Abstract
XCP1 is a xylem-specific papain-like cysteine peptidase in Arabidopsis. To determine whether XCP1 could be involved in tracheary element autolysis, promoter activity and localization of XCP1 were investigated using XCP1 promoter-β-glucuronidase fusions and immunofluorescence confocal microscopy. A tracheary element expression pattern was detected for XCP1. Results from confocal microscopy and biochemical subcellular fractionation indicated that XCP1 was localized in the vacuole. Ectopic expression of XCP1 resulted in a reduction in plant size in some lines and early leaf senescence, as indicated by early loss of leaf chlorophyll. Reduced plant size was correlated with higher levels of XCP1, as shown by immunoblot and peptidase activity gel analyses. The XCP1 prodomain exhibits exceptionally high similarity (greater than 80%) to the prodomains of papain and other papain-like enzymes isolated from papaya (Carica papaya) laticifers when compared with all other reported papain-like enzymes. The potential for XCP1 and papain to perform common functions as catalysts of autolytic processing following cell death due to programmed suicide or to wounding is discussed.
In plants, increased peptidase gene expression is associated with remobilization of nitrogen from senescing source tissues to storage or reproductive sinks and from seed protein reserves in support of germination and seedling growth (for review, see Granell et al., 1998; Beers et al., 2000). Increased peptidase activity is also linked to the carbon starvation response (Moriyasu and Ohsumi, 1996; Brouquisse et al., 1998). A detailed understanding of how peptidases function as necessary effectors of nitrogen remobilization during these processes is lacking.
Genetic evidence has contributed to an expanded view of plant peptidases that includes important roles as regulators of responses to environmental cues and plant hormones (for review, see Callis and Vierstra, 2000; Estelle, 2001). The ubiquitin-proteasome pathway is important in light- and auxin-mediated signaling pathways. For example, the level of Hy5, a transcription factor important in photomorphogenesis, is likely to be determined by COP1-mediated targeting to the proteasome (Osterlund et al., 2000). Turnover of IAA/AUX proteins may also be proteasome dependent (Worley et al., 2000; Estelle, 2001). Ser and Asp peptidases are important to cellular differentiation and plant-pathogen interactions. A subtilisin-like Ser peptidase gene, SDD1, is required for regulation of stomatal density and distribution (Berger and Altmann, 2000). In addition, activation tagging of a secreted Asp peptidase in Arabidopsis led to enhanced resistance to virulent strains of Pseudomonas syringae pv. tomato and P. syringae pv. Maculicola, and was correlated with constitutive expression of defense-related genes and elevated salicylic acid levels (R. Dixon and C. Lamb, personal communication).
Experimental models for studying tracheary element (TE) differentiation provide opportunities to investigate the potential of peptidases to act as regulators and effectors of an economically important differentiation process that terminates in cell death. Cultured zinnia (Zinnia elegans) mesophyll cells can be induced to differentiate to TEs in the presence of auxin and cytokinins (Kohlenbach and Schmidt, 1975; Fukuda and Komamine, 1980). Using this system, it has been determined that Ser and Cys proteolytic activities are increased during TE differentiation (Minami and Fukuda, 1995; Ye and Varner, 1996; Beers and Freeman, 1997; Woffenden et al., 1998). In addition, peptidase inhibitor studies have implicated Cys peptidases as essential catalysts of TE autolysis (Woffenden et al., 1998; McCann et al., 2000) and the 26S proteasome as a regulator of TE differentiation (Woffenden et al., 1998).
Although the zinnia system has yielded many novel insights specific to TE differentiation, it does not allow for the comprehensive investigation of all cell types within vascular tissues, i.e. TEs, fibers, and parenchyma in the xylem and sieve tube elements, companion cells, fibers, and parenchyma in the phloem. To facilitate xylem, phloem, and cambium gene discovery, Zhao et al. (2000) prepared cDNA libraries from xylem and bark of the root-hypocotyl of Arabidopsis. The distinct cell types within the secondary xylem and phloem all originate from the cambium, and hence these cDNA libraries are a rich source of genes that regulate vascular cell fate, including, but not limited to, peptidases with vascular tissue-specific roles.
Two papain-like Cys peptidases, XCP1 and XCP2, have been cloned from the Arabidopsis xylem cDNA library (Zhao et al., 2000). Northern-blot analysis has shown XCP1 to be detectable only in the xylem. Deduced amino acid sequences for XCP1 and XCP2 reveal the typical features of a papain-like zymogen, i.e. these peptidases are synthesized as prepropeptides that are processed to remove the pre and prodomains to yield the active form of the enzyme. An N-terminal signal sequence (predomain; Nakai and Kanehisa, 1992) predicts translocation of XCP1/XCP2 into the lumen of the ER. Once in the secretory pathway, it is likely that additional targeting information from the prodomain specifies the final destination that may include functionally distinct vacuoles (Holwerda et al., 1992; Swanson et al., 1998), the Golgi apparatus, or other endoplasmic reticulum (ER)-derived vesicles such as ricinosomes that accumulate during senescence (Schmid et al., 2001).
Tonoplast rupture in differentiating TEs (Groover and Jones, 1999; Kuriyama, 1999; Obara et al., 2001) allows for the mixing of vacuolar contents with the cytoplasm, and it is quickly followed by cell death (Groover et al., 1997) and degradation of the nucleus (Obara et al., 2001). Autolysis continues post mortem, presumably mediated by hydrolytic enzymes released from the vacuole. Localization of XCP1 and XCP2 within the vacuoles of TEs would be consistent with a role for these enzymes as peptidases that are effectors of autolysis, a critical final process in TE differentiation.
Using the putative promoters of XCP1 and XCP2 in promoter-β-glucuronidase (GUS) constructs, we have indirectly established that XCP1 and XCP2 are TE peptidases. This result was confirmed for XCP1 by immunofluorescence confocal microscopy using wild-type Arabidopsis seedlings incubated with anti-XCP1 antibodies. Results of confocal microscopy and subcellular fractionation experiments suggest vacuolar localization for XCP1. Ectopic expression of XCP1 resulted in early loss of leaf chlorophyll, and several transgenic lines produced stunted plants. Higher XCP1 levels were correlated with reduced plant size. Immunoblot and activity gel analyses of XCP1 revealed xylem-specific processing of XCP1 distinct from that observed for ectopically expressed XCP1.
RESULTS
XCP1 and XCP2 Promoter Activity
Previous work has shown XCP1 and XCP2 expression to be xylem specific (Zhao et al., 2000). To localize gene expression for XCP1 and XCP2 to a specific cell type within the xylem, we used the putative promoters for these genes to construct reporter gene fusions (XCP1-GUS and XCP2-GUS) for stable transformation of Arabidopsis. Our goal for XCP1/XCP2 was to use a 1- to 2-kb fragment, 5′ of the initiator Met to analyze for potential as a xylem-specific promoter. For the putative XCP1 promoter, however, the proximity of the nearest predicted open reading frame, upstream of the initiator Met for XCP1, limited the size of that promoter to 591 bps (see “Material and Methods”). In roots, hypocotyls, leaves (see exception below), stems, petals, sepals, styles, and siliques, GUS activity was limited to differentiating TEs for XCP1 and XCP2 promoters. In very young leaves only, staining resulting from GUS activity was also found at the base of trichomes located directly above TEs exhibiting GUS activity (data not shown). Whether this was due to gene expression in trichomes or to leakage of GUS or the blue GUS reaction product from autolysing TEs is not known.
