Abstract
The presence of members of the family Helicobacteraceae in the colon of children was investigated using PCR, denaturing gradient gel electrophoresis, and fluorescent in situ hybridization. The rate of detection of species belonging to the Helicobacteraceae family in children with inflammatory bowel disease (92%) and irritable bowel syndrome (100%) was found to be significantly higher than that in healthy children (25%) (P < 0.05).
Helicobacter species are gram-negative curved to spiral-shaped bacteria that naturally colonize the mucus layer of the gastrointestinal tract of a range of mammalian hosts (15). Based on their preferential site of colonization, species of the Helicobacter genus have been divided into gastric and enterohepatic subgroups. Helicobacters in the gastric group, represented by Helicobacter pylori, have been associated with gastritis and other gastric disorders (15, 16). Enterohepatic Helicobacter species, which colonize the intestine and hepatobiliary system, have been linked to chronic hepatic and intestinal diseases (9, 16) as well as inflammatory bowel disease (IBD) in immunodeficient rodents (10, 13). The role of Helicobacter species in human IBD is currently controversial (4, 5, 19).
In this study, we assessed the presence and spatial distribution of members of the Helicobacteraceae family (genera Helicobacter and Wolinella) in the colon of 21 children (16/21 male, aged 3 to 15 years) undergoing diagnostic colonoscopy and determined their association with colonic inflammation. The H. pylori status of all children, as assessed by histological examination and rapid urease testing of gastric biopsies, showed that 20/21 children were negative for H. pylori.
Three colonic biopsies were collected from each child at colonoscopy. One biopsy was fixed in 10% neutral formalin for routine histological examination, the second biopsy was fixed in Carnoy's fixative for fluorescent rRNA in situ hybridization (FISH) analysis and Alcian blue-periodic acid-Schiff (PAS) staining, and the third biopsy was placed directly into 600 μl of lysis buffer (Gentra Systems, Minneapolis, Minn.) for DNA extraction.
DNA was extracted from freshly collected biopsy tissues using the PUREGENE DNA extraction kit (Gentra Systems, Minneapolis, Minn.). The16S rRNA gene of members of the family Helicobacteraceae was amplified by PCR using primers H276f and H676r with modified thermal cycle conditions (18). Hot-start PCRs were performed in a 25-μl volume containing 10 pmol of each primer, 1× PCR buffer (Fisher Biotech), 200 nM concentrations of each nucleotide triphosphate, 1.5 mM MgCl2, and 200 to 250 ng of extracted DNA. Thermal cycling conditions were 94°C for 5 min and 35 cycles of 94°C for 5 s, 57°C for 5 s, and 72°C for 30 s. Twenty-microliter PCR products were examined on a 1.5% agarose gel. PCR-denaturing gradient gel electrophoresis (DGGE) analysis was conducted according to a method described previously by Grehan et al. (11). Individual DGGE bands were excised and sequenced.
Slides prepared from biopsies fixed in Carnoy's fixative were subjected to FISH analysis using three probes: probe Eu338 coupled with Cy3 to target all bacteria and probes HEL276/HEL715 coupled with fluorescein to target species of the family Helicobacteraceae (1, 16). Slides prepared from biopsy tissues fixed in Carnoy's fixative were also stained with Alcian blue-PAS to assess the mucus status (3, 7). Mucus thickness was measured between two intact crypts to ensure that the thickness was not influenced by the angle of sectioning. Ten random positions between the crypts in each biopsy were measured using the Adobe Photoshop 7.0 measuring tool.
Of the 21 children examined, 12 were diagnosed with IBD (11 with Crohn's disease [CD] and 1 with ulcerative colitis) and 5 were diagnosed with irritable bowel syndrome (IBS) (8). At the time of scheduled colonoscopy, the gastrointestinal symptoms of the remaining four children were no longer present; however, due to the anxiety of their parents, colonoscopy was still performed. All four children had suffered symptoms for a short duration (<3 months) and had normal histology. Based on these findings, they were classified as normal.
