Abstract
Traditionally, the capsular polysaccharide (CPS) antigen has been used to distinguish between the nine known serotypes of group B streptococcus (GBS) by classical antibody-antigen reactions. In this study, we used PCR for all CPSs and selected protein antigens, multilocus sequencing typing (MLST), and pulsed-field gel electrophoresis (PFGE) to molecularly characterize 92 clinical isolates identified as nontypeable (NT) by CPS-specific antibody-antigen reactivity. The PCR and MLST were performed on blinded, randomly numbered isolates. All isolates contained the cfb gene coding for CAMP factor. While most (56.5%) contained a single CPS-specific gene, 40 isolates contained either two or three CPS-specific genes. Type V CPS-specific gene was present in 66% of the isolates, and all serotypes except types IV, VII, and VIII were represented. Most (44.5%) of the isolates contained a single protein antigen gene (bca, bac, rib, alp1, or alp3), and the remaining isolates had multiple protein antigen genes. Of the 61 isolates that had the V CPS-specific gene, 48 (78.6%) had the alp3 gene. PFGE analysis classified the isolates into 21 profile groups, while MLST analysis divided the isolates into 16 sequence types. Forty-two (69%) of 61 isolates with the V CPS-specific gene were in PFGE profile group 4; 41 of these 42 were sequence type 1 by MLST. These data shed new light on the antigenic complexity of NT GBS isolates, information that can be valuable in the formulation of an effective GBS vaccine.
Streptococcus agalactiae (group B streptococcus [GBS]) is a common cause of neonatal sepsis and meningitis in the United States, and the incidence of GBS disease in nonpregnant adults and the elderly has increased in the last decade (10, 12). The phenotypic assessment of GBS strains by serotyping has traditionally been used in a clinical setting for classifying GBS isolates. Serotyping, based on the capsular polysaccharide (CPS) antigen, is important in understanding the distribution of GBS and aids in the formulation of a multivalent vaccine tailored to specific regions of the world (19, 25). The CPS is a major virulence factor with antiphagocytic function, and major advances have been made in the development of multivalent CPS-protein conjugate vaccines (2, 3, 31).
GBS is serologically classified into nine types—Ia, Ib, and II through VIII—based on the CPS produced (19, 40). When assessed by various techniques, GBS is found not to belong to homogeneous serotype groups, as considerable variation is found at the genetic and protein levels (14, 26). Genotypic methods, including pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST), have been used to genetically characterize and distinguish specific clones of GBS isolates (4, 15, 21, 35). Molecular serotyping by PCR of gene fragments of the variable region of the cps locus has been established (22). In addition, the distribution of surface proteins, such as C alpha protein, C beta protein, Rib, and the R proteins (R1 through R4), has been used to classify GBS isolates (16, 17, 26). Some of these proteins are highly associated with specific serotypes. For example, serotype Ia isolates express C alpha protein, serotype III isolates express Rib, and serotype V isolates are associated with R1 and R4 proteins (1, 14, 23).
Isolates that do not react with the standard CPS antisera are defined as nontypeable (NT). Approximately 2.9% of colonizing and 1.4% of invasive isolates in the United States are NT (5, 15, 16). Up to 12% of all Mexican GBS isolates were found to be NT in one study (30). Previous studies characterizing GBS isolates have found a relationship between DNA macrorestriction band patterns and serotype/protein profile, especially for type V/R1,R4 and NT/R1,R4 isolates (1). The same study also found remarkable heterogeneity within the NT/R4 group compared with typeable isolates (1). However, the distribution of molecular serotypes and surface protein antigen genes associated with NT isolates is not known. Associations found between protein profile, DNA macrorestriction band patterns, sequence type, and molecular serotypes will enhance our understanding of the epidemiology and pathogenesis of GBS infection. In this study, we characterized 92 NT isolates collected over 9 years and grouped them on the basis of expressed surface proteins and by a variety of genetic typing methods to gain a more complete analysis of these NT isolates.
(This research was presented in part at the XVI Lancefield International Symposium on Streptococci and Streptococcal Diseases, Cairns, Australia, 2005.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 92 NT GBS clinical isolates obtained between May 1993 and July 2002 were examined. They were recovered from subjects in Houston (n = 11), Pittsburgh (n = 68), Seattle (n = 12), and Denmark (n = 1). Eighty-seven were colonizing isolates from pregnant or nonpregnant women, and five were invasive isolates from neonates. The anatomic sources of these isolates were vagina (n = 46), rectum (n = 34), vagina/rectum (n = 7), blood (n = 3), and blood/cerebrospinal fluid (n = 2).