To illustrate this TE expression pattern for this report, we describe TE expression of XCP1-GUS and XCP2-GUS in leaves and roots. At the whole-leaf level, GUS activity, detected by histological staining, was visible in the vascular tissue at the base of the midrib and discontinuously throughout the leaf in vein segments corresponding to individual TEs for XCP1 and XCP2 promoters (Fig. 1). Both promoters specified GUS expression in TEs of primary and secondary xylem, shown here for XCP1 in root and leaf primary xylem (Fig. 1, B and D) and for XCP2 in root secondary xylem (Fig. 1C). Comparisons of the number of TEs showing GUS activity in leaves of similar age from XCP1-GUS versus XCP2-GUS plants (data not shown) indicated higher GUS expression in the XCP2-GUS plants, consistent with published quantitative reverse transcriptase-PCR data comparing XCP2 and XCP1 mRNA levels (Zhao et al., 2000). In the TE shown in Figure 1B, GUS activity appears to be compartmentalized within the cell, with areas lacking blue staining alternating with those showing GUS activity. This staining pattern may be a reflection of the cellular morphology resulting from vacuole or protoplast degeneration during TE differentiation.
Figure 1.
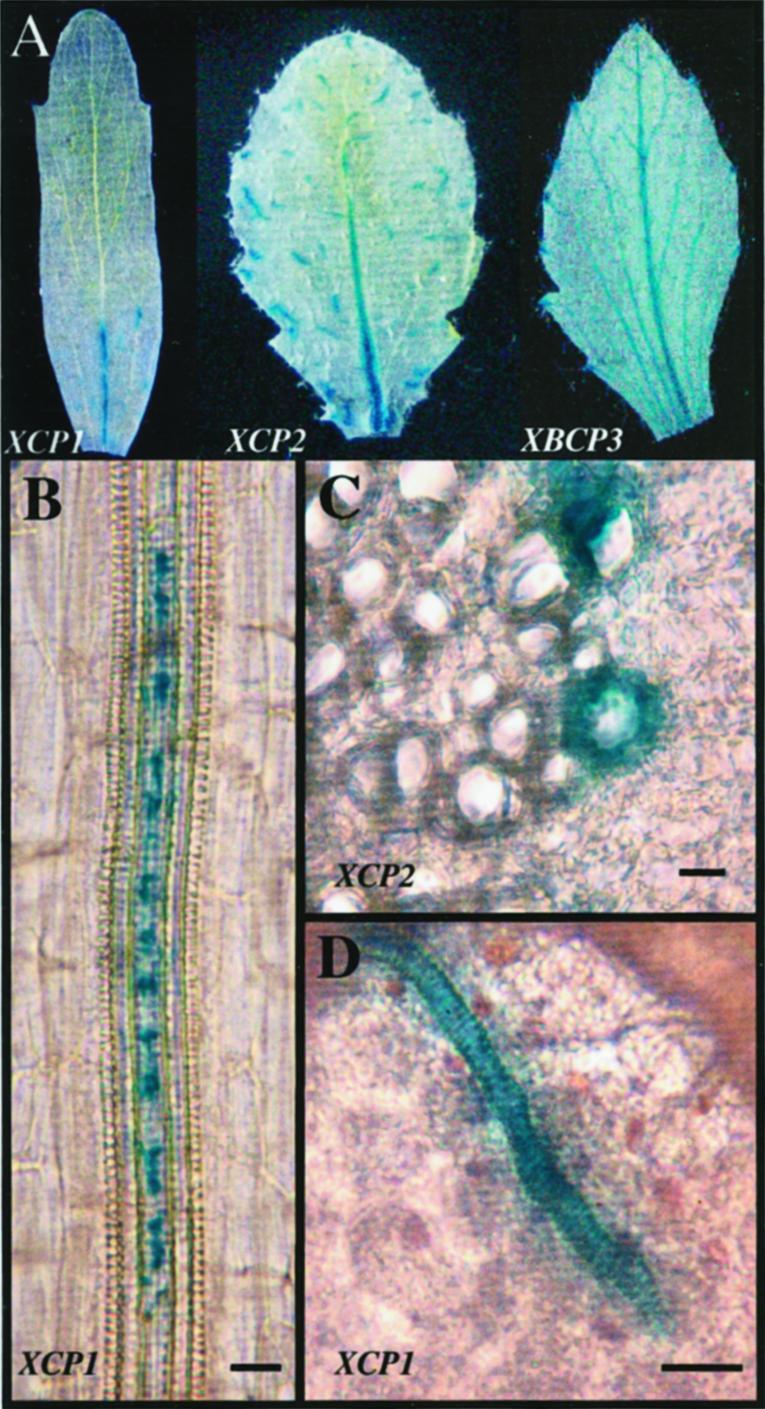
XCP1 and XCP2 promoters direct GUS expression in tracheary elements. A, GUS expression directed by promoters for XCP1, XCP2, and the control gene, XBCP3, is compared at the whole leaf level, illustrating that GUS activity is localized to specific areas within leaf vascular tissue for the XCP1 and XCP2 promoters. The XBCP3 promoter directs GUS expression in hydathodes and throughout the vascular system. GUS expression localized to tracheary elements is shown for XCP1 (B and D) in primary xylem and for XCP2 (C) in secondary xylem. The leaf shown in A for XCP1-GUS is a cauline leaf. Leaves shown for XCP2-GUS and XBCP3-GUS are rosette leaves. A and D were prepared using whole mounts of leaf tissue. B was prepared using a whole mount of the primary root from a 7-d-old seedling. C was prepared using a free-hand transverse section of root tissue from a 4-week-old plant. Bars = 20 μm.
TE-specific expression was consistent among transformed T1 plants and their progeny examined for 15 XCP1- and nine XCP2-GUS lines. To further demonstrate that TE-specific expression was due to the XCP1 and XCP2 promoters and not due to other effects of transgene insertion, a putative promoter for another vascular tissue papain-like peptidase, XBCP3 (xylem/bark Cys peptidase 3), isolated from xylem and bark cDNA libraries (Zhao et al., 2000), was also evaluated (Fig. 1A). GUS activity specified by the XBCP3 promoter was first visible in young leaves at hydathodes only (data not shown). As leaves expanded, XBCP3 promoter activity was increasingly apparent throughout the entire leaf vascular system, and activity at hydathodes persisted. In contrast to XCP1- and XCP2-GUS plants, XBCP3-GUS plants never exhibited GUS activity limited to differentiating TEs. The expression patterns observed for XCP1-, XCP2-, and XBCP3-GUS plants faithfully reflect the spatial distribution of these transcripts determined at the tissue level in northern-blot analyses and cDNA library screenings (Zhao et al., 2000) and demonstrate differential expression of Arabidopsis C1A family peptidases (Rawlings and Barrett, 1999) at the cell-type level.
