 × 10−6 (11); (ii) no mutations were observed when an unrelated GB virus/hepatitis G virus cDNA clone was amplified in triplicate and bidirectionally sequenced (975 nucleotides examined twice) (25), and (iii) the short length (541 bp) of the amplified products. Moreover, the probability, P, that the drastic C107R substitution (see below) would have merely occurred by chance is equal to 0.0005 (1/497 × 1/4), where 497 is the number of Taq-polymerized nucleotides (between primers) and 4 is the number of possible nucleotides at a given position.
This study shows a persistent infection with a mixture of both a so-called “wild-type” (1 clone) and S gene variants (5 clones) (Table 1). As the reported patient had never received hepatitis B immune globulin immunoprophylaxis or the HBsAg vaccine or been on antiviral therapy, it is suggested that surface variants might have emerged or been selected despite the host immune pressure. The source of his infection remains unknown at present. However, a putative role of his partner cannot be ruled out due to an eventual past occult infection (i.e., HBV DNA positive by PCR and HBsAg negative).
According to Ogura et al. (28), mutations within the a determinant during the natural course of infection are predominantly observed within the first loop (aa 107 to 138) (5, 6, 35), whereas those induced under immune pressure due to active and/or passive immunization are more frequently observed within the second loop (aa 139 to 147) (16, 29, 31, 34).
In this study, no amino acid changes occurred within the second loop, whereas three amino acid substitutions were observed within the first loop (C107R, T125A, and M133K) (Fig. 1, top). Such amino acid replacements might significantly/drastically affect the loop structure and its hydrophilicity profile (Fig. 2), since Cys, Thr, and Met residues exhibit an accessibility index of −10, 7.1, and 1.9, respectively, in contrast to that assigned for Arg, Ala, and Lys residues (9.8, 2.7, and 10, respectively). Furthermore, C107 is known to participate in the disulfide bonding of the S protein (Fig. 1) and is indispensable for secretion of the 20-nm particles (24). Its substitution by a large and positively charged amino acid, such as Arg, implies the impossibility of maintaining the correct conformation of the a determinant, thus preventing binding of neutralizing antibodies. A significant involvement of Cys replacements in the emergence of variants despite the presence of anti-HBs antibodies has recently been described (25), although it is the first time that this particular C107R substitution (chromatogram available upon request) is observed.
Mutations outside the MHR are frequent and tend to cluster in two regions around codons 44 to 49 and 152 to 213. The first region contains both an MHC-I-restricted T-cell epitope and a B-cell epitope, whereas the second region, at least up to amino acid 207, exhibits both MHC-II T helper epitopes (positions 136 to 155, 163 to 174, and 213 to 226) (2) as well as B-cell epitopes (14, 30). It was also reported (14) that changes within this second region, located immediately downstream of the a determinant, may alter the conformation of this immunogenic determinant. In agreement with this notion, Hou et al. (17) showed that amino acid insertions and deletions in this region abolish the binding to anti-HBs antibodies.
Considering that only a single mutation was observed in all five mutated clones within the surface antigen (I213L), a GenBank search was carried out to determine its frequency among worldwide HBV isolates. After alignment of genotype A S gene partial sequences (n = 35, including V00866, M57663, X02763, X51970, Z35717, E00010, X70185, Z72478, L13994, and S50225) (data not shown), it was observed that a Leu residue at position 213 was extremely unusual, since only one of them exhibited such replacement. Moreover, the observation of a Leu residue at position 213 has been reported in 1 of 37 (adw2) HBV-infected unvaccinated individuals, although it was observed in 1 ayw1 isolate (16). This intriguingly predominant substitution (observed in 5 of 6 clones of the HBV genotype A isolate herein reported) might suggest a putative role within the T helper epitope to evade CD4+ (HBV epitope)-specific cells. However, it is reportedly known that amino acids located at the extreme of a given MHC-II epitope have little, if any, effect on the peptide binding specificity. Likewise, such residues are not involved in TCR recognition. Alternatively, the existence of a rather extended humoral immune reactive area downstream of the MHR that includes residue 213 (as recently shown for residues 178 to 186) (30) or an eventual crucial influence of such I213L replacement on the conformational structure of the S protein should be further investigated. In any case, this and/or the remaining S gene changes could affect the S protein binding to anti-HBs antibodies, which might provide a putative explanation for the free cocirculation of both HBsAg and anti-HBs antibodies. Since the three-dimensional structure of the HBV surface protein is still unsolved (3), the prediction of conformational, biochemical, and functional effects as a consequence of these amino acid substitutions is merely speculative. Nevertheless, experiments involving site-directed mutagenesis and subsequent antigenic analysis of all the amino acid changes herein reported are in progress.