Double immunodiffusion reactions.
Five milliliters of cultures incubated for 7 h was inoculated into 50 ml of modified Todd-Hewitt broth and incubated overnight at 37°C. Cells were extracted with hot 0.2 N HCl, concentrated 5× in a Microcon YM-10 centrifugal filter device (Amicon, Bedford, MA), and serotyped by double immunodiffusion in agarose with hyperimmune rabbit antisera produced in-house to serotypes Ia, Ib, and II through VIII (15, 20). Isolates were defined as NT when no reaction of identity was obtained with any of the nine rabbit antisera. Similar reactions were also used to identify surface protein antigens C alpha, C beta, R1, R4, and group B protective surface protein (BPS) with the corresponding rabbit antisera (11, 17).
PFGE.
A rapid PFGE assay was performed on isolates according to a published protocol (4, 13, 38). Briefly, bacteria were embedded in agarose plugs and treated with mutanolysin and proteinase K to lyse the cell wall and precipitate the proteins. The DNA was digested in situ with SmaI restriction enzyme, and the resulting fragments were resolved by PFGE. The PFGE profiles of the isolates were compared visually with the band profiles of the prototype isolates according to Tenover et al.'s criteria (38). Each DNA profile group had a prototype with which all similar isolates were compared (5). Subgroup letter “a” was assigned in addition to the profile number if there was a one- or two-band difference from the prototype strain. An isolate with a three-band difference was assigned letter “b,” one with a four-band difference a “c,” and one with a five-band difference a “d.” Isolates with more than a five-band difference were considered to belong to a different profile group (1).
MLST.
Chromosomal DNA was extracted from cells by using QIAGEN′s genomic-tip kits. PCRs of the seven housekeeping genes (adhP, pheS, atr, glnA, sdhA, glcK, and tkt) were performed with primers described previously (21). The PCR products were then purified with Montage PCR centrifugal filter devices (Millipore, Bedford, MA). The amplicons were sequenced on both strands of DNA with internal nested primers and ABI Prism BigDye Terminators reaction mix. The reaction products were separated and detected with an ABI Prism 3700 DNA analyzer (Applied Biosystems). A consensus sequence was generated with the MacVector program (Accelrys, San Diego, CA) for the seven gene fragments. This sequence was then uploaded to the GBS database at http://pubmlst.org/sagalactiae/ to obtain an allele designation. The combination of alleles at the seven loci provided a sequence type (ST) for each isolate (21, 28, 39). The nucleotide sequences from each of the seven alleles were concatenated, resulting in a single large sequence representing that strain. The genetic relatedness of the strains was then assessed by cluster analysis with the unweighted pair group method with arithmetic averages (UPGMA) algorithm. The phylogenetic reconstruction was carried out with 500 replications by bootstrapping, which uses a resampling approach to estimate the confidence that can be assigned to particular nodes in the generated tree.
Molecular serotyping.
PCRs were performed with primer combinations shown previously to amplify regions of the CPS locus (Ia, Ib, III, IV, V, and VI) described by Kong et al. (22) and primers for CPS loci II, VII, and VIII kindly provided by Lawrence Madoff, Channing Laboratory (unpublished data). DNA samples from the Channing Laboratory prototype strains of the nine serotypes listed in reference panel 1 of reference 22 were used as positive controls. In addition, the cfb (CAMP factor) gene was used as a control for GBS DNA (33). Molecular serotyping was accomplished by amplifying each gene fragment uniquely associated with one of the nine serotypes. Using the chromosomal DNA, all 92 isolates and controls were amplified with a Ia polysaccharide-specific primer, and then all 92 isolates were amplified with a Ib polysaccharide-specific primer and so on. A band of the expected size seen on the gel was scored as positive for that serotype.
Surface protein genes.
PCRs were performed with primer combinations shown previously to amplify regions of the bca (C alpha protein), bac (C beta protein), rib (Rib protein), alp1 (C alpha-like protein 1), and alp3 (C alpha-like protein 3) genes (23). In addition, the cfb gene was used as a control for GBS DNA (33). DNA from the prototype strains H36B (bca and bac), M781 (rib), 515 (alp1), and CJB111 (alp3) was used as a positive control for the presence of surface protein genes. The PCRs were performed on blinded, randomly numbered isolates.