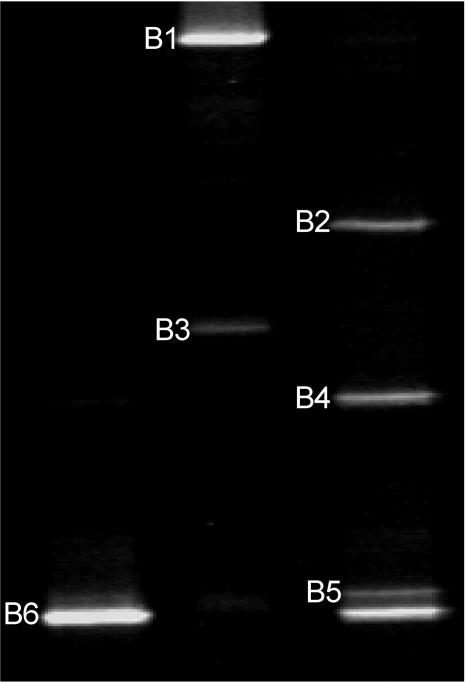
Members of the Helicobacteraceae were detected in 11/12 children with IBD and 5/5 children with IBS, which was significantly higher than in healthy children (1/4 children) (P < 0.05; Fisher's exact test); there was no significant difference between the IBD and IBS groups. DNA from 14 biopsies (nine children with IBD, four children with IBS, and one healthy child) that was positive for Helicobacteraceae by PCR was available for DGGE analysis. Four of the biopsies examined showed a single DGGE band, while in the remaining 10 biopsies, multiple DGGE bands varying from two to four bands were observed. Representative DGGE banding profiles are shown in Fig. 1. No specific DGGE profile was associated with either IBD or IBS. Sequencing of the individual DGGE bands generated a DNA sequence of 392 bp, and a BLAST search of GenBank revealed that they were similar to a range of species from the family Helicobacteraceae (GenBank accession numbers DQ218043 to DQ218048).
FIG. 1.
Representative DGGE banding patterns observed in this study. Bands at positions B1 to B6 were sequenced. B1, B2, B3, and B4 were similar to Helicobacter ganmani (99%), W. succinogenes (99%), Helicobacter hepaticus (100%), and H. pylori (99%), respectively. B5 was similar to Helicobacter bilis (100%) and Helicobacter cinaedi (100%). B6 was similar to Helicobacter trogontum and “Helicobacter rappini” (proposed name) (100%).
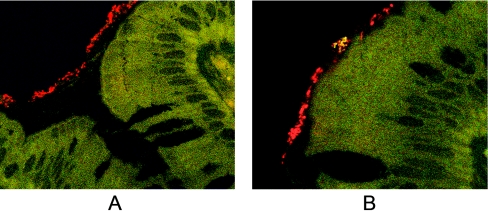
Fourteen biopsy specimens, 10 from children with CD and 4 from children with IBS, were available for FISH analysis and mucus staining. For 7/14 biopsy samples, EUB338-positive signals (all bacteria) were detected in the mucus layer but not in the crypts. Signals that were positive for Helicobacteraceae were detected in the mucus layer of four children, two with IBS and two with CD. Representative results of the FISH analysis are shown in Fig. 2.
FIG. 2.
FISH analysis of sections cut from biopsy samples. (A) Bacteria were seen only in the mucus layer and not in the intestinal crypts. (B) Positive signal of the Helicobacteraceae. The general bacterial probe EUB338 was coupled with Cy3, and the probes HEL276/HEL715, which target species of the family Helicobacteraceae, were coupled with fluorescein. Organisms that were not members of the family Helicobacteraceae are shown in red due to hybridization with EUB338 only. Organisms from the family Helicobacteraceae appear yellow, as they hybridize with both the EUB338 (red) probe and the HEL276/HEL715(green) probes. Magnification, ×1,000.