Bearing in mind the overlapping structure of the S and P genes of HBV (Fig. 1), amino acid substitutions within the S gene might or might not account for amino acid substitutions in the P gene and vice versa. In this study, analysis of the overlapping fragment of HBV polymerase showed several amino acid changes. One of the observed mutations (codon 489) resulted in the premature termination of the Pol protein (Table 1). It is assumed that since certain mutations in the P gene may impair virus replication to a different extent, a minor population of intact genomes should be present to help the formation of viral particles by complementation (38).
As expected for patients not treated with lamivudine, the active site of the RT enzyme exhibited the wild-type amino acid sequence (YMDD) (Fig. 1), where Met is usually replaced by either Val or Ile in lamivudine-resistant isolates.
In addition, it is suggested that the circulation of some of the observed mutated HBV epitopes might represent a potential population risk, since neither current hepatitis B vaccines nor hepatitis B immune globulin effectively prevents the liver disease associated with some of them. Moreover, some of the recorded HBs antigen variants may influence the accuracy of the results obtained with currently used diagnostic tests, as reported with previously studied mutants (9, 12, 13, 19, 20, 30, 37), since some of them may prove undetectable, although others may not result in a complete loss of antigenicity (17).
In brief, this study underlines the unusual simultaneous detection of both humoral and cellular mutated HBV epitopes during the natural course of a chronic HBV infection despite the presence of anti-HBs antibodies. The extremely rare C107R replacement (reviewed in references 32 and 37) and the I213L unusual substitution (alone and combined with the herein reported exchanges) deserve to be further explored in future in vitro studies of HBsAg-anti-HBs antibody interactions, since they may be immunogenic, although with a changed specificity (42).
Acknowledgments
We are indebted to A. M. Andreetta, J. Trinks, and María de los Ángeles Oubiña for excellent technical assistance. We are truly grateful to María Victoria Illas for enhancing readability.
This study was partly supported by the following grants: BID 1201/OC-AR-PICT 10.871 from the National Agency for Scientific and Technological Promotion (ANPCyT), PIP 842 and PIP 6.065 from the National Research Council (CONICET), and UBACYT M057 from the University of Buenos Aires.
REFERENCES
- 1.Basuni, A. A., L. Butterworth, G. Cooksley, S. Locarnini, and W. F. Carman. 2004. Prevalence of HBs Ag mutants and impact of hepatitis B infant immunisation in four Pacific Island countries. Vaccine 22:2791-2799. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, T., K. Weinberger, and W. Jilg. 2002. Variants of two major T cell epitopes within the hepatitis B surface antigen are not recognized by specific T helper cells of vaccinated individuals. Hepatology 35:455-465. [DOI] [PubMed] [Google Scholar]
- 3.Berting, A., J. Hahnen, M. Kröger, and W. H. Gerlich. 1995. Computer-aided studies on the spatial structure of the small hepatitis B surface protein. Intervirology 38:8-15. [DOI] [PubMed] [Google Scholar]
- 4.Blitz, L., F. H. Pujol, P. D. Swenson, L. Porto, R. Atencio, M. Araujo, L. Costa, D. C. Monsalve, J. R. Torres, H. A. Fields, S. Lambert, C. Van Geyt, H. Norder, L. O. Magnius, J. M. Echevarria, and L. Stuyver. 1998. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J. Clin. Microbiol. 36:648-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carman, W. F. 1998. Infections associated with medical intervention: hepatitis viruses and HGV. Br. Med. Bull. 54:731-748. [DOI] [PubMed] [Google Scholar]
- 6.Carman, W. F., A. Owsianka, L. A. Wallace, B. C. Dow, and D. J. Multimer. 1999. Antigenic characterisation of pre- and post-liver transplant hepatitis B surface antigen. Sequences from patients treated with Hepatitis B immune globulin. J. Hepatol. 31:195-201. [DOI] [PubMed] [Google Scholar]
- 7.Carman, W. F., C. Trautwein, F. J. van Deursen, K. Colman, E. Dornan, G. McIntyre, J. Waters, V. Kliem, R. Muller, H. C. Thomas, and M. P. Manns. 1996. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology 24:489-493. [DOI] [PubMed] [Google Scholar]