RESULTS
PFGE profiles of the isolates.
The macrorestriction profiles of the 92 NT isolates were classified into 21 DNA profile groups. There were 10 clusters ranging from 2 to 44 isolates. Sixty-three isolates were classified in DNA profile groups 1 to 4 (1). The largest group (44 [47.8%] isolates) belonged to profile group 4, consisting of isolates with similar banding patterns in four subgroups designated 4a through 4d. Eleven isolates were not clustered and had unique macrorestriction profiles. A summary of protein expression and PFGE profiles is shown in Table 1. Isolates that expressed C alpha protein only (n = 12) were distributed among seven PFGE groups: 1, 11, 12, 13, 15, 16, and 33. Eighteen isolates expressing C alpha and C beta proteins were distributed among seven PFGE groups: 2, 8, 18, 19, 32, 34, and 35. Only two isolates expressed BPS protein, and these isolates belonged to PFGE group 32b and ST-10.
TABLE 1.
Distribution of PFGE groups and sequence types based on surface protein profile
| Protein profile | No. of isolates | DNA macrorestriction profile no.a | STa |
|---|---|---|---|
| C alpha only | 12 | 1a3, 152, (1, 1d, 11, 12a, 13b, 16, 33)1 | 235, 223, 12, (7, 44)1 |
| C alpha + C beta | 18 | 195, 2a4, 323, 2b2, (8, 18, 34, 35)1 | 89, 124, 103, (239, 240)1 |
| C alpha + C beta + BPS | 2 | 32b2 | 102 |
| None | 20 | 4a7, 45, 4d3, (1a, 4b, 13b, 16, 21b)1 | 115, 222, (23, 28, 238)1 |
| R4 | 13 | 34, 3a3, 282, (4a, 9b, 20, 22)1 | 175, 194, (1, 31, 44, 237)1 |
| R1 and R4 | 27 | 421, 4a4, 4c2 | 127 |
Superscripts indicate the number of isolates in the PFGE profile groups and sequence types. Each set of numbers in parentheses accounts for one profile group or ST.
Relatedness of GBS isolates by sequence typing.
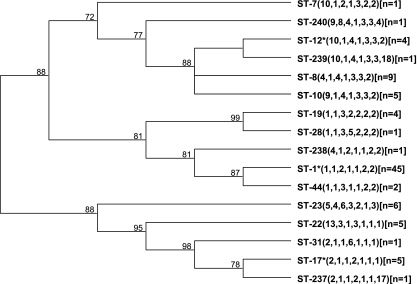
MLST divided the 92 isolates into 16 sequence types. Of these 16 STs, 4 (STs 237 to 240) were newly identified and deposited in the GBS MLST database. BURST (version 1.0; http://www.mlst.net) analysis grouped the STs into four clonal group complexes (group 1 to 4) and four singletons that were not part of any complex (21). Group 1 consisted of STs 8, 10, 12, and 239; group 2 had STs 19 and 28; group 3 had STs 1, 44, and 238; and group 4 had STs 17, 31, and 237. The genetic relatedness between the STs is shown in Fig. 1. Groups 1, 3, and 4 had STs 12, 1, and 17 as the ancestral genotype, and the other STs in the complex were single- and double-locus variants of the ancestral genotype. The ST-1 complex had the most isolates (n = 48), followed by the ST-12 complex with 19 isolates, and seven STs were represented by single isolates.
FIG. 1.
UPGMA dendrogram showing the genetic relationship between the 16 STs. The allelic profile of each ST is shown in parentheses next to the ST designation. The number of isolates found in each ST is shown in brackets. BURST analysis identified four clonal group complexes: group 1 had STs 8, 10, 12, and 239; group 2 had STs 19 and 28; group 3 had STs 1, 44, and 238; and group 4 had STs 17, 31, and 237. The ancestral genotype of the three groups (12, 1, and 17) is denoted by an asterisk. The numbers at the nodes represent the percentage of times each node was represented in the bootstrap analysis.
Surface protein antigens and their genes.