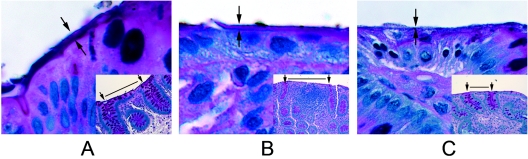
While an intact layer of mucus was observed in all biopsy samples from children with IBS (Fig. 3A), in children with CD, the mucus layer appeared thinner (Fig. 3B) or in some cases partially lost (Fig. 3C). The thickness of the mucus layer in children with CD was 0.84 μm ± 0.68 μm (mean ± standard deviation), and the mean thickness of the mucus layer in children with IBS was 3.30 μm ± 1.24 μm. The mucus thickness in the CD group was found to be significantly thinner than that in the IBS group (P < 0.001; unpaired t test).
FIG. 3.
Alcian blue-PAS-stained sections cut from biopsy samples from children with CD and IBS. A shows the intestinal mucus layer in a patient with IBS; B and C show a thin layer and the absence of an intestinal mucus layer, respectively, in biopsy samples from two patients with CD.
The rate of detection of members of the family Helicobacteraceae by PCR (11/12 in the IBD group, 5/5 in the IBS group, and 1/4 in healthy children) in our study was significantly higher than that previously reported (5, 19). While this may be explained by the fact that our study examined children and other studies examined adults, two technical factors may also have contributed to our high detection rate. First, we observed that for successful detection of these organisms by PCR, sufficient template DNA is essential (200 ng to 250 ng DNA). Second, we extracted DNA from freshly collected biopsy samples; thus, no freezing and thawing was involved, a procedure that is likely to affect the detection of bacteria present in low numbers.
Separation of the amplified 16S rRNA gene by DGGE showed that a high percentage (71%) of biopsy samples had more than one band. Sequencing of the DGGE bands showed that multiple species of the family Helicobacteraceae were present. This finding is consistent with a recent South African study that reported that between two and five Campylobacter-like and Helicobacter-like organisms were frequently recovered from stools of diarrheic children (14). In addition, Haggerty et al. previously reported the detection of multiple non-H. pylori helicobacters in DNA extracted from feces of young children (12).
As FISH-positive signals are based upon the binding of specific labeled probes to 16S rRNA gene molecules, the presence of viable organisms is essential. Positive FISH signals for Helicobacteraceae were observed in the mucus layers of 4 of the 14 PCR-positive children, (2/4 with IBS and 2/10 with CD), thus providing evidence that these organisms are viable. While this may be seen as a low rate of detection, this finding is not unexpected. The low numbers of target bacteria and the slow growth rate of these organisms would have resulted in a reduction in the detection of these organisms. As Helicobacter species colonize the mucus layer of the intestinal tract, the status of mucus in the biopsies is also likely to affect the detection of these organisms by FISH. Examination of the mucus layer revealed that in many of the CD children, the mucus layer was depleted or partially lost at sites of inflammation, with the thickness being significantly less than that for children with IBS. Such findings suggest that future studies aimed at the detection of Helicobacteraceae in the intestinal tract using biopsies from patients with IBD should optimally include biopsies from both inflamed and noninflamed areas.
To our knowledge, detection of organisms closely related to Wolinella succinogenes in the colon of children represents the first documented report of this organism inhabiting the gastrointestinal tract of humans. W. succinogenes, initially isolated from the rumen of cattle, has been considered to be a nonpathogenic organism (20). However, complete genome sequencing of W. succinogenes has revealed that this organism shares 1,269 of its 2,046 genes with H. pylori and Campylobacter jejuni, many of which were identified as virulence factors (2). Interestingly, W. succinogenes was recently detected in gastric biopsies from a sea lion with gastritis (17). Our finding of W. succinogenes in the intestinal tract of children with gastrointestinal symptoms suggests that the status of this organism as a nonpathogen may need to be reevaluated.
In summary, for the first time, we have detected viable organisms similar to members of the family Helicobacteraceae in the intestinal tract of children and have demonstrated that these organisms colonize the mucus layers of the human intestine. Given that Helicobacter species can induce IBD in immunodeficient mice, the high prevalence of Helicobacter species in the biopsy samples of children with IBD and IBS should prompt further studies to investigate their possible role in human IBD.