Immunofluorescence Confocal Microscopy for Cell Type and Subcellular Localization
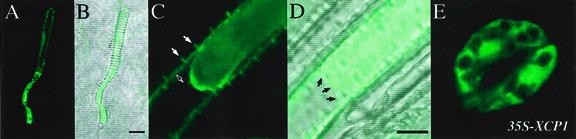
For direct evidence of XCP1 localization, leaves, flowers, and roots of Arabidopsis seedlings were prepared for detection of XCP1 by immunofluorescence. Observation of fluorescence in wild-type Arabidopsis organs confirmed the localization of XCP1 to TEs as observed for XCP1-GUS plants (Fig. 2, A and C). Superimposing a transmitted light image on the confocal images showing XCP1 labeling demonstrates localization within TEs, which may be identified by their helical or sclerified cell walls (Fig. 2, B and D). Seedlings incubated with the fluorescently labeled secondary antibody alone showed non-specific labeling only at the cut ends of the organs, i.e. no TEs were labeled (data not shown). Extravascular labeling not associated with cut ends was observed only when XCP1 was expressed ectopically (35S-XCP1 plants), e.g. in root cortex cells (data not shown) and guard cells (Fig. 2E). The labeling of non-TEs in 35S-XCP1 plants indicates that if XCP1 were present in cells other than TEs of wild-type plants, it would be detected by our methods, i.e. TE-specific labeling is not due to chance.
Figure 2.
Immunofluorescence detection of XCP1 in TEs in wild-type Arabidopsis. XCP1 in TEs in sepals (A) and in roots from 7-d-old seedlings (C) was detected using affinity-purified anti-XCP1 antibody and secondary goat anti-rabbit antibody conjugated to alexa fluor 488. Labeling shows XCP1 localized in a vacuole (C) adjacent to an intact unlabeled vacuole in the same cell (D, black arrows). Labeled XCP1 is also associated with the TE cell wall perforations (C, solid white arrows) and possibly with the plasma membrane (C, open white arrow). Only when XCP1 was ectopically expressed was labeling specifically detected in cell types other than TEs such as the guard cells shown in E. Bars = 10 μm.
Consistent with data from XCP1-GUS experiments, only a few TEs within the organs prepared for confocal microscopy were detected using anti-XCP1 antibody. Although precise subcellular localization of XCP1 was not possible for most labeled TEs, perhaps reflecting normal TE protoplast degeneration (Fig. 2A), one exception to apparent protoplast disorganization was noted. Figure 2C shows an XCP1-containing vacuole possessing an intact membrane at one end. Using zinnia leaves fixed for electron microscopy, Burgess and Linstead (1984) also reported finding TEs containing vacuoles with one end intact and one end merged with the cytosol. The long (up to 300 μm) sclerified TEs in primary roots of young seedlings were found to contain two or three vacuoles, and the end of the labeled vacuole shown in Figure 2C is in contact with the end of an unlabeled vacuole (Fig. 2D, black arrows). The presence of another intact vacuole in this TE indicates that this membrane-limited labeling is confined to a vacuole and is not distributed throughout a contracting protoplast. The presence of unlabeled and labeled vacuoles in the same cell may reflect specialization of vacuoles in TEs. Vacuole specialization in aleurone cells undergoing programmed cell death has been reported (Swanson et al., 1998). Fluorescence was frequently associated with the cell wall in mature TEs, as evident by labeling in cell wall perforations (Fig. 2C, solid white arrows). Labeling on the inner surface of the cell wall may also indicate XCP1 associated with the plasma membrane (Fig. 2C, open white arrow). Labeling of the cell wall may reflect leakage of XCP1 from TE protoplasts or the secretion of some fraction of XCP1 during TE differentiation.
Localization of XCP1 in 35S-XCP1 Plants
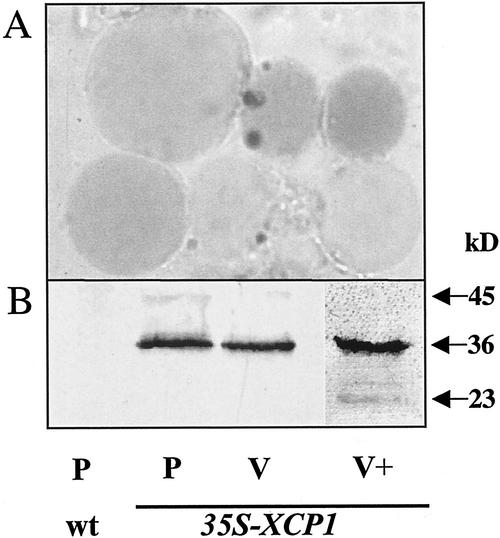
We also determined whether ectopically expressed XCP1 was targeted to vacuoles. Vacuoles, stained with neutral red to aid detection, were prepared from protoplasts isolated from wild-type and 35S-XCP1 plants (Fig. 3A). Observation of vacuoles being released following lysis of protoplasts revealed the presence of a single large vacuole for each protoplast (data not shown). Counts of contaminating protoplasts that copurified with vacuoles indicated 3% contamination, whereas marker enzyme assays for malate dehydrogenase as a negative marker versus α-mannosidase as a positive marker revealed a 12.5% contamination level for extravacuolar proteins (data not shown). The immunoblot depicted in Figure 3B shows XCP1 levels in protoplasts versus vacuoles. The 36-kD polypeptide is the primary form detected by immunoblot analysis of protoplasts from 35S-XCP1 plants and is equal to the predicted molecular mass of proXCP1. A larger polypeptide barely detectable at 45 kD may be preproXCP1. The 45-kD protein is 5 kD greater than the predicted mass for preproXCP1. One potential N-glycosylation site at Asn180 is present, suggesting that glycosylation may lead to a larger form than predicted from the unmodified polypeptide. As expected, wild-type protoplasts contained no detectable XCP1. A comparison of extracts from equal numbers of vacuoles and protoplasts from 35S-XCP1 plants (Fig. 3B) revealed approximately equal levels of XCP1, indicating that nearly all protoplast XCP1 is located in the vacuole. Similar immunoblot results were obtained from two independent vacuole isolation experiments. Doubling the vacuolar protein level loaded for immunoblot analysis revealed the presence of additional immunoreactive proteins with the most prominent, after the 36-kD protein, migrating as a 23-kD protein, the expected sized for the mature active form of XCP1 (Fig. 3B). The presence of an additional, barely detectable immunoreactive protein migrating between the 36- and 23-kD forms suggests that post-translational processing more complex than a single proteolytic cleavage of the proprotein may be required for XCP1 maturation.
Figure 3.
Ectopically expressed XCP1 localizes to vacuoles. A, Vacuoles purified from protoplasts prepared from leaves of line S2, 35S-XCP1 plants were stained with neutral red to aid in detection during purification. B, Immunoblot analysis using anti-XCP1 antibody shows that XCP1 is not detectable in protoplasts isolated from leaves of wild-type plants (P, wt). Extracts prepared from equal numbers (1.4 × 105) of protoplasts (P, 35S-XCP1) or vacuoles (V, 35S-XCP1) contained nearly identical levels of XCP1 at 36 kD. Loading 2-fold more vacuoles (V+, 35S-XCP1) revealed immunoreactivity at 23 kD, the expected mass for mature XCP1, in addition to the 36-kD form.