Of the five surface groups of proteins tested immunologically, the largest group of isolates (n = 27) expressed the R1 and R4 proteins, and all belonged to ST-1 and DNA profile group 4 (Table 1). Two isolates expressed C alpha, C beta, and BPS proteins and belonged to DNA profile group 32b. Isolates expressing C alpha protein were associated with multiple DNA profile groups or sequence types. Isolates that did not express any surface proteins tested (n = 20) were significantly associated (85%) with DNA profile group 4 and the ST-1 complex (Table 1). Of the five surface protein genes tested, 41 isolates (44.5%) had a single surface protein gene, while 6 isolates (6.5%) had two different combinations of four genes (Table 2). Overall, 50 (54.4%) isolates had the alp3 gene, and all 27 isolates expressing the R1 and R4 proteins had alp3. Seventeen of the 20 isolates that did not express any surface proteins had the alp3 gene, which suggests that this gene is either conditionally expressed or restricted to certain lineages. Twelve of the 13 (92.3%) isolates that expressed R4 (Rib) protein were associated with the rib gene (37). All isolates (n = 18) expressing both C alpha and C beta proteins had bca and bac genes (Table 2).
TABLE 2.
Distribution of surface protein genes and surface protein profiles of the 92 NT isolates
| Protein gene(s) | No. of isolates that have the gene for:
|
Total | |||||
|---|---|---|---|---|---|---|---|
| C alpha only | C alpha + C beta | C alpha + C beta + BPS | None | R4 | R1 and R4 | ||
| bca | 1 | 2 | 3 | ||||
| rib | 9 | 9 | |||||
| alp1 | 2 | 2 | |||||
| alp3 | 11 | 1 | 15 | 27 | |||
| bca + bac | 3 | 14 | 2 | 19 | |||
| bac + alp3 | 7 | 7 | |||||
| bac + alp1 | 2 | 2 | |||||
| bac + rib | 2 | 2 | |||||
| alp1 + alp3 | 1 | 2 | 1 | 4 | |||
| bca + bac + alp1 | 2 | 2 | |||||
| bca + bac + alp3 | 1 | 1 | 2 | 1 | 5 | ||
| bca + bac + rib | 1 | 1 | 1 | 3 | |||
| bac + alp1 + alp3 | 1 | 1 | |||||
| bca + bac + alp1 + alp3 | 1 | 1 | 2 | 4 | |||
| bca + bac + rib + alp3 | 1 | 1 | 2 | ||||
| Total | 12 | 18 | 2 | 20 | 13 | 27 | 92 |
Molecular serotyping identifies multiple cps-specific genotypes.
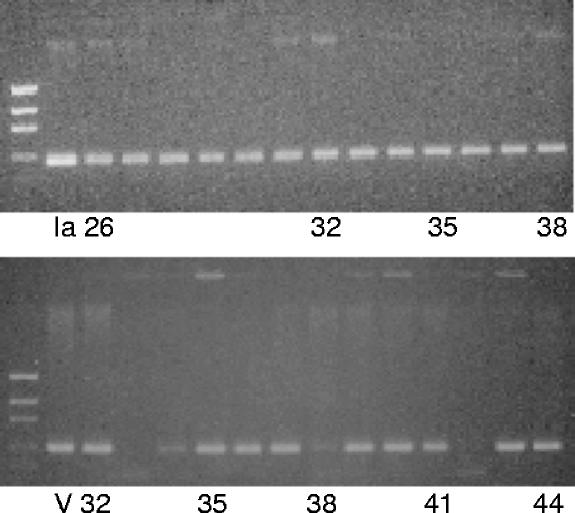
All molecular serotypes except IV, VII, and VIII were represented in the 92 NT isolates (Table 3). Fifty-two isolates (56.5%) had a single CPS-specific gene, 33 isolates (35.9%) had two CPS-specific genes, and 7 isolates (7.6%) had three CPS-specific genes. Representative gels showing isolates with Ia CPS- and V CPS-specific genes are shown in Fig. 2. The type V CPS-specific gene was present in 61 (66%) isolates (Table 3), of which 48 (78.6%) had the alp3 gene. Forty-two of the 61 isolates (69%) were in PFGE profile group 4, and 41 of these isolates were ST-1. Of the nine different combinations of multiple molecular serotypes found, type Ia or V was found in each of the nine groups. Isolates with molecular serotypes Ib, III, and V were associated most frequently with sequence type complexes 8, 17, and 1, respectively. Isolates with the dual types Ia and V had more variation in the PFGE profile groups, sequence types, and surface protein gene combinations than any other group (Table 3). In addition, isolates with the dual surface protein genes bca and bac had the most variation in the distribution of molecular serotypes as well as the sequence types. All isolates (n = 8) with type Ib were associated with bca and bac genes, and all isolates (n = 9) with type III were associated with the rib gene.
TABLE 3.
