Abstract
Streptococcus strains from animal and human sources identified biochemically as Streptococcus porcinus were investigated by 16S rRNA gene sequencing. The nine human strains isolated between 1997 and 2005 formed a single cluster with more than 2.1% dissimilarity with S. porcinus strains from animal sources. A novel species, Streptococcus pseudoporcinus sp. nov., is proposed.
The genus Streptococcus is a large group with currently more than 60 species, many of which were described in the last decade (6). The recent use of molecular characterization techniques may in part explain the sudden increase in the number of species, and one can assume that this trend will continue.
The species Streptococcus porcinus was described in 1984 (2) and is commonly associated with various pathological infections in swine (2, 12). Isolation of S. porcinus from humans has rarely been reported and is mostly from the female genitourinary tract (3, 5, 9). In fact, because of its biochemical similarities with Streptococcus agalactiae (commonly associated with female genitourinary tract infection or colonization) and serological cross-reactivity with group B streptococcal reagent (5, 11), human infections or colonization by S. porcinus may have been underestimated. In the province of Québec, Canada, nine strains isolated from human clinical specimens were identified as Streptococcus porcinus by conventional physiological tests described previously (5, 10).
These strains, obtained from three Montreal hospitals, were isolated between 1997 and 2005 from the genitourinary tracts of nine female patients (21 to 49 years old, mean = 30) originating from different countries. Table 1 shows phenotypic characteristics of the nine human strains as well as seven animal strains isolated from swine between 1995 and 2005 in the province of Québec. Past reports (3, 11) described all S. porcinus strains of human origin as being acetoin (Voges-Proskauer [VP]) positive and pyrrolidonyl arylamidase (PYR) positive or weakly positive, whereas group B streptococci were reported to be VP and PYR negative. In the present investigation, human strains were positive for VP and PYR in 56% and 33% of the cases, respectively, compared to 100% and 57% for animal strains.
TABLE 1.
Phenotypic characteristics of human and animal strains of Streptococcus biochemically identified as S. porcinus
| Test | % of strains of indicated type with positive results
|
|
|---|---|---|
| Human (n = 9) | Animal (n = 7) | |
| Hydrolysis of hippurate | 0 | 0 |
| VP reaction | 56 | 100 |
| PYR test | 30 | 57 |
| Leucine aminopeptidase test | 100 | 100 |
| Acid production from: | ||
| Mannitol | 100 | 100 |
| Sorbitol | 100 | 100 |
Recently, we subjected these 16 strains to 16S rRNA gene sequencing for molecular characterization and identification purposes. In addition, the 16S rRNA gene sequences of 12 Streptococcus ATCC type strains were sequenced and included in the study. For sequencing purposes, bacteria were disrupted with zirconium/silica beads by using a bead beater (Biospec Inc.). Genomic DNA was extracted using a QIAamp DNA mini kit (QIAGEN Inc.). The 16S rRNA gene was amplified by PCR using primers Ai (AGRGTTYGATYCTGGCTCAGGAYG) and rJ (GGTTACCTTGTTACGACTT) (4, 8). The 1,500-bp amplified fragment was purified by using the Minelute filtration system (QIAGEN Inc.) and sequenced on both strands using the Ai, rJ, D, E, and rE primers (7).
DNA sequences were determined with an ABI 3100 sequencer using a BigDye sequencing kit (Applied Biosystems). The sequences were subjected to a BLAST analysis and aligned with the ClustalW program. Phylogenetic analysis was performed using the Lasergene software V6.1 (DNAstar).