Effects of Ectopic Expression of XCP1 on Plant Growth and Senescence
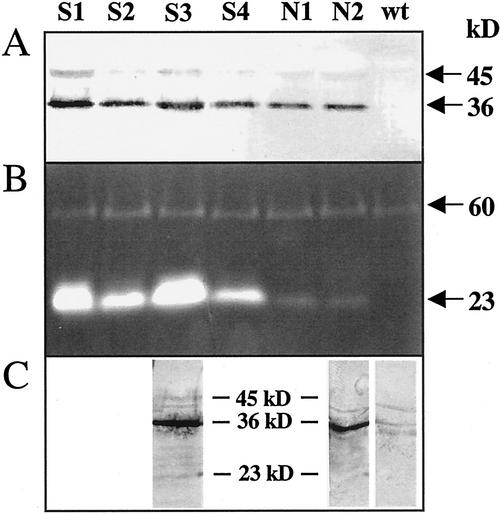
Several lines of 35S-XCP1 transgenics produced plants that were noticeably smaller (S lines) than wild-type plants, whereas other lines yielded apparently normal-sized plants (N lines). When biomass was determined for S3, S5, and S6 lines and N1 and N2 lines, grams fresh weight of 3-week-old S line plants ranged from 20% to 50% less than that of N line plants (Table I). Biomass of 3-week-old N line plants did not differ from that of control plants. S and N lines yielded plants that began to lose chlorophyll from leaves at an earlier age than control plants, with extractable chlorophyll at 6 weeks ranging from 91% to 42% of that for control plants for the seven S and N lines tested (Table I). Early chlorophyll loss did not appear to correlate with the reduced plant size phenotype, however, as N line plants yielded some of the lowest chlorophyll values, whereas line S5 chlorophyll was closest, at 91%, to that of control plants. Immunoblot (Fig. 4A) and activity gel (Fig. 4B) analyses using S3 plants described in Table I and additional S lines (S1, S2, and S4) for comparison with N1 and N2 plants indicated that reduced plant size was correlated with increased levels of immunoreactive XCP1 (36 kD) and peptidase activity at 23 kD, the mass predicted for mature, active XCP1. For all 35S-XCP1 lines, XCP1 abundance and activity was above that detectable in extracts from wild-type plants (Fig. 4A) or from control lines transformed with the same vector containing an unrelated sequence (data not shown).
Table I.
Impact of 35S-XCP1 transgene on biomass of 3-week-old plants and extractable chlorophyll from 6-week-old plants
| Line | Plant Biomass | Chlorophyll Extracted |
|---|---|---|
| 100 g fresh wt plant−1 | mg chlorophyll L−1 | |
| C | 1.25 (100) | 130.9 (100) |
| S3 | 0.82 (65) | 60.3 (46) |
| S5 | 0.63 (50) | 118.6 (91) |
| S6 | 0.99 (79) | 88.6 (68) |
| N1 | 1.18 (94) | 70.4 (54) |
| N2 | 1.30 (104) | 64.5 (49) |
| N3 | ND | 84.5 (65) |
| N4 | ND | 55.1 (42) |
C, Control, plants transformed with pCB302 containing an unrelated sequence; ND, not determined. Values shown for chlorophyll are means of three replicates from a single extract for each line. Values in parentheses are percentages relative to control plants.
Figure 4.
Analysis of XCP1 protein and peptidase activity in Arabidopsis plants ectopically expressing XCP1 reveals higher levels of XCP1 in stunted plant lines (S lines) compared with normal-sized plant lines (N lines). Immunoblot analysis (A) using anti-XCP1 antibody shows higher levels of XCP1 at 45 kD (putative preproXCP1) and 36 kD (putative proXCP1) positively correlated with peptidase activity (B) at 23 kD (putative mature XCP1). Protein levels used for each lane of the immunoblot and activity gel in A and B were 30 and 60 μg, respectively. Immunoblot analysis using anti-XCP1 antibody and 60 μg of protein from representative S and N lines (C) reveals immunoreactivity at 23 kD for the S3 line but not for N2 or wild-type plants.
Immunoreactive protein at 23 kD was not detectable when S and N lines were evaluated using 30 μg of protein (Fig. 4A). However, in general agreement with results using vacuolar extracts, increasing the protein level (to 60 μg) revealed the presence of a 23-kD protein (Fig. 4C). The 23-kD protein was visible in S3 line extracts but not in extracts from N2 or wild-type plants, consistent with the relative levels of active 23-kD peptidase in these lines (Fig. 4B). Increasing the level of protein for the immunoblot of wild-type leaf extract to 60 μg revealed the presence of a low level of endogenous XCP1 (36 kD). Consistent with our findings with higher levels of vacuolar protein (Fig. 3), additional barely detectable immunoreactive proteins were observed when the 60-μg blot was compared with the 30-μg blot (Fig. 4, A versus C). These additional proteins were most abundant in the S3 extract and are within the molecular mass range defined by preproXCP and the predicted mature XCP1, suggesting that they may be post-translational processing intermediates of the XCP1 zymogen. The unrelated 60-kD peptidase detectable in wild-type and all 35S-XCP1 extracts (Fig. 4B) serves as a loading control for the immunoblots and activity gel shown in Figure 4.
Identification of Ectopically Expressed XCP1 and Autocatalytic Processing of XCP1
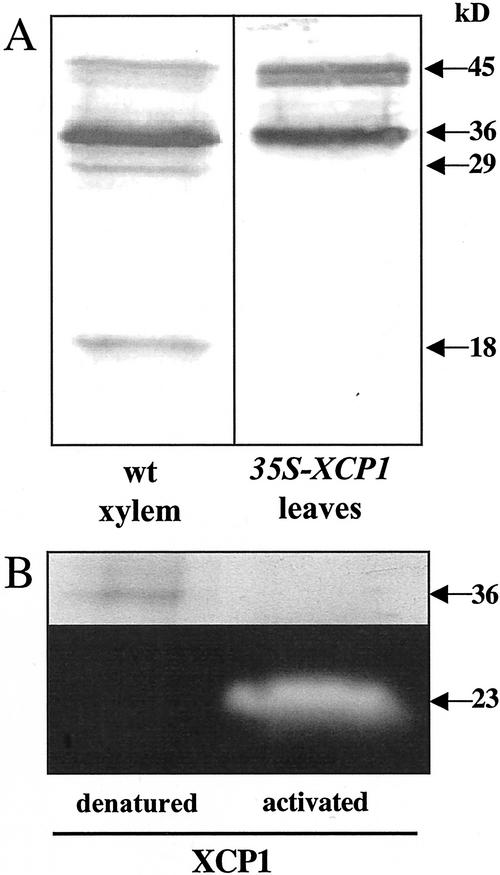
The XCP1 profiles detected by immunoblot analysis indicate that differences exist between XCP1 processing in 35S-XCP1 plants and that in wild-type xylem (Fig. 5A). In addition to the major immunoreactive 45- and 36-kD proteins common to wild-type xylem and 35S-XCP1 extracts, the xylem extract contains additional major immunoreactive polypeptides detectable at 29 and 18 kD, perhaps reflecting xylem-specific processing of XCP1. The absence of detectable levels of immunoreactivity at 23-kD in the xylem extract suggests that accumulation of the 23-kD form of XCP1 reported here (Figs. 3 and 4) may be a unique or at least more predominant product of ectopic XCP1 processing, whereas additional processing to produce an 18-kD form may be the ultimate fate of XCP1 in the xylem. It is interesting that Cys peptidase activity migrating as an 18-kD protein was also extracted from Arabidopsis xylem but not from phloem (Zhao et al., 2000).
Figure 5.
