Abstract
A new molecular subtyping approach was developed which is based on the amplification and sequencing of a repetitive region of the P1 gene of Mycoplasma pneumoniae. It allows the differentiation of all known subtypes and variants of M. pneumoniae as well as the identification of new subtypes directly in clinical samples to characterize endemic and epidemic M. pneumoniae infections.
Mycoplasma pneumoniae is a common agent of infections of the human respiratory tract, with clinical courses ranging from mild forms of pharyngitis and tracheobronchitis to severe cases of interstitial pneumonia (11). Epidemiological studies have shown that M. pneumoniae is responsible for up to 50% of community-acquired pneumonias (7, 15, 26). Epidemic outbreaks were registered in time intervals between 3 and 7 years (16, 22), leading to the hypothesis that different subtypes of M. pneumoniae with variable antigenic properties are circulating in the human population (6, 10). Although from a genetic point of view M. pneumoniae can be considered a highly uniform species, isolates could be assigned to two subtypes differing significantly in the sequences of only four adherence-related proteins (18, 21, 23). Among these, the 170-kDa protein P1 was identified as a major adhesin and an immunodominant protein of M. pneumoniae (9, 19). The P1 gene (MPN141) (2) contains copies of repetitive elements (repMp4 and repMp2/3) (20) with subtype-specific sequence differences (Fig. 1). Recently, two variants of M. pneumoniae were described which showed sequence variations in only the repMp2/3 element of the P1 gene (4, 12). Epidemiological information regarding the subtypes of M. pneumoniae infections is based on the molecular typing of limited numbers of isolated strains (1, 3, 5, 13, 22, 25). More detailed knowledge is hampered by the fact that culture of M. pneumoniae is time-consuming (up to 21 days) and is not a widely used method in routine bacteriological practice compared to serology, direct antigen tests, or PCR (17, 24, 26).
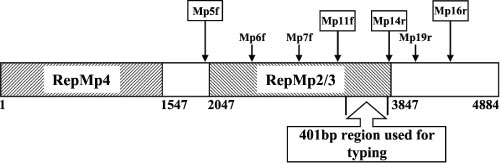
FIG. 1.
Schematic illustration of the P1 gene (MPN141) of M. pneumoniae and location of the investigated 401-bp part (nucleotide positions 3379 to 3779) of the repetitive element repMp2/3. The nucleotide positions are in relation to the start of the P1 gene of the subtype 1 strain M. pneumoniae M129 (8). Locations of the primers are indicated by arrows.
Here we present a method for the characterization of subtypes and variants of M. pneumoniae in DNA samples from 73 respiratory tract specimens (sputa, bronchoalveolar lavage fluid, and throat washing fluid) collected in the years 2003-2005 in a German network of competence (CAPNETZ) established to investigate the etiology of community-acquired pneumonia (27). The collection was complemented with 35 PCR-positive specimens (nasopharyngeal and pharyngeal swabs) from outpatients in Switzerland sampled in the years 2004-2005.
DNAs in all samples were isolated with a QIAamp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations. The subtyping protocol is based on a nested PCR to amplify part of the repMp2/3 element of the P1 gene of M. pneumoniae, followed by sequencing. The primer pairs (ThermoHybaid, Ulm, Germany) used for the first amplification and nested PCR are summarized in Table 1. First amplifications were done in a final volume of 25 μl containing 2.5 μl 10× PCR buffer (Promega, Madison, WI), 2.5 μl deoxynucleoside triphosphates (200 mM each; HYBAID-AGS, Heidelberg, Germany), 0.125 μl AmpliTaq (Promega, Madison, WI), 1.0 μl of each primer (10 pmol), and 2.5 μl template. The reaction mix was heated for 3 min at 94°C, and the DNA was amplified by 35 cycles, using the following settings: 60 s at 94°C, 60 s at 58°C, and 3 min at 72°C, followed by a final 10-min extension at 72°C. A 1-μl portion of the first-round PCR product was used for nested PCR. The amplification program was set as described above, but with a cycle number of 30. Under the same conditions, a seminested PCR with the product of the first amplification and primers Mp5f and Mp19r was carried out to amplify the complete repetitive element repMp2/3. PCR products were analyzed in a 1.5% agarose gel, purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany), and sequenced using a BigDye Terminator cycle sequencing kit and an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, CA).
TABLE 1.