- 8.Chen, W. N., and C. J. Oon. 1999. Mutation “hot spot” in HLA class I-restricted T-cell epitope on hepatitis B surface antigen in chronic carriers and hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 262:757-761. [DOI] [PubMed] [Google Scholar]
- 9.Chiou, H. L., T. S. Lee, J. Kuo, Y. C. Mau, and M. S. Ho. 1997. Altered antigenicity of ′a′ determinant variants of hepatitis B virus. J. Gen. Virol. 78:2639-2645. [DOI] [PubMed] [Google Scholar]
- 10.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathogenesis. Annu. Rev. Immunol. 13:29-60. [DOI] [PubMed] [Google Scholar]
- 11.Cline, J., J. C. Braman, and H. H. Hogrefe. 1996. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24:3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coleman, P. F., Y. C. J. Chen, and I. K. Mushahwar. 1999. Immunoassay detection of hepatitis B surface antigen mutants. J. Med. Virol. 59:19-24. [DOI] [PubMed] [Google Scholar]
- 13.Gerlich, W. H. 2004. Diagnostic problems caused by HBs Ag mutants-a consensus report of an expert meeting. Intervirology 47:310-313. [DOI] [PubMed] [Google Scholar]
- 14.Ghany, M. G., B. Ayola, F. G. Villamil, R. G. Gish, S. Roitter, J. M. Vierling, and A. S. F. Lok. 1998. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology 28:213-222. [DOI] [PubMed] [Google Scholar]
- 15.Günther, S., G. Sommer, F. von Breunig, A. Iwanska, T. Kalinina, M. Sternec, and H. Will. 1998. Amplification of full-length hepatitis B virus genomes from samples from patients with low levels of viremia: frequency and functional consequences of PCR-induced mutations. J. Clin. Microbiol. 36:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hino, K., Y. Katoh, E. Vardas, J. Sim, K. Okita, and W. F. Carman. 2001. The effect of introduction of universal childhood hepatitis B immunization in South Africa on the prevalence of serologically negative hepatitis B virus infection and the selection of immune escape variants. Vaccine 19:3912-3918. [DOI] [PubMed] [Google Scholar]
- 17.Hou, J., P. Karayiannis, J. Waters, K. Luo, C. Liang, and H. Thomas. 1995. A unique insertion in the S gene of surface antigen-negative hepatitis B virus Chinese carriers. Hepatology 21:273-278. [DOI] [PubMed] [Google Scholar]
- 18.Hsu, H. Y., M. H. Chang, S. H. Liaw, Y. H. Ni, and H. L. Chen. 1999. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology 30:1312-1317. [DOI] [PubMed] [Google Scholar]
- 19.Ireland, J. H., B. O′Donnell, A. A. Basuni, J. D. Kean, L. A. Wallace, G. K. K. Lau, and W. F. Carman. 2000. Reactivity of 13 in vitro expressed hepatitis B surface antigen variants in 7 commercial diagnostic assays. Hepatology 31:1176-1182. [DOI] [PubMed] [Google Scholar]
- 20.Jolivet-Reynaud, C., M. Lésenéchal, B. O′Donnell, L. Becquart, A. Foussadier, F. Forge, N. Battail-Poirot, X. Lacoux, W. Carman, and M. Jolivet. 2001. Localization of hepatitis B surface antigen epitopes present on variants and specifically recognised by anti-hepatitis B surface antigen monoclonal antibodies. J. Med. Virol. 65:241-249. [DOI] [PubMed] [Google Scholar]
- 21.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 22.Lindh, M., A. S. Anderson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographic origin of hepatitis B virus. Large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 23.Liu, C. J., J. H. Kao, W. Y. Shau, P. J. Chen, M. Y. Lai, and D. S. Chen. 2002. Naturally occurring hepatitis B surface gene variants in chronic hepatitis B virus infection: correlation with viral serotypes and clinical stages of liver disease. J. Med. Virol. 68:50-59. [DOI] [PubMed] [Google Scholar]
- 24.Mangold, C. M. T., and R. E. Streeck. 1993. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J. Virol. 67:4588-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathet, V. L., M. Feld, L. Espinola, D. O. Sánchez, V. Ruiz, O. Mandó, G. Carballal, J. F. Quarleri, F. D′Mello, C. R. Howard, and J. R. Oubiña. 2003. Hepatitis B virus S gene mutants in a patient with chronic active hepatitis with circulating anti-HBs antibodies. J. Med. Virol. 69:18-26. [DOI] [PubMed] [Google Scholar]
- 26.Mizukoshi, E., J. Sidney, B. Livingston, M. Ghany, J. H. Hoofnagle, A. Sette, and B. Rehermann. 2004. Cellular immune responses to the hepatitis B virus polymerase. J. Immunol. 173:5863-5871. [DOI] [PubMed] [Google Scholar]