Distribution of molecular serotypes based on sequence types and PFGE profiles
| Molecular serotype | Sequence typea | PFGE profilea | No. of isolates |
|---|---|---|---|
| Ia | 191, 232 | 1a2, 221 | 3 |
| Ib | 122, 86 | 2a2, 341, 193, 351, 2b1 | 8 |
| II | 11, 441, 221 | 4c1, 12a1, 13b1 | 3 |
| III | 191, 441, 311, 174, 2371 | 34, 3a2, 9b1, 201 | 8 |
| V | 2401, 101, 191, 126 | 414, 4a9, 4d3, 181, 281, 32b1 | 29 |
| VI | 11 | 151 | 1 |
| Ia + Ib | 2391, 121, 81 | 2a2, 191 | 3 |
| Ia + II | 101, 281, 11, 221 | 4c1, 13b1, 21b1, 321 | 4 |
| Ia + III | 171 | 3a1 | 1 |
| Ia + V | 71, 101, 191, 2381, 113, 234 | 11, 1a2, 1d1, 410, 4a3, 4b1, 281, 32b1, 331 | 21 |
| Ib + V | 121, 81 | 2b1, 81 | 2 |
| II + V | 101, 221 | 161, 321 | 2 |
| Ia + Ib + V | 81 | 191 | 1 |
| Ia + II + V | 101, 11, 222 | 41, 111, 161, 321 | 4 |
| Ia + V + VI | 12 | 41, 151 | 2 |
Superscripts indicate the number of isolates in the PFGE profile groups and sequence types.
FIG. 2.
PCR assay on selected nontypeable group B streptococcus performed with type Ia capsular polysaccharide-specific primer (top panel) and type V capsular polysaccharide-specific primer (bottom panel). Isolates 32 and 34 to 37 were scored positive for possessing genes for both type Ia and type V capsular polysaccharides. Ia and V denote the control DNA from Ia strain 090 and type V strain CJB111. The PCR product sizes were 354 bp and 374 bp for type Ia and type V capsular polysaccharide-specific genes, respectively.
Relationship between molecular serotype, sequence type, and PFGE profile.
ST-1 complex isolates had the most molecular serotypes represented, and molecular serotype III was associated with the ST-17 complex (Table 3). Isolates with molecular serotype Ib were significantly associated with the ST-12 complex, and isolates with serotype V were found in all STs except the ST-17 complex, which predominately contained serotype III isolates. For the five invasive isolates in the collection studied, no significant associations were found with ST and PFGE profile groups. Overall, a few strong associations were found between PFGE profile groups and STs. For example, all isolates (n = 46) with PFGE profile group 4 or group 3 (n = 7) belonged to the ST-1 complex or the ST-17 complex, respectively (Table 1). In addition, all isolates with PFGE profile groups 2 (n = 6), 19 (n = 5), and 32 (n = 5) were associated with the ST-12 complex, and the singleton ST-23 had PFGE profile group 1 (n = 6) isolates (Table 1). Isolates with the ST-1 complex had the highest distribution of surface protein gene combinations. Consistent with previous findings with typeable GBS (21), NT isolates with bca and bac genes were significantly associated with the ST-12 complex and also expressed C alpha and C beta proteins. All strains in our study with molecular type Ib were associated with the ST-12 complex. ST-23 was associated with the alp1 gene, and most isolates with the rib gene were distributed predominantly among ST-19 and ST-17 complexes.
DISCUSSION
The data obtained from this study describe the various combinations of surface proteins produced by NT isolates in relation to other DNA-based markers, such as sequence type, PFGE profile group, and molecular serotype. DNA-based typing methods are useful tools in epidemiologic studies of infectious diseases. A variety of typing methods are required to cover all aspects of variation posed by GBS. These methods will be useful for classifying NT isolates for epidemiological surveillance associated with vaccine trials. Overall, the data from our analysis of 92 NT GBS isolates collected over 9 years mostly from different parts of the United States indicate that there is considerable genetic and surface protein heterogeneity among NT isolates. This fact is highlighted by our finding of multiple molecular serotypes, 16 sequence types including 4 new types, 21 PFGE profile groups, and 15 surface protein gene combinations. Several nonrandom associations between PFGE profile groups and sequence types, molecular serotypes and STs or PFGE profiles, and surface protein expression and PFGE profiles or STs were identified. For example, we found that molecular serotype Ib was associated with the ST-12 complex and that all isolates within this complex produced C alpha and C beta proteins and had both bca and bac genes, suggesting a homogenous population structure among serotype Ib isolates. This finding corroborates and extends a previous finding that bac gene-positive strains that were epidemiologically unrelated were genetically homogenous based on PFGE patterns and ribotyping (9).