Primers used for amplification and characterization of repMp2/3 of the P1 gene (MPN141) of M. pneumoniae in respiratory tract samples
| Primer purpose | Primer name | Sequence (5′-3′) | Nucleotide positiona | Size (bp) of productb |
|---|---|---|---|---|
| First amplification | Mp5f | TTG ACA AGA CCG TCC AAT CC | 1992-2011 | 2,465 |
| Mp16r | TTG GTT GGG TAT CTT GAT CAG G | 4456-4435 | ||
| Nested PCR | Mp11f | CCT CGT TGT CAG TGG CAC C | 3315-3333 | 533 |
| Mp14r | GCT GGG TGG AAT GGT CTG TAC | 3847-3827 | ||
| Seminested PCR | Mp19r | GTT TCC GTC ACT CGT GCT TG | 4038-4019 | 2,047 |
| Sequencing | Mp6f | GGG AAT GGG TAC AGG TAT GGC | 2434-2454 | |
| Mp7f | AGG CCA CCA ACT ACA CCA A | 2882-2900 |
From the start of the P1 gene.
Corresponding to the sequence of the subtype 1 strain M. pneumoniae M129 (8).
A 401-bp region (nucleotide positions 3379 to 3779 from the start of the P1 gene in M. pneumoniae M129) (8) was selected for comparison of the nucleotide sequences of the investigated samples with the published P1 sequences of M. pneumoniae strain M129 (subtype 1, accession number M18639), strain 1842 (subtype 2, AF290002), variant 1 (AF290000), and variant 2 (AB024618). As indicated in Fig. 2, the sequences show clear differences between the known subtypes and variants of M. pneumoniae. In samples with the novel repMp2/3 sequence, the complete repetitive element (nucleotide positions 2047 to 3847 from the start of the P1 gene in M. pneumoniae M129) (8, 20) was sequenced, using the primers Mp5f, Mp6f, Mp7f, and Mp19r (Table 1; Fig. 1). Sequence analysis was performed with the SeqMan and MegAlign tools and protein translation was done with the EditSeq tool of the program DNAStar (DNASTAR Inc., Madison, WI), using the Mycoplasma coding table.
FIG. 2.
Differences between the amino acid sequences of the P1 proteins of subtypes 1 (ST1) and 2 (ST2) and variants 1 (V1) and 2 (V2) of M. pneumoniae in the investigated 401-bp part of the repetitive element repMp2/3. Amino acid positions of the P1 proteins are indicated according to the published sequences (4, 8, 12, 23). The sequence of the new variant was characterized as V2b.
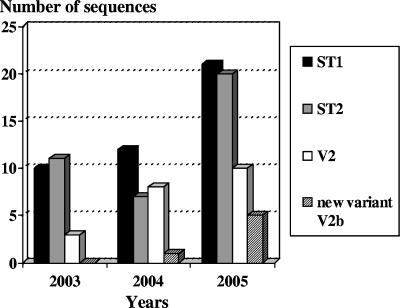
Figure 3 summarizes the sequencing results for the 108 amplicons obtained by subtyping PCR targeting the repetitive element repMp2/3 of the P1 gene of M. pneumoniae in clinical samples. No differences between the samples from Germany and Switzerland concerning the relative proportions of subtypes and variants were detected. Overall, 43 sequences could be assigned to subtype 1 (39.8%), 38 could be assigned to subtype 2 (35.2%), and 21 were identical to the repMp2/3 sequence of variant 2 isolates (19.4%). Sequences typical for variant 1 strains could not be found during the period 2003-2005. The DNAs in six samples (representing 5.6% of all samples) revealed a repMp2/3 element of the P1 gene which was not identified in previously characterized isolates of M. pneumoniae. The following identities of the nucleotide/amino acid sequences of the novel repMp2/3 element with the published complete repMp2/3 sequences in the P1 genes of other subtypes and variants of M. pneumoniae were calculated: 91.1/90.0% (subtype 1, strain M129), 93.9/92.9% (subtype 2, strain 1842), 92.3/91.4% (variant 1), and 96.7/95.7% (variant 2). Identities of the repMp2/3 elements of the P1 genes in the six differing patient samples to the 10 repMp2/3 nucleotide sequences characterized for M. pneumoniae strain M129 (8, 20) varied between 25.1 (repMp2/3-815) and 91.1% (repMp2/3-817).
FIG. 3.