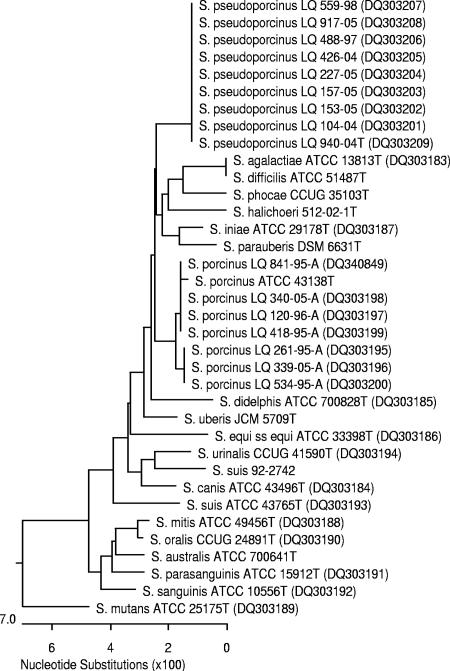
The phylogenetic tree confirms that all of the isolates are related to the genus Streptococcus. The nine human strains exhibit 100% similarity/0% dissimilarity and form a single cluster that exhibits more than 2.1% dissimilarity with the other Streptococcus species. The animal strains (including the type strain ATCC 43138) form another cluster that exhibits between 2.1 and 2.4% dissimilarity with the nine human strains (Fig. 1).
FIG. 1.
Phylogenetic tree, based on comparative analysis of the 16S rRNA gene sequences, showing the relationships of Streptococcus pseudoporcinus sp. nov. with other Streptococcus species. GenBank accession numbers are shown in parentheses.
Table 2 presents sequence dissimilarities between 16S rRNA gene sequences of one representative strain of human isolates (LQ 940-04T) and the closest type strains of Streptococcus species. The lowest scores were for S. porcinus (2.5%), Streptococcus iniae (2.7%), and Streptococcus uberis (2.7%). Considering the 1.0% value the limit in species definition (1), the nine human strains can be considered representative of a new species, namely, Streptococcus pseudoporcinus sp. nov., in reference to the similarity of its biochemical profile to that of S. porcinus. Strain LQ 940-04T was chosen as the type strain.
TABLE 2.
Similarities and dissimilarities among 16S rRNA sequences of Streptococcus species type strains
| Species | % Similarity or dissimilarity to indicated species
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. iniae | S. didelphis | S. canis | S. agalactiae | S. uberis | S. difficilis | S. parauberis | S. halichoeri | S. porcinus | LQ 940-04T | |
| S. iniae | 95.6 | 96.4 | 96.5 | 96.7 | 96.5 | 97.7 | 96.1 | 96.3 | 97.2 | |
| S. didelphis | 4.3 | 95.2 | 95.3 | 96.5 | 95.3 | 95.7 | 95.5 | 96.4 | 96.4 | |
| S. canis | 3.5 | 5.0 | 96.0 | 97.2 | 96.0 | 96.4 | 96.2 | 96.4 | 96.1 | |
| S. agalactiae | 3.3 | 4.9 | 4.2 | 95.4 | 100.0 | 96.0 | 95.9 | 96.2 | 96.5 | |
| S. uberis | 3.1 | 3.6 | 2.9 | 4.8 | 95.4 | 97.0 | 95.1 | 97.1 | 97.4 | |
| S. difficilis | 3.3 | 4.9 | 4.2 | 0.0 | 4.8 | 96.0 | 95.9 | 96.2 | 96.5 | |
| S. parauberis | 2.2 | 4.5 | 3.7 | 4.1 | 3.0 | 4.1 | 95.6 | 96.5 | 96.5 | |
| S. halichoeri | 3.7 | 4.5 | 3.9 | 4.2 | 5.0 | 4.2 | 4.5 | 96.3 | 96.2 | |
| S. porcinus | 3.6 | 3.7 | 3.7 | 3.9 | 3.0 | 3.9 | 3.6 | 3.7 | 97.5 | |
| LQ 940-04T | 2.7 | 3.7 | 4.0 | 3.6 | 2.7 | 3.6 | 3.6 | 3.8 | 2.5 | |
In their study conducted on 25 human and 16 nonhuman strains (mostly swine), Duarte et al. (3) reported high similarities of whole-cell protein profiles among S. porcinus strains from different sources. However, using the randomly amplified polymorphic DNA-PCR and pulsed-field gel electrophoresis analyses, the authors identified two main clusters, I and II. The human isolates were included in cluster I, whereas the nonhuman ones were included in cluster II. The authors determined the existence of a few clonal groups of S. porcinus, adapted to the human host.