Acknowledgments
This work was approved by the Research Ethics Committee of the University of New South Wales and the South East Sydney Area Health Service-Eastern Section, Sydney, Australia (ethics no. 03/163).
This study was funded by a faculty research grant from the University of New South Wales, Sydney, Australia.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bancroft, J. D., and A. Stevens. 1996. Theory and practice of histological techniques, 4th ed. Churchill Livingstone, Nottingham, United Kingdom.
- 4.Bell, S. J., S. A. Chisholm, R. J. Owen, S. P. Borriello, and M. A. Kamm. 2003. Evaluation of Helicobacter species in inflammatory bowel disease. Aliment. Pharm. Ther. 18:481-486. [DOI] [PubMed] [Google Scholar]
- 5.Bohr, U. R., B. Glasbrenner, A. Primus, A. Zagoura, T. Wex, and P. Malfertheiner. 2004. Identification of enterohepatic Helicobacter species in patients suffering from inflammatory bowel disease. J. Clin. Microbiol. 42:2766-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, V., G. Crocetti, M. Grehan, L. Zhang, S. Danon, A. Lee, and H. Mitchell. 2005. Visualization of Helicobacter species within the murine cecal mucosa using specific fluorescence in situ hybridization. Helicobacter 10:114-124. [DOI] [PubMed] [Google Scholar]
- 7.Cook, H. C. 1974. Manual of histological demonstration techniques, 4th ed. Butterworth, London, United Kingdom.
- 8.Drossman, D. A. 2005. What does the future hold for irritable bowel syndrome and the functional gastrointestinal disorders? J. Clin. Gastroenterol. 39:S251-S256. [DOI] [PubMed] [Google Scholar]
- 9.Fox, J. G. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic disease. Gut 50:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox, J. G., L. Yan, B. Shames, J. Campbell, J. C. Murphy, and X. Li. 1996. Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus. Infect. Immun. 64:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grehan, M., G. Tamotia, B. Robertson, and H. Mitchell. 2002. Detection of Helicobacter colonization of the murine lower bowel by genus-specific PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 68:5164-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haggerty, T. D., S. Perry, L. Sanchez, G. Perez-Perez, and J. Parsonnet. 2005. Significance of transiently positive enzyme-linked immunosorbent assay results in detection of Helicobacter pylori in stool samples from children. J. Clin. Microbiol. 43:2220-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kullberg, M. C., J. M. Ward, P. L. Gorelick, P. Caspar, S. Hieny, A. Cheever, D. Jankovic, and A. Sher. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12 and gamma interferon-dependent mechanism. Infect. Immun. 66:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lastovica, A. J., and E. L. Roux. 2003. Optimal detection of Campylobacter spp. in stools. J. Clin. Pathol. 56:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, A. 1989. Helicobacter pylori and helicobacter-like organisms in animals: overview of mucus colonising organisms, p. 259-275. In B. Rathbone and V.Heatley (ed.), Campylobacter pylori and gastroduodenal disease, 2nd ed. Blackwell Scientific Publications, Oxford, United Kingdom.
- 16.O'Rourke, J. L., M. Grehan, and A. Lee. 2001. Non-pylori Helicobacter species in humans. Gut 49:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oxley, A. P. A., M. Powell, and D. B. McKay. 2004. Species of the family Helicobacteraceae detected in an Australian sea lion (Neophoca cinerea) with chronic gastritis. J. Clin. Microbiol. 42:3505-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riley, L. K., C. L. Franklin, R. R. Hook, Jr., and C. Besch-Williford. 1996. Identification of murine helicobacters by PCR and restriction enzyme analyses. J. Clin. Microbiol. 34:942-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streutker, C. J., C. N. Bernstein, V. L. Chan, R. H. Riddell, and K. Croitoru. 2004. Detection of species-specific Helicobacter ribosomal DNA in intestinal biopsy samples from a population-based cohort of patients with ulcerative colitis. J. Clin. Microbiol. 42:660-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolin, M. J., E. A. Wolin, and N. J. Jacobs. 1961. Cytochrome-producing anaerobic vibrio, Vibrio succinogenes, sp. n. J. Bacteriol. 81:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]