Previous studies on the population structure of GBS using MLST analysis has shown that isolates with certain STs, such as 1, 17, 19, and 23, most commonly cause human infections (6, 8, 27). Isolates belonging to these four ST complexes have been recovered globally and were also found in our study; together they constitute about 72% of the total isolates (21). These isolates, which include both colonizing and invasive strains, have diversifying capsular serotypes, which suggests that under certain opportunistic conditions they can cause invasive disease. ST variation has also been shown within a single serotype and multiple serotypes associated with a single ST (21), which is suggestive of capsular switching through recombination at the variable region of the cps locus (21, 27). The data from this study have extended this observation by showing that multiple molecular serotypes are associated with a given ST and, conversely, several genotypes under a given molecular serotype. Dual and triple molecular serotypes could have arisen from recombination of serotype-specific capsular genes, since the cps locus is highly conserved at the ends and contains a serotype-specific variable gene region in the middle (7). An NT phenotype could simply arise by such a recombination event, resulting in the disruption of the genes in the cps locus. In a previous study on the distribution of GBS serotypes in Boston, dual serotypes were identified in 4.4% of 114 isolates by probing whole cells with serotype-specific antisera (32). An interesting finding was that each dual serotype isolate was positive for serotype V (32). Surprisingly, 80% of the isolates with dual and triple molecular serotypes in the current study were positive for type V, which indicates that certain lineages of GBS isolates with molecular serotype V are more competent and have a greater propensity to acquire DNA, which can generate more genetic and antigenic diversity.
When PFGE profile groups were compared with the MLST data, certain clusters of isolates with similar PFGE profiles (1 through 4) were found to belong to the same ST complex (23, 12, 17, or 1), suggesting clonality among these isolates. Nearly 78% of the isolates expressing R1 and R4 proteins in this study were classified within PFGE profile group 4, suggesting that the NT/R1,R4 isolates are genetically homogeneous. In addition, all of these isolates were of ST-1 and had the alp3 gene. This finding confirms and extends a previous observation from a recent study that examined the genetic relatedness between NT/R1,R4 and V/R1,R4 isolates by PFGE and found that there was a genotypic shift from PFGE profile group 4 to 4a among V/R1,R4 isolates but not among NT/R1,R4 isolates, as 75% of both NT/R1,R4 and V/R1,R4 isolates were classified within PFGE prototype profile group 4 (1).
Previous studies have shown that certain surface antigenic proteins are frequently associated with a given serotype, and such associations are utilized in classifying GBS isolates (1, 16, 23). Associations between molecular serotypes and protein antigens may represent specific evolutionary lineages and could lack functional significance, but an interesting hypothesis predicts that immune selection has favored strains with specific combinations of protein antigens because it improves their fitness (18). The presence of surface protein genes does not always correlate with the expression of the corresponding protein, since 20 isolates with seven different combinations of surface protein gene profiles did not express any proteins. More extensive analysis of various surface proteins and their genes is required to further classify the diversifying subtypes found among NT GBS isolates. Future epidemiologic surveillance using the molecular methods described here will assess the extent of clonal divergence or homogeneity within subgroups of NT GBS isolates.
The molecular genetic basis for the NT phenotype is not known. NT isolates with a single molecular serotype are known to harbor substitution mutations and insertion sequence elements in the cps biosynthetic genes, although these mutations have not been formally demonstrated to cause the nontypeable phenotype (34, 36). A vaccine based on CPS will not be effective in eliciting optimal protection from NT GBS infection, since NT isolates produce very low levels of capsule or have modified capsular structures that do not react to antiserum obtained from any of the nine known serotypes. However, NT and different serotypes of GBS are known to express a variety of surface proteins in various combinations involved in the protective immune response (24, 26). The five proteins analyzed in this study have been shown to be involved in eliciting a protective immunologic response in animal models and serve as useful candidates for serotype-independent, protein-based vaccines (26). Alternatively, as shown in a recent study involving multistrain genome analysis, the use of appropriate combinations of protective antigens as vaccines may be the solution for preventing NT GBS infections (29).
Acknowledgments
This work was supported by NIH-NIAID contract N01-AI-25495.