Our findings are in agreement with those of Duarte et al. (3), but our study reveals that some human clinical isolates may in fact belong to the new species Streptococcus pseudoporcinus. However, 16S sequencing is a necessary tool to differentiate S. porcinus from S. pseudoporcinus. The nine human strains of Streptococcus isolated from the genitourinary tract of women in the province of Québec and identified biochemically as Streptococcus porcinus were ultimately identified as Streptococcus pseudoporcinus by 16S rRNA gene sequencing. To our knowledge, we present the first molecular characterization by 16S rRNA gene sequencing of biochemically identified S. porcinus.
Description of Streptococcus pseudoporcinus sp. nov.
Cells of Streptococcus pseudoporcinus sp. nov. are spherical to ovoid, gram positive, nonmotile, and generally arranged in short chains. On blood agar, its colonies are generally small and smooth, with entire margins, and β hemolytic, showing a large zone of complete hemolysis. There is slight to normal growth at 10°C, but mostly no growth at 45°C, in 7 days. Growth in 6.5% NaCl broth is obtained in 24 to 48 h. Catalase and benzidine tests are negative. No extracellular polysaccharide is produced. Fermentative metabolism is used. Acid is usually produced from glucose, glycerol, maltose, mannitol, ribose, salicin, sorbitol, sucrose, and trehalose but not from lactose or melibiose. Arginine and esculin, but not starch, bile esculin, urea, or hippurate, are usually hydrolyzed. Leucine, aminopeptidase test results are positive. VP reaction is variable. PYR is often negative. The species does not react with Lancefield group A, B, C, D, F, or G antisera (Streptex by Murex). A vancomycin screen (30-μg disks; Oxoid) indicated sensitivity.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were submitted to GenBank, and the obtained accession numbers are shown in Fig. 1. All of the accession numbers in Fig. 1 are newly established in our study.
Acknowledgments
We are very grateful to the bacteriology team for their technical assistance and to Roland Brousseau from the Biotechnology Research Institute and Manon Lorange for their critical reading of the manuscript.
REFERENCES
- 1.Clarridge, J. E., III. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, M. D., J. A. E. Farrow, V. Katic, and O. Kandler. 1984. Taxonomic studies on streptococci of serological groups E, P, U and V: description of Streptococcus porcinus sp. nov. Syst. Appl. Microbiol. 5:402-413. [Google Scholar]
- 3.Duarte, R. S., R. R. Barros, R. R. Facklam, and L. M. Teixeira. 2005. Phenotypic and genotypic characteristics of Streptococcus porcinus isolated from human sources. J. Clin. Microbiol. 43:4592-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facklam, R., J. Elliott, N. Pigott, and A. R. Franklin. 1995. Identification of Streptococcus porcinus from human sources. J. Clin. Microbiol. 33:385-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Facklam, R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harf-Monteil, C., A. L. Fleche, P. Riegel, G. Prevost, D. Bermond, P. A. Grimont, and H. Monteil. 2004. Aeromonas simiae sp. nov., isolated from monkey faeces. Int. J. Syst. Evol. Microbiol. 54:481-485. [DOI] [PubMed] [Google Scholar]
- 8.Janvier, M., and P. A. Grimont. 1995. The genus Methylophaga, a new line of descent within phylogenetic branch gamma of proteobacteria. Res. Microbiol. 146:543-550. [DOI] [PubMed] [Google Scholar]
- 9.Martin, C., V. Fermeaux, J. L. Eyraud, and Y. Aubard. 2004. Streptococcus porcinus as a cause of spontaneous preterm human stillbirth. J. Clin. Microbiol. 42:4396-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruoff, K. L., R. A. Whiley, and D. Beighton. 2003. Streptococus, p. 405-421. In P. R. Murray, E. J. Baro, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington D.C.
- 11.Thompson, T., and R. Facklam. 1997. Cross-reactions of reagents from streptococcal grouping kits with Streptococcus porcinus. J. Clin. Microbiol. 35:1885-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wessman, G. E. 1986. Biology of the group E streptococci: a review. Vet. Microbiol. 12:297-328. [DOI] [PubMed] [Google Scholar]