Time-dependent distribution of sequences typical for subtype 1 (ST1), subtype 2 (ST2), variant 2 (V2), and the new variant V2b of M. pneumoniae (n = 108 [73 samples from Germany and 35 samples from Switzerland]).
The results of different studies based on the characterization of isolated strains confirmed the parallel occurrence of the two subtypes of M. pneumoniae in the human population (5, 10, 22). The investigation of limited numbers of isolates showed the dominance of one subtype, followed by an increase in the other subtype during the next years. This led to an alternation of specific subtype accumulations or outbreaks (5). Experimental results supposed a subtype-specific host immune response which might be responsible for changes in protective host immunity and for the switch of the dominant subtype (6, 10).
The sequences of the new repMp2/3 element (Fig. 2) of the P1 gene were identical in clinical samples taken from six epidemiologically unrelated patients at different locations and times, demonstrating the occurrence of the variants in specimens from Germany and Switzerland. According to the sequence of the P1 gene, the M. pneumoniae strain can be considered variant (6). Sequencing of the repMp4 element of these samples resulted in complete homology with the subtype 2-specific sequence (23; data not shown), indicating a one-step recombination in the repetitive elements of the P1 gene. This fact is in agreement with the sequences of variants 1 (4) and 2 (12), which also demonstrate differences only in the repMp2/3 element of the P1 gene. The sequence should therefore be classified as variant 2b, whereas the former variant 2 has to be categorized as variant 2a. Further investigations are necessary to determine the antigenic properties of the variable sequences in the P1 gene and their influence on the immune response of the host.
Seventy-five percent of the investigated sequences could be assigned to the two subtypes, which showed almost equal distributions over 3 years. In a recent study on subtyping of more than 100 M. pneumoniae isolates from different countries, 3% of the strains belonged to variant 2a (5). Among 218 strains from pneumonia patients in Japan, nearly 2% (four strains) were variant 2a isolates (12). In the present study, a significantly larger proportion (19%) of the sequences could be assigned to variant 2a, indicating a more frequent circulation of these strains in the investigated population.
This study is the first to subtype M. pneumoniae directly in respiratory tract specimens from patients with community-acquired pneumonia by a molecular approach which includes all known variants (14). The results are important for an actual overview of the predominant M. pneumoniae strains, giving the opportunity to recognize changes of the subtype/variant composition which probably indicate the starting point of an increased number of respiratory tract infections. Furthermore, the molecular, culture-independent investigation of clinical material provides an early indication of the emergence of new combinations of repetitive elements in the P1 gene with the potential for spreading in the human population. The novel repetitive element repMp2/3 in the P1 genes in respiratory tract specimens from six patients confirmed the possibility of ongoing recombination processes in M. pneumoniae for which the molecular basis is currently not understood.
Nucleotide sequence accession number.
The sequence of the novel repMp2/3 element reported in this study is listed in the GenBank nucleotide sequence data bank under accession number DQ383277.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft (JA 399/10-1) and the BMBF network of competence (CAPNETZ).
REFERENCES
- 1.Cousin-Allery, A., A. Charron, B. de Barbeyrac, G. Fremy, J. S. Jensen, H. Renaudin, and C. Bebear. 2000. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 124:103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dandekar, T., M. Huynen, J. T. Regula, B. Ueberle, C. U. Zimmermann, M. A. Andrade, T. Doerks, L. Sánchez-Pulido, B. Snel, M. Suyama, Y. P. Yuan, R. Herrmann, and P. Bork. 2000. Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorigo-Zetsma, J. W., J. Dankert, and S. A. J. Zaat. 2000. Genotyping of Mycoplasma pneumoniae clinical isolates reveals eight P1 subtypes within two genomic groups. J. Clin. Microbiol. 38:965-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorigo-Zetsma, J. W., B. Wilbrink, J. Dankert, and S. A. J. Zaat. 2001. Mycoplasma pneumoniae P1 type 1- and type 2-specific sequences within the P1 cytadhesin gene of individual strains. Infect. Immun. 69:5612-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumke, R., I. Catrein, E. Pirkl, R. Herrmann, and E. Jacobs. 2003. Subtyping of Mycoplasma pneumoniae isolates based on extended genome sequencing and on expression profiles. Int. J. Med. Microbiol. 292:513-525. [DOI] [PubMed] [Google Scholar]