XCP1 processing in wild-type xylem is different from that in 35S-XCP1 plants, and the 36-kD form of XCP1 can be activated in vitro. A, Polypeptides detected with anti-XCP1 antibody at 36 and 45 kD are common to wild-type xylem and 35S-XCP1 plants. Immunoreactivity at 29 and 18 kD is unique to xylem. B, In vitro autocatalytic processing of the 36-kD form of XCP1 excised from an SDS-PAGE gel resulted in proteolytic activity at 23 kD, and the loss of protein at 36 kD. For the denatured control, degradation and activation of the 36-kD protein was not observed.
For independent confirmation of the identity of the most prominent (36 kD) protein detected in anti-XCP1 immunoblots (Fig. 4A), ectopically expressed XCP1 was partially purified from line S2 plants (see “Materials and Methods”) for determination of peptide mass by matrix-assisted laser desorption ionization-mass spectrometry (MALDI-MS). Following SDS-PAGE of partially purified XCP1, the 36-kD protein was excised and digested with trypsin. Analysis by MALDI-MS yielded three detectable peaks at 716, 1,606, and 2,109 daltons that corresponded to the peptides NSWGPR, ALAHQPVSVAIEASGR, and VTISGYEDVPENDDESLKV, respectively, uniquely from XCP1. In contrast to preparations from 35S-XCP1 plants where the 36-kD polypeptide accounted for the majority of the protein in the preparation, partial purification of protein from control plants transformed with the pCB302 vector containing an unrelated sequence did not yield a large quantity of protein at 36 kD. This comparison indicates that the results obtained for MALDI-MS analysis are specific to ectopic XCP1 and do not reflect endogenous TE XCP1 levels.
To demonstrate that the 36-kD protein identified by MALDI-MS as XCP1 could be processed to yield the 23-kD active enzyme detected in extracts from 35S-XCP1 plants, an in vitro activation of the 36-kD protein was performed. For the experiment shown in Figure 5B, partially purified XCP1 was resolved using SDS-PAGE. Following incubation for 3 h in XCP1 activation buffer (Zhao et al., 2000), the region of the gel corresponding to 36-kD (well isolated from the 23-kD region of the gel) was excised and its contents were extracted from the gel slice and resolved again by SDS-PAGE, followed by activity gel analysis. This in vitro peptidase activation experiment resulted in the loss of detectable protein at 36 kD and the presence of proteolytic activity at 23 kD. These results using XCP1 purified from Arabidopsis are consistent with those reported previously using recombinant XCP1 purified from Escherichia coli showing apparent autocatalytic processing of XCP1 (Zhao et al., 2000). A control gel slice not exposed to activation buffer and boiled prior to activity gel analysis retained the 36-kD polypeptide (Fig. 5B), indicating that processing of proXCP1 was enzyme dependent. Considered together, the in vitro processing of the 36-kD form of XCP1 to produce a 23-kD peptidase and the presence of immunoreactive protein at 23 kD in leaf and vacuolar extracts from 35S-XCP1 plants indicate that XCP1 is responsible for the 23-kD peptidase activity detected in 35S-XCP1 plants.
DISCUSSION
Little is known about the ≥28 predicted papain-like Cys peptidases encoded by the Arabidopsis genome. Expression for genes SAG12 (Gan and Amasino, 1995) and SAG2/AtALEU (Hensel et al., 1993; Ahmed et al., 2000) is associated with senescence, whereas two other genes for C1A peptidases, rd21a and rd19a, are induced by drought stress (Koizumi et al., 1993). Our laboratory previously reported the xylem-specific expression of the Arabidopsis papain-like peptidase XCP1. To our knowledge, in this report, we have provided the first experimental evidence that XCP1 is localized to TE vacuoles. This report also provides confirmation that the cDNA library constructed from Arabidopsis xylem (Zhao et al., 2000) is a valuable tool that facilitates isolation of the TE-specific members of large peptidase or other multigene families.
A comparison of the immunoblot profile of ectopically expressed XCP1 with that of XCP1 from wild-type xylem revealed distinct XCP1 fingerprints. Both samples contained immunoreactive protein at 45 kD, perhaps due to heavy trafficking of XCP1 through the secretory pathway leading to high steady-state levels of the prepropeptide. Immunolocalization of chymopapain to the ER membranes of differentiating laticifers has been reported (Ying et al., 1994) and may indicate the presence of high levels of unprocessed chymopapain zymogen in these latex-containing xylem cells. In the transgenic plants used for this report, 35S-driven expression of XCP1 could lead to saturation of secretory pathway components with preproXCP1 and to detectable levels of this form. The immunoreactive polypeptides at 36 and 23 kD and the peptidase activity at 23 kD correspond to the predicted sizes of proXCP1 (36 kD) and mature XCP1 (23 kD), using homology with papain to predict the cleavage sites (Groves et al., 1996). That the xylem extract contains an additional form of XCP1, at 18 kD, that comigrates with a previously reported xylem-specific peptidase activity (Zhao et al., 2000) suggests that processing of XCP1 in TEs requires TE-specific factors. Numerous Cys, Ser, Asp, and metallopeptidases are associated with xylem differentiation in Arabidopsis (E. Beers and C. Zhao, unpublished data), and one or more of these activities may be required to generate the xylem-specific immunoreactive 18-kD polypeptide reported here. The physiological significance of any xylem-specific forms of XCP1 relative to TE autolysis or other aspects of differentiation is not known.
Although we observed a correlation between reduced seedling size and high XCP1 activity as shown in Figure 4, in vitro activity may not accurately reflect in planta activity of XCP1. If the prodomain is cleaved from the propeptide in an acidic vacuole, its continued noncovalent association with mature XCP1 within the vacuole may be inhibitory (Taylor et al., 1995). SDS-PAGE in preparation for activity gels would result in the resolution of the cleaved prodomain and mature XCP1, leading to subsequent in vitro activity. As an alternative to phenotype linked to increased peptidase activity, the reduced plant size and early plant senescence phenotypes of the 35S-XCP1 plants (Table I) may be the result of stress linked to saturation of key secretory pathway components by high levels of XCP1.
Peptidase prodomains are important regulators of targeting (Holwerda et al., 1992; Ahmed et al., 2000), folding, and activity (Mach et al., 1994; Tao, et al., 1994; Taylor et al., 1995). The prodomains (excluding the highly divergent 23-amino acid C terminus; Taylor et al., 1995) of XCP1, XCP2, and the zinnia TE papain-like peptidase, p48–17 (Ye and Varner, 1996), are exceptional among reported papain-like Cys peptidases in that they exhibit the highest degree of similarity with the prodomains of papaya (Carica papaya) proteinases I (papain), II (chymopapain), III (caricain), and IV (glycyl endopeptidase) (mean of 82% similarity for TE peptidases compared with papain proteinases I–IV). In addition, the TE peptidases possess a propeptide N-terminal consensus sequence [D/E][F/Y]SI[V/L]GY, of unknown function, shared with papaya laticifer proteinases I, III, and IV. (Chymopapain possesses a similar but more divergent DFYTVGY N-terminal sequence.) This motif occupies the same relative position as the NPIR domain known to be important for vacuolar targeting of aleurain (Holwerda et al., 1992) and AtALEU (Ahmed et al., 2000), and it may be part of an N-terminal propeptide targeting signal.