We thank Carol J. Baker and Sharon Hillier for providing some of the isolates. We also thank Dennis L. Kasper for advice and guidance and Lawrence C. Madoff and Meghan Gilmore for providing primers and helpful advice.
REFERENCES
- 1.Amundson, N. R., A. E. Flores, S. L. Hillier, C. J. Baker, and P. Ferrieri. 2005. DNA macrorestriction analysis of nontypeable group B streptococcal isolates: clonal evolution of nontypeable and type V isolates. J. Clin. Microbiol. 43:572-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, C. J., and M. S. Edwards. 2003. Group B streptococcal conjugate vaccines. Arch. Dis. Child. 88:375-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, C. J., L. C. Paoletti, M. A. Rench, H.-K. Guttormsen, M. S. Edwards, and D. L. Kasper. 2004. Immune response of healthy women to 2 different group B streptococcal type V capsular polysaccharide-protein conjugate vaccines. J. Infect. Dis. 189:1103-1112. [DOI] [PubMed] [Google Scholar]
- 4.Benson, J. A., and P. Ferrieri. 2001. Rapid pulsed-field gel electrophoresis method for group B streptococcus isolates. J. Clin. Microbiol. 39:3006-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, J. A., A. E. Flores, C. J. Baker, S. L. Hillier, and P. Ferrieri. 2002. Improved methods for typing nontypeable isolates of group B streptococci. Int. J. Med. Microbiol. 292:37-42. [DOI] [PubMed] [Google Scholar]
- 6.Bisharat, N., D. W. Crook, J. Leigh, R. M. Harding, P. N. Ward, T. J. Coffey, M. C. Maiden, T. Peto, and N. Jones. 2004. Hyperinvasive neonatal group B streptococcus has arisen from a bovine ancestor. J. Clin. Microbiol. 42:2161-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cieslewicz, M. J., D. Chaffin, G. Glusman, D. Kasper, A. Madan, S. Rodrigues, J. Fahey, M. R. Wessels, and C. E. Rubens. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect. Immun. 73:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, H. D., N. Jones, T. S. Whittam, S. Elsayed, N. Bisharat, and C. J. Baker. 2004. Multilocus sequence typing of serotype III group B streptococcus and correlation with pathogenic potential. J. Infect. Dis. 189:1097-1102. [DOI] [PubMed] [Google Scholar]
- 9.Dmitriev, A., Y. Y. Hu, A. D. Shen, A. Suvorov, and Y. H. Yang. 2001. Chromosomal analysis of group B streptococcal clinical strains: bac gene-positive strains are genetically homogenous. FEMS Microbiol. Lett. 208:93-98. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, J. A., K. D. Farmer, and R. R. Facklam. 1998. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J. Clin. Microbiol. 36:2115-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdogan, S., P. K. Fagan, S. R. Talay, M. Rohde, P. Ferrieri, A. E. Flores, C. A. Guzmán, M. J. Walker, and G. S. Chhatwal. 2002. Molecular analysis of group B protective surface protein, a new cell surface protective antigen of group B streptococci. Infect. Immun. 70:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farley, M. M. 2001. Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33:556-561. [DOI] [PubMed] [Google Scholar]
- 13.Fasola, E., C. Livdahl, and P. Ferrieri. 1993. Molecular analysis of multiple isolates of the major serotypes of group B streptococci. J. Clin. Microbiol. 31:2616-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrieri, P., C. J. Baker, S. L. Hillier, and A. E. Flores. 2004. Diversity of surface protein expression in group B streptococcal colonizing & invasive isolates. Indian J. Med. Res. 119:191-196. [PubMed] [Google Scholar]
- 15.Ferrieri, P., D. S. Cho, C. Livdahl, C. E. Rubens, and A. E. Flores. 1997. DNA restriction profiles of nontypeable group B streptococcal clinical isolates, p. 343-346. In T. Horaud, A. Bouvet, R. Leclercq, H. de Montclos, and M. Sicard (ed.), Streptococci and the host. Proceedings of the 13th Lancefield International Symposium on Streptococci and Streptococcal Diseases. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 16.Ferrieri, P., and A. E. Flores. 1997. Surface protein expression in group B streptococcal invasive isolates, p. 635-637. In T. Horaud, A. Bouvet, R. Leclercq, H. de Montclos, and M. Sicard (ed.), Streptococci and the host. Proceedings of the 13th Lancefield International Symposium on Streptococci and Streptococcal Diseases. Plenum Press, New York, N.Y.