- 6.Dumke, R., I. Catrein, R. Herrmann, and E. Jacobs. 2004. Preference, adaptation and survival of Mycoplasma pneumoniae subtypes in an animal model. Int. J. Med. Microbiol. 294:149-155. [DOI] [PubMed] [Google Scholar]
- 7.Foy, H. M. 1993. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin. Infect. Dis. 17:37-46. [DOI] [PubMed] [Google Scholar]
- 8.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, P. C., C. H. Huang, H. M. Collier, and W. A. Clyde, Jr. 1983. Demonstration of antibodies to Mycoplasma pneumoniae attachment protein in human sera and respiratory secretions. Infect. Immun. 41:437-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs, E., M. Vonski, K. Oberle, O. Opitz, and K. Pietsch. 1996. Are outbreaks and sporadic respiratory infections by Mycoplasma pneumoniae due to two distinct subtypes? Eur. J. Clin. Microbiol. Infect. Dis. 15:38-44. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs, E. 1997. Mycoplasma infections of the human respiratory tract. Wien. Klin. Wochenschr. 109:574-577. [PubMed] [Google Scholar]
- 12.Kenri, T., R. Taniguchi, Y. Sasaki, N. Okazaki, M. Narita, K. Izumikawa, M. Umetsu, and T. Sasaki. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67:4557-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong, F., S. Gordon, and G. L. Gilbert. 2000. Rapid-cycle PCR for detection and typing of Mycoplasma pneumoniae in clinical specimens. J. Clin. Microbiol. 38:4256-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layani-Milon, M. P., I. Gras, M. Valette, J. Luciani, J. Stagnara, M. Aymard, and B. Lina. 1999. Incidence of upper respiratory tract Mycoplasma pneumoniae infections among outpatients in Rhone-Alpes, France, during five successive winter periods. J. Clin. Microbiol. 37:1721-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lind, K., M. W. Benzon, J. S. Jensen, and W. A. Clyde, Jr. 1997. A seroepidemiological study of Mycoplasma pneumoniae infections in Denmark over the 50-year period 1946-1995. Eur. J. Epidemiol. 13:581-586. [DOI] [PubMed] [Google Scholar]
- 17.Loens, K., D. Ursi, H. Goossens, and M. Ieven. 2003. Molecular diagnosis of Mycoplasma pneumoniae respiratory tract infections. J. Clin. Microbiol. 41:4915-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razin, S., and E. Jacobs. 1992. Mycoplasma adhesion. J. Gen. Microbiol. 138:407-422. [DOI] [PubMed] [Google Scholar]
- 20.Ruland, K., R. Wenzel, and R. Herrmann. 1990. Analysis of three different repeated DNA elements present in the P1 operon of Mycoplasma pneumoniae: size, number and distribution on the genome. Nucleic Acids Res. 18:6311-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruland, K., R. Himmelreich, and R. Herrmann. 1994. Sequence divergence in the ORF6 gene of Mycoplasma pneumoniae. J. Bacteriol. 176:5202-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki, T., T. Kenri, N. Okazaki, M. Iseki, R. Yamashita, M. Shintani, Y. Sasaki, and M. Yayoshi. 1996. Epidemiological study of Mycoplasma pneumoniae infections in Japan based on PCR-restriction fragment length polymorphism of the P1 cytadhesin gene. J. Clin. Microbiol. 34:447-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su, C. J., A. Chavoya, S. F. Dallo, and J. B. Baseman. 1990. Sequence divergence of the cytadhesin gene of Mycoplasma pneumoniae. Infect. Immun. 58:2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Templeton, K. E., S. A. Scheltinga, A. W. Graffelman, J. M. van Schie, J. W. Crielaard, P. Sillekens, P. J. van den Broek, H. Goossens, M. F. C. Beersma, and E. C. J. Claas. 2003. Comparison and evaluation of real-time PCR, real-time nucleic acid sequence-based amplification, conventional PCR, and serology for diagnosis of Mycoplasma pneumoniae. J. Clin. Microbiol. 41:4366-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ursi, D., M. Heven, H. van Bever, W. Quint, H. G. M. Niesters, and H. Goossens. 1994. Typing of Mycoplasma pneumoniae by PCR-mediated DNA fingerprinting. J. Clin. Microbiol. 32:2873-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17:697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welte, T., N. Suttorp, and R. Marre. 2004. CAPNETZ—community-acquired pneumonia competence network. Infection 32:234-238. [DOI] [PubMed] [Google Scholar]