Despite the long history of biochemical characterizations and industrial applications of papain and other Cys peptidases isolated from papaya, biological roles for these enzymes have not been clearly defined. Papain and its paralogs in papaya are localized in ER-derived vesicles of laticifers located in the xylem and phloem (Ying et al., 1994). That a maize Cys peptidase and the prodomain of papaya proteinase IV may function to inhibit insect digestive processes (Visal et al., 1998; Pechan et al., 2000) is consistent with a potential plant protection role for some papain-like enzymes. Papaya papain-like enzymes may also catalyze latex coagulation to promote wound sealing following insect feeding (El Moussaoui et al., 2001). Perhaps the degeneration and leakage of protoplast contents during the maturation of TEs represents a genetically programmed “wound” that requires XCP1/XCP2-mediated processing similar to that mediated by papain excreted from laticifers after damage to papaya tissue by herbivores.
In this report, we describe XCP1 as a TE peptidase sharing exceptional prodomain sequence similarity with papain, a laticifer peptidase. That TEs and laticifers are xylem cell types in papaya suggests that some peptidase genes expressed in TEs may have been recruited for defensive roles as excreted peptidases in papaya. This important new function may have occurred through mutations resulting in subtle alterations in subcellular trafficking and/or temporal expression patterns of papain-like enzymes in xylem cells, and may have also required duplication and further specialization of genes originally encoding TE-specific papain-like enzymes. Comparisons of laticifers and TEs with regard to expression and trafficking of their potentially orthologous C1A peptidases may reveal novel features of specialized plant vacuoles that function as vectors of peptidases balanced between intracellular autolysis and plant defense.
MATERIALS AND METHODS
Plant Material
For isolation of secondary xylem, Arabidopsis (ecotype Columbia) plants were grown at a density of four to six plants per 10-cm pot, as described in Zhao et al. (2000). Arabidopsis plants used for Agrobacterium tumefaciens-mediated transformations, protoplast, and vacuole preparations and leaf protein extraction were grown at higher density (approximately 50–75 plants pot−1).
XCP1-GUS Plants
A 591-bp region from Arabidopsis chromosome four, i.e. the region flanked by the stop codon of the predicted preceding gene and the initial Met of XCP1 (AF191027), was amplified by PCR from Arabidopsis genomic DNA using primers that incorporated an SpeI site (underlined) at the 5′ end (5′-GCACTAGTGTGTTTGCACTTTGCAGG-3′) and a NcoI site at the 3′ end (5′-GCCATGGCCAAATTTGTTCACTGAG-3′). The resulting PCR product was cloned into a pGEM-T Easy vector (Promega, Madison WI), and subsequently into pSG506 vector (Gan and Amasino, 1995) as a NotI/NcoI fragment, replacing the SAG12 promoter. An SpeI/BamHI digest of the resulting vector was used to subclone the XCP1 promoter-GUS-Mas-terminator cassette from pSG506 into the binary vector, pCB302 (Xiang et al., 1999). The resulting XCP1-GUS binary vector was used to transform A. tumefaciens, strain GV310. Four-week-old Arabidopsis plants were infected with transformed A. tumefaciens, according to the vacuum infiltration protocol of Bechtold and Pelletier (1998). Transgenic plants expressing the selectable marker bar were identified using 0.4% (v/v) Finalé (Aventis, Strasbourg, France).
XCP2-GUS and XBCP3-GUS Plants
A 1.98-kb region upstream of the XCP2 (AF191028) initial Met was amplified from genomic DNA by PCR using a sense primer (5′-TCTAGAACCGTCGTCGCAGGTTAATA-3′) incorporating an XbaI site at the 5′ end, and an antisense primer (5′-GGATCCAAAGAGCCGTTTGAGTACGT-3′) incorporating a BamHI site. The resulting PCR product was cloned into pGEM-T Easy. The resulting vector was digested with XbaI, blunted using the Klenow fragment of DNA polymerase I, and the promoter was released from the vector using BamHI. The binary vector pBI121 (Jefferson et al., 1987) was prepared for ligation with the promoter by digestion with HindIII, treatment with Klenow, and digestion with BamHI. For XBPCP3-GUS, the 950-bp putative promoter for XBCP3 (AF388175) was amplified from genomic DNA using the sense primer (5′-AAGCTTAAGGATGTATTTTTTTTATC-3′) including an existing HindIII site, and an antisense primer (5′-GGATCCTGTAATTTTTGTTTGTTAGAATC-3′) adding a BamHI site, and was subcloned into pBI121 as a HindIII/BamHI fragment from a pGEM-T Easy vector. XBCP3 and XCP2 promoters replaced the cauliflower mosaic virus 35S promoter in pBI121. XCP2-GUS and XBCP3-GUS binary vectors were used to transform A. tumefaciens and Arabidopsis as described for the XCP1-GUS. Transgenic plants were selected by spraying kanamycin (50 μg mL−1) directly on 10-d-old T1 seedlings.
35S-XCP1 Plants
XCP1 was amplified from a xylem cDNA library (Zhao et al., 2000) by PCR using the sense primer (5′-TTGGCCATGGCTTTTTCTGCACCA-3′) incorporating an NcoI site, and the antisense primer (5′-AGATCTATCTATCACTTGGTCT-3′) incorporating a BglII site. The resulting product was cloned into pGEM-T Easy and from there into the vector pAVA121 (von Armin et al., 1998) as a NcoI/BglII fragment in fusion with the cauliflower mosaic virus 35S promoter. The expression cassette from pAVA121 was subcloned into pCB302 using a PstI digest. Transformation and selection of transgenics were performed as described for the XCP1-GUS plants.
Analysis of Plant Biomass and Chlorophyll Content
Biomass of above ground tissue (T2, herbicide-resistant plants) for selected S and N lines of 35S-XCP1 plants was determined using 3-week-old plants grown under identical light, nutrient, and watering regimes. Plants were grown at a population density of 80 to 110 plants in 10-cm pots. For chlorophyll measurements, 3 to 5 g of tissue from herbicide-resistant 6-week-old plants was homogenized using a mortar and pestle and a 2:1, 80% (v/v) acetone:tissue ratio. Following centrifugation, chlorophyll in the supernatant was determined as in Beers et al. (1992). Three replicates, as three aliquots from a single preparation, were evaluated.
Protein Extraction
Protein for immunoblots and activity gels was extracted from leaves, protoplasts, vacuoles, and secondary xylem by grinding tissue in 100 mm sodium phosphate, pH 7.2, with 7 mm 2-mercaptoethanol, and 20 μm leupeptin. Secondary xylem was powdered in liquid nitrogen prior to buffer extraction. The homogenate was clarified by centrifugation at 14,000g for 15 min at 4°C. Protein concentrations were determined using bicinchoninic acid (Sigma, St. Louis). In some cases, extracts were concentrated on YM10 membranes (Millipore, Bedford, MA) prior to protein quantification.
Immunoblot and Activity Gel Analyses
Immunoblots were prepared according to Woffenden et al. (1998) using an affinity-purified antibody raised against purified poly-His-tagged XCP1 (Zhao, et al., 2000). Activity gels were prepared as described in Beers and Freeman (1997).