- 17.Flores, A. E., and P. Ferrieri. 1989. Molecular species of R-protein antigens produced by clinical isolates of group B streptococci. J. Clin. Microbiol. 27:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta, S., M. C. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2:437-442. [DOI] [PubMed] [Google Scholar]
- 19.Hickman, M. E., M. A. Rench, P. Ferrieri, and C. J. Baker. 1999. Changing epidemiology of group B streptococcal colonization. Pediatrics 104:203-209. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, N., J. F. Bohnsack, S. Takahashi, K. A. Oliver, M. S. Chan, F. Kunst, P. Glaser, C. Rusniok, D. W. Crook, R. M. Harding, N. Bisharat, and B. G. Spratt. 2003. Multilocus sequence typing system for group B streptococcus. J. Clin. Microbiol. 41:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Molecular profiles of group B streptococcal surface protein antigen genes: relationship to molecular serotypes. J. Clin. Microbiol. 40:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Ichiman, H. Ohtsuka, M. Kaku, L. C. Paoletti, P. Ferrieri, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, G., M. Stålhammar-Carlemalm, and T. Areschoug. 2005. Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin. Microbiol. Rev. 18:102-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luan, S.-L., M. Granlund, M. Sellin, T. Lagergård, B. G. Spratt, and M. Norgren. 2005. Multilocus sequence typing of Swedish invasive group B streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J. Clin. Microbiol. 43:3727-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 99:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maione, D., I. Margarit, C. D. Rinaudo, V. Masignani, M. Mora, M. Scarselli, H. Tettelin, C. Brettoni, E. T. Iacobini, R. Rosini, N. D'Agostino, L. Miorin, S. Buccato, M. Mariani, G. Galli, R. Nogarotto, V. N. Dei, F. Vegni, C. Fraser, G. Mancuso, G. Teti, L. C. Madoff, L. C. Paoletti, R. Rappuoli, D. L. Kasper, J. L. Telford, and G. Grandi. 2005. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science 309:148-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palacios, G. C., E. K. Eskew, F. Solorzano, and S. J. Mattingly. 1997. Decreased capacity for type-specific-antigen synthesis accounts for high prevalence of nontypeable strains of group B streptococci in Mexico. J. Clin. Microbiol. 35:2923-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paoletti, L. C., and D. L. Kasper. 2003. Glycoconjugate vaccines to prevent group B streptococcal infections. Expert Opin. Biol. Ther. 3:975-984. [DOI] [PubMed] [Google Scholar]
- 32.Paoletti, L. J., J. Bradford, and L. C. Paoletti. 1999. A serotype VIII strain among colonizing group B streptococcal isolates in Boston, Massachusetts. J. Clin. Microbiol. 37:3759-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picard, F. J., and M. G. Bergeron. 2004. Laboratory detection of group B Streptococcus for prevention of perinatal disease. Eur. J. Clin. Microbiol. Infect. Dis. 23:665-671. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy, S. V., P. Ferrieri, L. C. Madoff, A. E. Flores, and L. C. Paoletti. 2004. Immunologic and genetic analysis of atypical serotype V group B streptococcus, abstr. D-1733, p. 160. Program Abstr. 44th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, D.C.
- 35.Rolland, K., C. Marois, V. Siquier, B. Cattier, and R. Quentin. 1999. Genetic features of Streptococcus agalactiae strains causing severe neonatal infections, as revealed by pulsed-field gel electrophoresis and hylB gene analysis. J. Clin. Microbiol. 37:1892-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sellin, M., C. Olofsson, S. Håkansson, and M. Norgren. 2000. Genotyping of the capsule gene cluster (cps) in nontypeable group B streptococci reveals two major cps allelic variants of serotypes III and VII. J. Clin. Microbiol. 38:3420-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, B. L., A. Flores, J. Dechaine, J. Krepela, A. Bergdall, and P. Ferrieri. 2004. Gene encoding the group B streptococcal protein R4, its presence in clinical reference laboratory isolates & R4 protein pepsin sensitivity. Indian J. Med. Res. 119:213-220. [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urwin, R., and M. C. J. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 40.Zaleznik, D. F., M. A. Rench, S. Hillier, M. A. Krohn, R. Platt, M.-L. T. Lee, A. E. Flores, P. Ferrieri, and C. J. Baker. 2000. Invasive disease due to group B streptococcus in pregnant women and neonates from diverse population groups. Clin. Infect. Dis. 30:276-281. [DOI] [PubMed] [Google Scholar]