- 27.Ni, F., D. Fang, R. Gan, Z. Li, S. Duan, and Z. Xu. 1995. A new immune escape mutant of hepatitis B virus with an Asp to Ala substitution in aa144 of the envelope major protein. Res. Virol. 146:397-407. [DOI] [PubMed] [Google Scholar]
- 28.Ogura, Y., M. Kurosaki, Y. Asahina, N. Enomoto, F. Maruno, and C. Sato. 1999. Prevalence and significance of naturally occurring mutations in the surface and polymerase genes of hepatitis B virus. J. Infect. Dis. 180:1444-1451. [DOI] [PubMed] [Google Scholar]
- 29.Oon, C. J., W. N. Chen, S. Koh, and G. K. Lim. 1999. Identification of hepatitis B surface antigen variants with alterations outside the a determinant in immunized Singapore infants. J. Infect. Dis. 179:259-263. [DOI] [PubMed] [Google Scholar]
- 30.Paulij, W. P., L. M. Wit, C. M. G. Sünnen, M. H. van Roosmalen, A. Petersen-van Ettekoven, M. P. Cooreman, and R. A. Heijtink. 1999. Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. J. Gen. Virol. 80:2121-2126. [DOI] [PubMed] [Google Scholar]
- 31.Protzer-Knolle, U., U. Naumann, R. Bartenschlager, T. Berg, U. Hopf, K. H. Meyer zum Buschenfelde, P. Neuhaus, and G. Gerken. 1998. Hepatitis B virus with antigenically altered hepatitis B surface antigen is selected by high-dose hepatitis B immune globulin after liver transplantation. Hepatology 27:254-263. [DOI] [PubMed] [Google Scholar]
- 32.Pumpens, P., E. Grens, and M. Nassal. 2002. Molecular epidemiology and immunology of Hepatitis B virus infection-an update. Intervirology 45:218-232. [DOI] [PubMed] [Google Scholar]
- 33.Rehermann, B., and M. Nascimbeni. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Immunol. 5:215-218. [DOI] [PubMed] [Google Scholar]
- 34.Schilling, R., S. Ijaz, M. Davidoff, J. Yee Lee, S. Locarnini, R. Williams, and N. V. Naoumov. 2003. Endocytosis of hepatitis B immune globulin into hepatocytes inhibits the secretion of hepatitis B virus surface antigen and virions. J. Virol. 77:8882-8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, B. C., S. H. Kim, H. Kim, Y. H. Ying, H. J. Kim, Y. J. Kim, J. H. Yoon, H. S. Lee, C. Y. Cha, Y. H. Hook, and B. J. Kim. 2005. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J. Med. Virol. 76:194-202. [DOI] [PubMed] [Google Scholar]
- 36.Tai, P., D. Banik, G. Lin, S. Pai, M. H. Lin, G. Yuoh, S. Che, S. H. Hsu, T. C. Chen, T. T. Kuo, C. S. Lee, C. S. Yang, and C. Shih. 1997. Novel and frequent mutations of hepatitis B virus coincide with a major histocompatibility complex class I-restricted T-cell epitope of the surface antigen. J. Virol. 71:4852-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber, B. 2005. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J. Clin. Virol. 32:102-112. [DOI] [PubMed] [Google Scholar]
- 38.Weinberger, K. M., G. Zoulek, T. Bauer, S. Böhm, and W. Jilg. 1999. A novel deletion mutant of hepatitis B virus surface antigen. J. Med. Virol. 58:105-110. [DOI] [PubMed] [Google Scholar]
- 39.Weinberger, K. M., T. Bauer, S. Böhm, and W. Jilg. 2000. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBs Ag) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBs Ag in serum. J. Gen. Virol. 81:1165-1174. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, J. N., D. J. Nokes, and W. F. Carman. 1999. The predictive pattern of emergence of vaccine-resistant hepatitis B: a cause for concern? Vaccine 17:973-978. [DOI] [PubMed] [Google Scholar]
- 41.Yamamoto, K., M. Horikita, F. Tsuda, K. Itoh, Y. Akahane, S. Yotsumoto, H. Okamoto, Y. Miyakawa, and M. Mayumi. 1994. Naturally occurring escape mutants of hepatitis B virus with mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J. Virol. 68:2671-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng, X., K. M. Weinberger, R. Gehrke, M. Isogawa, G. Hilken, T. Kemper, Y. Xu, D. Yang, W. Jilg, M. Roggendorf, and M. Lu. 2004. Mutant hepatitis B virus surface antigens (HBs Ag) are immunogenic but may have a changed specificity. Vaccine 392:454-464. [DOI] [PubMed] [Google Scholar]
- 43.zu Putlitz, J., R. E. Lanford, R. I. Carlson, L. Notvall, S. M. de la Monte, and J. R. Wands. 1999. Properties of monoclonal antibodies directed against hepatitis B virus polymerase protein. J. Virol. 73:4188-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]