Immunofluorescence Confocal Microscopy
Fixation and labeling of roots and flowers from Arabidopsis for confocal microscopy were performed as described by Wymer et al. (1999). Roots from 3-d-old seedlings or the terminal flower cluster from 4-week-old Arabidopsis plants were fixed in freshly prepared 4% (w/v) formaldehyde in PEM 50:5:5 (50 mm PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.9, 5 mm EGTA, and 5 mm MgSO4) and 0.2% (v/v) IGEPAL CA-630 (Sigma) for 1 h at room temperature (rt). The roots/flower clusters were rinsed with PEM 50:5:5 and were allowed to dry at rt until moisture was no longer visible on the tissue surface. The tissue was digested for 10 min at rt in 0.025% (w/v) cellulase R10, 0.025% (w/v) cellulase R5, 0.0125% (w/v) pectolyase, 0.0125% (w/v) macerozyme (Karlan Research Products, Santa Rosa, CA), and 1% (w/v) Driselase (Sigma) in PEM 50:5:5, followed by rinsing six times with 0.2% (v/v) IGEPAL in PEM 50:5:5 and before air-drying. The tissue was incubated in blocking solution (3% [w/v] bovine serum albumin [BSA] and 0.2% [v/v] IGEPAL in PEM 50:5:5) for 90 min at rt prior to incubation overnight at 4°C with anti-XCP1 antibody, preabsorbed against a crude protein extract from mature leaves, in blocking solution. The next morning, the roots/flower clusters were washed in blocking solution for 1 h at rt, with the solution changed three times, prior to incubation with secondary goat anti-rabbit antibody conjugated to alexa fluor 488 (Molecular Probes, Eugene, OR) for 2 h at 37°C. The labeled tissue was finally washed with 0.2% (v/v) IGEPAL in PEM 50:5:5 over the next 48 h with several buffer changes and it was stored at 4°C. The tissue was mounted for microscopy using a LSM 510 confocal microscope (Zeiss, Jena, Germany).
Protoplast and Vacuole Preparation
Protoplasts were prepared from leaves of 3- to 4-week-old Arabidopsis plants according to Abel and Theologis (1998) with slight modifications. The leaves were rinsed four times with sterile distilled water, blotted dry on filter paper, and chopped with a razor blade (approximate final size of 9 mm2). The leaf tissue was then incubated in 0.5 m mannitol as a preplasmolysis step. After 1 h at rt, the mannitol solution was replaced with 30 mL of protoplasting solution (Ahmed et al., 2000; 0.4 m mannitol, 1% [w/v] cellulase R-10, 0.5% [w/v] macerozyme, 0.5% [w/v] BSA, 30 mm CaCl2, 5 mm 2-mercaptoethanol, and 5 mm MES [2-(N-morpholino)ethanesulfonic acid], pH 5.7). The chopped leaf tissue was vacuum infiltrated with protoplasting buffer and was incubated at rt with gentle agitation (100 rpm) for 3 to 4 h or until the majority of protoplasts were released. The digested material was filtered through an 83-μm screen to separate protoplasts from leaf tissue. Filtered protoplasts were pelleted and washed once with 0.5 m mannitol, 5 mm MES (pH 6.0), and 1 mm CaCl2. Protoplasts were counted on a hemocytometer. Approximately 80% of the protoplasts were used for isolating vacuoles according to Ahmed et al. (2000) using a 4% (w/v) Ficoll step to separate the vacuoles from pelletable debris.
Malate Dehydrogenase and α-Mannosidase Enzyme Assays
The NADH-malate dehydrogenase assays of protoplast and vacuole extracts were performed using final concentrations of 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid], pH 7.5, 338 μm NADH, and 1 mm oxaloacetate (Beers et al., 1992). The α-mannosidase assays were performed according to Ahmed et al. (2000) using a TKO 100 mini-fluorometer (Hoeffer, San Francisco, CA). The concentrations of the assay buffer were 0.1% (w/v) BSA, 100 mm potassium acetate, pH 5.0, and 1.42 mg mL−1 4-methylumbelliferyl-α-d-mannopyranoside (Sigma). Three replicates, as three aliquots from a single protoplast/vacuole preparation, were evaluated.
Partial Purification of XCP1 from 35S-XCP1 Plants
Nonsenescing leaves from 3-week-old plants were homogenized in citrate-phosphate buffer (22 mm citric acid, 55 mm dibasic sodium phosphate, pH 5.5, 7 mm 2-mercaptoethanol, and 20 μm leupeptin) (3:1, buffer:tissue). Following centrifugation at 14,000g for 10 min, the supernatant was transferred to a clean tube and was incubated on ice for 6 h to promote protein precipitation. Insoluble protein was pelleted by centrifugation at 14,000g for 15 min, and the resulting supernatant was concentrated approximately 5-fold using YM10 concentrators prior to overnight incubation at −80°C for further protein precipitation. Following pelleting of the newly precipitated proteins by centrifugation at 14,000g for 15 min, the supernatant was applied to a Sephadex G-100 column equilibrated with 100 mm sodium phosphate buffer, pH 7.2, 7 mm 2-mercaptoethanol, and 20 μm leupeptin. Fractions (1 mL) showing the highest levels of immunoreactivity with anti-XCP1 antibody, by immunoblot analysis, were pooled, concentrated and stored at −80°C for further analysis.
MALDI-MS Identification of XCP1
The partially purified XCP1 was resolved using SDS-PAGE and was stained with Coomassie. The 36-kD protein corresponding to the molecular mass of the major immunoreactive protein was excised and subjected to digestion in-gel with modified trypsin (Promega) without prior alkylation. The trypsinized polypeptide was analyzed on a Kompact Seq MALDI-time of flight mass spectrometer (Kratos, Chestnut Ridge, NY) using internal calibration (modified trypsin peaks at 842.5 and 2211.1 m/z). The resulting peptide masses were submitted to the PepIdent program (http://www.expasy.org/tools/peptident.html) where the search was limited to the Arabidopsis genome.
In Vitro Processing of proXCP1
XCP1 was partially purified from 35S-XCP1 plants as described for this report, but excluding the gel filtration step. Following resolution by SDS-PAGE, the polyacrylamide gel containing duplicate samples of partially purified XCP1 was divided in two; one-half was stored at −80°C, whereas the other was incubated in activation buffer (Beers and Freeman, 1997) for 3 h. Following activation, a gel slice corresponding to 36 kD was excised and protein was extracted by homogenization in SDS-PAGE sample buffer. The corresponding 36-kD slice was also excised from the gel stored at −80°C, extracted in SDS-PAGE sample buffer, and boiled to denature the 36-kD proXCP1. Both protein extracts were then processed for peptidase activity gel analysis according to Beers and Freeman (1997).
ACKNOWLEDGMENTS
We thank Earl Petzold for expert technical assistance and Steve Hunsucker for the in-gel digestions and MALDI-time of flight analysis. We also thank Dr. Ray F. Evert for providing preprints, Dr. Elizabeth Grabau for critical reading of the manuscript, and Dr. Richard Helm for helpful discussions concerning MALDI-MS.
Footnotes
This research was supported by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (project no. 9801401).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.010514.
LITERATURE CITED
- Abel S, Theologis A. Transient gene expression in protoplasts of Arabidopsis thaliana. In: Marinez-Zapater JM, Salinas J, editors. Arabidopsis Protocols. Totowa, NJ: Humana Press; 1998. pp. 209–218. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Rojo E, Kovaleva V, Venkataraman S, Dombrowski J, Matsuoka K, Raikhel N. The plant vacuolar sorting receptor AtELP is involved in transport of NH(2)-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol. 2000;149:1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Beers EP, Freeman TB. Proteinase activity during tracheary element differentiation in Zinnia mesophyll cultures. Plant Physiol. 1997;113:873–880. doi: 10.1104/pp.113.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers EP, Moreno TN, Callis J. Subcellular localization of ubiquitin and ubiquinated proteins in Arabidopsis thaliana. J Biol Chem. 1992;267:15432–15439. [PubMed] [Google Scholar]
- Beers EP, Woffenden BJ, Zhao C. Plant proteolytic enzymes: possible roles during programmed cell death. Plant Mol Biol. 2000;44:399–415. doi: 10.1023/a:1026556928624. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Brouquisse R, Gaudillere JP, Raymond P. Induction of carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol. 1998;117:1281–1291. doi: 10.1104/pp.117.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J, Vierstra RD. Protein degradation in signaling. Curr Opin Plant Biol. 2000;3:381–386. doi: 10.1016/s1369-5266(00)00100-x. [DOI] [PubMed] [Google Scholar]
- Burgess J, Linstead P. Comparison of tracheary element differentiation in intact leaves and isolated mesophyll cells of Zinnia elegans. Micron Microscop Acta. 1984;15:153–160. [Google Scholar]
- El Moussaoui A, Nijs M, Paul C, Wintjens R, Vincentelli M, Azarkan M, van Looze Y. Revisiting the enzymes stored in the laticifers of Carica papaya in the context of their possible participation in the plant defense mechanism. Cell Mol Life Sci. 2001;58:556–570. doi: 10.1007/PL00000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M. Proteases and cellular regulation in plants. Curr Opin Plant Biol. 2001;4:254–260. doi: 10.1016/s1369-5266(00)00169-2. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Komamine A. Direct evidence for cytodifferentiation to tracheary elements without intervening mitosis in a culture of single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 1980;65:61–64. doi: 10.1104/pp.65.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Granell A, Cercos M, Carbonell J. Plant cysteine proteinases in germination and senescence. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. New York: Academic Press; 1998. [Google Scholar]
- Groover A, DeWitt N, Heidel A, Jones A. Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma. 1997;196:197–211. [Google Scholar]
- Groover A, Jones AM. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 1999;119:375–384. doi: 10.1104/pp.119.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M, Taylor M, Scott M, Cummings N, Pickersgill R, Jenkins J. The prosequence of procaricain forms an α-helical domain that prevents access to the substrate-binding cleft. Structure. 1996;4:1193–1203. doi: 10.1016/s0969-2126(96)00127-x. [DOI] [PubMed] [Google Scholar]
- Hensel LL, Grbic V, Baumgarten DA, Bleecker AB. Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell. 1993;5:553–564. doi: 10.1105/tpc.5.5.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holwerda BC, Padgett HS, Rogers JC. Proaleurain vacuolar targeting is mediated by short contiguous peptide interactions. Plant Cell. 1992;4:307–318. doi: 10.1105/tpc.4.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlenbach HW, Schmidt B. Cytodifferenzierung in form einer direkten umwandlung isolierter mesophyllzellen zu tracheiden. Z Pflanzenphysiol. 1975;75:369–374. [Google Scholar]
- Koizumi M, Yamaguchi-Shinozaki K, Tsuji H, Shinozaki K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene. 1993;129:175–182. doi: 10.1016/0378-1119(93)90266-6. [DOI] [PubMed] [Google Scholar]
- Kuriyama H. Loss of tonoplast integrity programmed in tracheary element differentiation. Plant Physiol. 1999;121:763–774. doi: 10.1104/pp.121.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach L, Mort JS, Glossl J. Noncovalent complexes between the lysosomal proteinase cathepsin B and its propeptide account for stable, extracellular, high molecular mass forms of the enzyme. J Biol Chem. 1994;269:13036–13040. [PubMed] [Google Scholar]
- McCann MC, Stacey NJ, Roberts K. Targeted cell death in xylogenesis. In: Bryant JA, Huges SG, Garland JM, editors. Programmed Cell Death in Animals and Plants. Oxford: BIOS Scientific Publishers; 2000. pp. 193–201. [PubMed] [Google Scholar]
- Minami A, Fukuda H. Transient and specific expression of a cysteine endoproteinase associated with autolysis during differentiation of Zinnia mesophyll cells into tracheary elements. Plant Cell Physiol. 1995;36:1599–1606. [PubMed] [Google Scholar]
- Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–1241. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa H. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Kuriyama H, Fukuda H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in zinnia. Plant Physiol. 2001;125:615–626. doi: 10.1104/pp.125.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Pechan T, Ye L, Chang Y, Mitra A, Lin L, Davis FM, Williams WP, Luthe DS. A unique 33-kD cysteine proteinase accumulates in response to larval feeding in maize genotypes resistant to fall armyworm and other Lepidoptera. Plant Cell. 2000;12:1031–1040. doi: 10.1105/tpc.12.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 1999;27:325–331. doi: 10.1093/nar/27.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Simpson DJ, Sarioglu H, Lottspeich F, Gietl C. The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:5353–5358. doi: 10.1073/pnas.061038298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson SJ, Bethke PC, Jones RL. Barley aleurone cells contain two types of vacuoles: characterization of lytic organelles by use of fluorescent probes. Plant Cell. 1998;10:685–698. doi: 10.1105/tpc.10.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K, Stearns NA, Dong J, Wu QL, Sahagian GG. The proregion of cathepsin L is required for proper folding, stability, and ER exit. Arch Biochem Biophys. 1994;311:19–27. doi: 10.1006/abbi.1994.1203. [DOI] [PubMed] [Google Scholar]
- Taylor MA, Baker KC, Briggs GS, Connerton IF, Cummings NJ, Pratt KA, Revell DF, Freedman RB, Goodenough PW. Recombinant pro-regions from papain and papaya proteinase IV are selective high affinity inhibitors of the mature papaya enzymes. Protein Eng. 1995;8:59–62. doi: 10.1093/protein/8.1.59. [DOI] [PubMed] [Google Scholar]
- Visal S, Taylor MA, Michaud D. The proregion of papaya proteinase IV inhibits Colorado potato beetle digestive cysteine proteinases. FEBS Lett. 1998;434:401–405. doi: 10.1016/s0014-5793(98)01018-7. [DOI] [PubMed] [Google Scholar]
- von Arnim, Deng XW, Stacey MG. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- Woffenden BJ, Freeman TB, Beers EP. Proteasome inhibitors prevent tracheary element differentiation in Zinnia mesophyll cell cultures. Plant Physiol. 1998;118:419–430. doi: 10.1104/pp.118.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signaling. Plant J. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Wymer CL, Bevan AF, Boudonck K, Lloyd CW. Confocal microscopy of plant cells. Methods Mol Biol. 1999;122:103–130. doi: 10.1385/1-59259-722-x:103. [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. A mini binary vector series for plant transformation. Plant Mol Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Ye Z-H, Varner JE. Induction of cysteine and serine proteinases during xylogenesis in Zinnia elegans. Plant Mol Biol. 1996;30:1233–1246. doi: 10.1007/BF00019555. [DOI] [PubMed] [Google Scholar]
- Ying Z, Ben-ren J, Bing Y. Laticifer ultrastructural and immunocytochemical studies of papain in Carica papaya. Acta Bot Sinica. 1994;36:497–501. [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP. Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 2000;123:1185–1196. doi: 10.1104/pp.123.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]